QTL Pyramiding and Its Use in Breeding for Increasing the Phytoextraction Efficiency of Soil Cd via High-Cd-Accumulating Rice
Abstract
:1. Introduction
2. Results
2.1. Agronomic Traits and Cd Accumulation Ability of Parental Varieties
2.2. Development of TJN25-11 by a Three-Way Cross
2.3. QTL Analysis
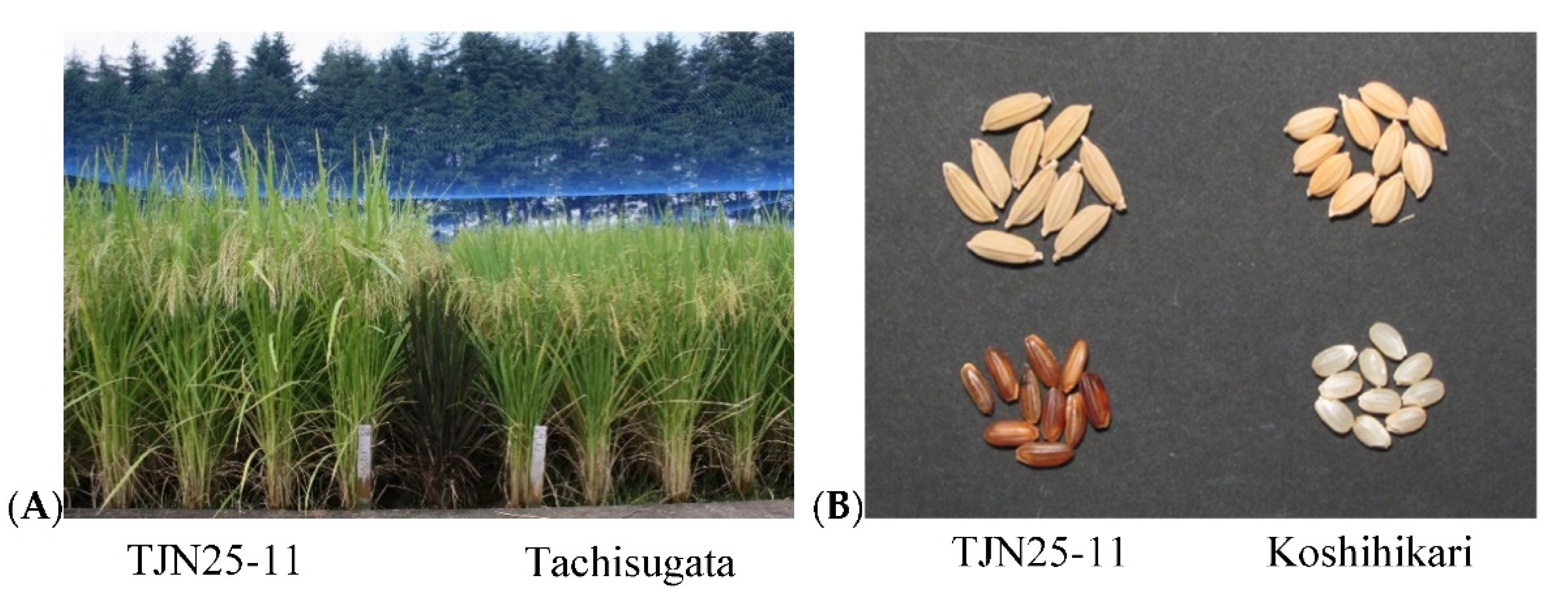
2.4. Characteristics of TJN25-11
2.4.1. Agronomic Traits of TJN25-11
2.4.2. Cd Accumulation Ability of TJN25-11
2.4.3. Cd-Phytoextraction Efficiency of TJN25-11
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Field Tests
4.3. Evaluation of Agronomic Traits
4.4. Cd Analysis
4.5. QTL Analysis of Straw Cd Concentrations
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, L.; Cui, X.; Cheng, H.; Chen, F.; Wang, J.; Zhao, X.; Lin, C.; Pu, X. A review of soil cadmium contamination in China including a health risk assessment. Environ. Sci. Pollut. Res. 2015, 22, 16441–16452. [Google Scholar] [CrossRef] [PubMed]
- CODEX Alimentarius. General Standard for Contaminants and Toxins in Food and Feed. CXS 193-1995. 2019. Available online: https://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FStandards%252FCXS%2B193-1995%252FCXS_193e.pdf (accessed on 18 July 2022).
- Arao, T.; Ishikawa, S.; Murakami, M.; Abe, K.; Maejima, Y.; Makino, T. Heavy metal contamination of agricultural soil and countermeasures in Japan. Paddy Water Environ. 2010, 8, 247–257. [Google Scholar] [CrossRef]
- Ishikawa, S.; Ishimaru, Y.; Igura, M.; Kuramata, M.; Abe, T.; Senoura, T.; Hase, Y.; Arao, T.; Nishizawa, N.K.; Nakanishi, H. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc. Natl. Acad. Sci. USA 2012, 109, 19166–19171. [Google Scholar] [CrossRef] [Green Version]
- Ibaraki, T.; Kuroyanagi, N.; Murakami, M. Practical phytoextraction in cadmium-polluted paddy fields using a high cadmium accumulating rice plant cultured by early drainage of irrigation water. Soil Sci. Plant Nutr. 2009, 55, 421–427. [Google Scholar] [CrossRef]
- Zhao, F.J.; Tang, Z.; Song, J.J.; Huang, X.Y.; Wang, P. Toxic metals and metalloids: Uptake, transport, detoxification, phytoremediation, and crop improvement for safer food. Mol. Plant 2022, 15, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Ebbs, S.D.; Lasat, M.M.; Brady, D.J.; Cornish, J.; Gordon, R.; Kochian, L.V. Phytoextraction of cadmium and zinc from a contaminated soil. J. Environ. Qual. 1997, 26, 1424–1430. [Google Scholar] [CrossRef]
- Ishikawa, S.; Ae, N.; Murakami, M.; Wagatsuma, T. Is Brassica juncea a suitable plant for phytoremediation of cadmium in soils with moderately low cadmium contamination? - Possibility of using other plant species for Cd-phytoextraction. Soil Sci. Plant Nutr. 2006, 52, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.; Ae, N.; Ishikawa, S. Phytoextraction of cadmium by rice (Oryza sativa L.), soybean (Glycine max (L.) Merr.), and maize (Zea mays L.). Environ. Pollut. 2007, 145, 96–103. [Google Scholar] [CrossRef]
- Murakami, M.; Nakagawa, F.; Ae, N.; Ito, M.; Arao, T. Phytoextraction by rice capable of accumulating Cd at high levels: Reduction of Cd content of rice grain. Environ. Sci. Technol. 2009, 43, 5878–5883. [Google Scholar] [CrossRef]
- Takahashi, R.; Ito, M.; Kawamoto, T. The Road to Practical Application of Cadmium Phytoremediation Using Rice. Plants 2021, 10, 1926. [Google Scholar] [CrossRef]
- Uraguchi, S.; Mori, S.; Kuramata, M.; Kawasaki, A.; Arao, T.; Ishikawa, S. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 2009, 60, 2677–2688. [Google Scholar] [CrossRef] [Green Version]
- Miyadate, H.; Adachi, S.; Hiraizumi, A.; Tezuka, K.; Nakazawa, N.; Kawamoto, T.; Katou, K.; Kodama, I.; Sakurai, K.; Takahashi, H.; et al. OsHMA3, a P-1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011, 189, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, D.; Koyama, E.; Yamaji, N.; Ma, J.F. Physiological, genetic, and molecular characterization of a high-Cd-accumulating rice cultivar, Jarjan. J. Exp. Bot. 2011, 62, 2265–2272. [Google Scholar] [CrossRef]
- Abe, T.; Ito, M.; Takahashi, R.; Honma, T.; Sekiya, N.; Shirao, K.; Kuramata, M.; Murakami, M.; Ishikawa, S. Breeding of a practical rice line “TJTT8” for phytoextraction of cadmium contamination in paddy fields. Soil Sci. Plant Nutr. 2017, 63, 388–395. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Kuramata, M.; Iwasaki, M.; Honma, T.; Ibaraki, T.; Yamamoto, T.; Yano, M.; Murakami, M.; Ishikawa, S. “MJ3” and “MA22”, new high-cadmium-accumulating practical rice lines with non-shattering derived from gamma ray mutation. Breed. Res. 2013, 15, 17–24. (In Japanese) [Google Scholar] [CrossRef]
- Takahashi, R.; Ito, M.; Katou, K.; Sato, K.; Nakagawa, S.; Tezuka, K.; Akagi, H.; Kawamoto, T. Breeding and characterization of the rice (Oryza sativa L.) line “Akita 110” for cadmium phytoremediation. Soil Sci. Plant Nutr. 2016, 62, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.; Ito, M.; Kato, K.; Kodama, I.; Shibata, S.; Sato, K.; Matsumoto, S.; Kawamoto, T. Breeding and characterization of the high cadmium-accumulating rice line ‘Akita 119’. Breed. Sci. 2020, 70, 631–636. [Google Scholar] [CrossRef]
- Ueno, D.; Kono, I.; Yokosho, K.; Ando, T.; Yano, M.; Ma, J.F. Identification of a novel QTL for shoot Cd accumulation in rice. UC Davis Dep. Plant Sci. 2009. Available online: https://escholarship.org/uc/item/2bb1f8fb (accessed on 18 July 2022).
- Abe, T.; Taguchi-Shiobara, F.; Kojima, Y.; Ebitani, T.; Kuramata, M.; Yamamoto, T.; Yano, M.; Ishikawa, S. Detection of a QTL for accumulating Cd in rice that enables efficient Cd phytoextraction from soil. Breed. Sci. 2011, 61, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Xue, D.W.; Chen, M.C.; Zhang, G.P. Mapping of QTLs associated with cadmium tolerance and accumulation during seedling stage in rice (Oryza sativa L.). Euphytica 2009, 165, 587–596. [Google Scholar] [CrossRef]
- Ueno, D.; Kono, I.; Yokosho, K.; Ando, T.; Yano, M.; Ma, J.F. A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytol. 2009, 182, 644–653. [Google Scholar] [CrossRef]
- Ohta, H.; Nemoto, H.; Ando, I.; Kato, H.; Sato, H.; Hirabayashi, H.; Takeuchi, Y.; Ishii, T.; Maeda, H.; Imbe, T.; et al. “Tachisugata”, a new rice cultivar for whole crop silage use. Bull. Natl. Inst. Crop. Sci. 2010, 11, 67–84, (In Japanese with English Summary). [Google Scholar]
- Furukawa, T.; Maekawa, M.; Oki, T.; Suda, I.; Iida, S.; Shimada, H.; Takamure, I.; Kadowaki, K.I. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 2007, 49, 91–102. [Google Scholar] [CrossRef]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCouch, S.R.; Teytelman, L.; Xu, Y.B.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.Y.; Maghirang, R.; Li, Z.K.; Xing, Y.Z.; et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef]
- International Rice Genome Sequencing Project; Sasaki, T. The map-based sequence of the rice genome. Nuture 2005, 436, 793–800. [Google Scholar] [CrossRef]
- Ishikawa, S.; Makino, T.; Ito, M.; Harada, K.; Nakada, H.; Nishida, I.; Nishimura, M.; Tokunaga, T.; Shirao, K.; Yoshizawa, C.; et al. Low-cadmium rice (Oryza sativa L.) cultivar can simultaneously reduce arsenic and cadmium concentrations in rice grains. Soil Sci. Plant Nutr. 2016, 62, 327–339. [Google Scholar] [CrossRef] [Green Version]
- Lander, E.; Abrahamason, J.; Barlow, A.; Daly, M.; Lincoln, S.; Newburg, L.; Green, P. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]

| Variety | Days to Heading | Culm Length | Lodging Degree | Shattering Behavior | Cd Concentration | Total Amount of Cd Accumulation in Aerial Parts | ||
|---|---|---|---|---|---|---|---|---|
| Straw | Grain | |||||||
| (cm) | (0–5) | (s-r) | (mg kg−1) | (mg kg−1) | (mg m−2) | |||
| Field A | Jarjan | 110 | 109 b | 0.0 | s | 10.0 a ± 6.0 | 8.7 a ± 8.1 | 8.8 a |
| Nepal 555 | 104 | 118 a | 4.0 | s | 8.6 a ± 6.3 | 2.0 ab ± 1.5 | 4.7 ab | |
| Tachisugata | 107 | 94 c | 0.0 | r | 4.0 a ± 3.0 | 1.3 b ± 1.0 | 2.1 b | |
| Field B | Jarjan | 115 | 129 a | 0.0 | s | 14.5 a ± 3.2 | 4.9 a ± 1.0 | 13.7 a |
| Nepal 555 | 107 | 126 a | 3.0 | s | 10.6 b ± 2.7 | 2.2 b ± 0.5 | 9.8 b | |
| Tachisugata | 111 | 101 b | 0.0 | r | 7.2 b ± 1.5 | 2.2 b ± 0.4 | 6.6 c | |
| Chr. | QTL | Marker Interval | Nearest Marker | LOD * | R2 ** | AE *** |
|---|---|---|---|---|---|---|
| 2 | qHCd2 | RM1211-RM5303 | RM1385 | 4.57 | 18.8 | 1.05 |
| 6 | qHCd6 | RM3414-RM2615 | RM8258 | 5.76 | 22.8 | −1.17 |
| Variety | Days to Heading | Culm Length | Straw Weight | Panicle Weight | Lodging Degree | |
|---|---|---|---|---|---|---|
| (cm) | (g m−2) | (g m−2) | (0–5) | |||
| Field A | TJN25-11 | 117 | 94 a ± 3.2 | 992 a ± 60 | 332 a ± 39 | 0 |
| TJTT8 | 116 | 81 b ± 1.3 | 922 a ± 9 | 237 a ± 53 | 0 | |
| Cho-ko-koku | 112 | 82 b ± 3.2 | 651 b ± 89 | 594 b ± 110 | 2 | |
| Field B | TJN25-11 | 123 | 102 a ± 4.1 | 777 a ± 75 | 150 a ± 18 | 0 |
| TJTT8 | 121 | 94 a ± 1.8 | 784 a ± 69 | 137 a ± 12 | 0 | |
| Cho-ko-koku | 115 | 100 a ± 4.6 | 613 b ± 40 | 328 b ± 48 | 3 |
| Variety | Cd Concentration | Amount of Cd That Accumulated * | Total Amount of Cd that Accumulated in Aerial Parts ** | |||
|---|---|---|---|---|---|---|
| Straw | Panicle | Straw | Panicle | |||
| (mg kg−1) | (mg kg−1) | (mg m−2) | (mg m−2) | (mg m−2) | ||
| Field A | TJN25-11 | 24.5 a ± 2.9 | 37.4 a ± 5.5 | 24.2 a ± 1.7 | 12.3 a ± 0.7 | 36.5 a ± 2.2 |
| TJTT8 | 19.7 a ± 2.6 | 26.4 a ± 4.3 | 18.2 ab ± 2.4 | 6.1 b ± 0.3 | 24.3 b ± 2.3 | |
| Cho-ko-koku | 18.2 a ± 3.4 | 11.2 b ± 4.7 | 12.0 b ± 3.3 | 6.3 b ± 1.4 | 18.3 b ± 4.8 | |
| Field B | TJN25-11 | 21.1 a ± 1.4 | 27.9 a ± 2.4 | 16.3 a ± 1.7 | 4.2 a ± 0.2 | 20.5 a ± 1.6 |
| TJTT8 | 15.0 b ± 1.3 | 15.8 b ± 2.4 | 11.8 b ± 1.8 | 2.2 b ± 0.4 | 14.0 b ± 2.0 | |
| Cho-ko-koku | 12.8 b ± 0.7 | 5.7 c ± 0.4 | 7.9 c ± 0.8 | 1.9 b ± 0.4 | 9.7 c ± 1.1 | |
| Variety | Soil Cd Concentration (0.1 M HCl Extractable) | Average Amount of Cd in the Soil | Total Amount of Cd That Accumulated in Aerial Parts (F) *** | Phytoextraction Efficiency | |||||
|---|---|---|---|---|---|---|---|---|---|
| Before Planting (A) | After Planting (B) | Before Planting (C) * | After Planting (D) ** | Reduction (E = C − D) | Actual Reduction Rate of Soil Cd # | Theoretical Reduction Rate of Soil Cd $ | |||
| (mg kg−1) | (mg kg−1) | (mg m−2) | (mg m−2) | (mg m−2) | (mg m−2) | (%) | (%) | ||
| Field A | TJN25-11 | 0.33 ± 0.05 | 0.26 ± 0.02 | 49.5 | 39.0 | 10.5 | 36.5 | 21.2 | 73.7 |
| TJTT8 | 0.34 ± 0.06 | 0.30 ± 0.06 | 51.0 | 45.0 | 6.0 | 24.3 | 11.8 | 47.6 | |
| Cho-ko-koku | 0.32 ± 0.06 | 0.30 ± 0.05 | 48.0 | 45.0 | 3.0 | 18.3 | 6.3 | 38.1 | |
| Field B | TJN25-11 | 0.51 ± 0.00 | 0.40 ± 0.06 | 76.5 | 60.0 | 16.5 | 20.5 | 21.6 | 26.8 |
| TJTT8 | 0.53 ± 0.04 | 0.47 ± 0.08 | 79.5 | 70.5 | 9.0 | 14.0 | 11.3 | 17.6 | |
| Cho-ko-koku | 0.50 ± 0.06 | 0.46 ± 0.07 | 75.0 | 69.0 | 6.0 | 9.7 | 8.0 | 12.9 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abe, T.; Ito, M.; Takahashi, R.; Honma, T.; Kuramata, M.; Ishikawa, S. QTL Pyramiding and Its Use in Breeding for Increasing the Phytoextraction Efficiency of Soil Cd via High-Cd-Accumulating Rice. Plants 2022, 11, 2178. https://doi.org/10.3390/plants11162178
Abe T, Ito M, Takahashi R, Honma T, Kuramata M, Ishikawa S. QTL Pyramiding and Its Use in Breeding for Increasing the Phytoextraction Efficiency of Soil Cd via High-Cd-Accumulating Rice. Plants. 2022; 11(16):2178. https://doi.org/10.3390/plants11162178
Chicago/Turabian StyleAbe, Tadashi, Masashi Ito, Ryuichi Takahashi, Toshimitsu Honma, Masato Kuramata, and Satoru Ishikawa. 2022. "QTL Pyramiding and Its Use in Breeding for Increasing the Phytoextraction Efficiency of Soil Cd via High-Cd-Accumulating Rice" Plants 11, no. 16: 2178. https://doi.org/10.3390/plants11162178
APA StyleAbe, T., Ito, M., Takahashi, R., Honma, T., Kuramata, M., & Ishikawa, S. (2022). QTL Pyramiding and Its Use in Breeding for Increasing the Phytoextraction Efficiency of Soil Cd via High-Cd-Accumulating Rice. Plants, 11(16), 2178. https://doi.org/10.3390/plants11162178






