The ABCISIC ACID INSENSITIVE (ABI) 4 Transcription Factor Is Stabilized by Stress, ABA and Phosphorylation
Abstract
:1. Introduction
2. Results
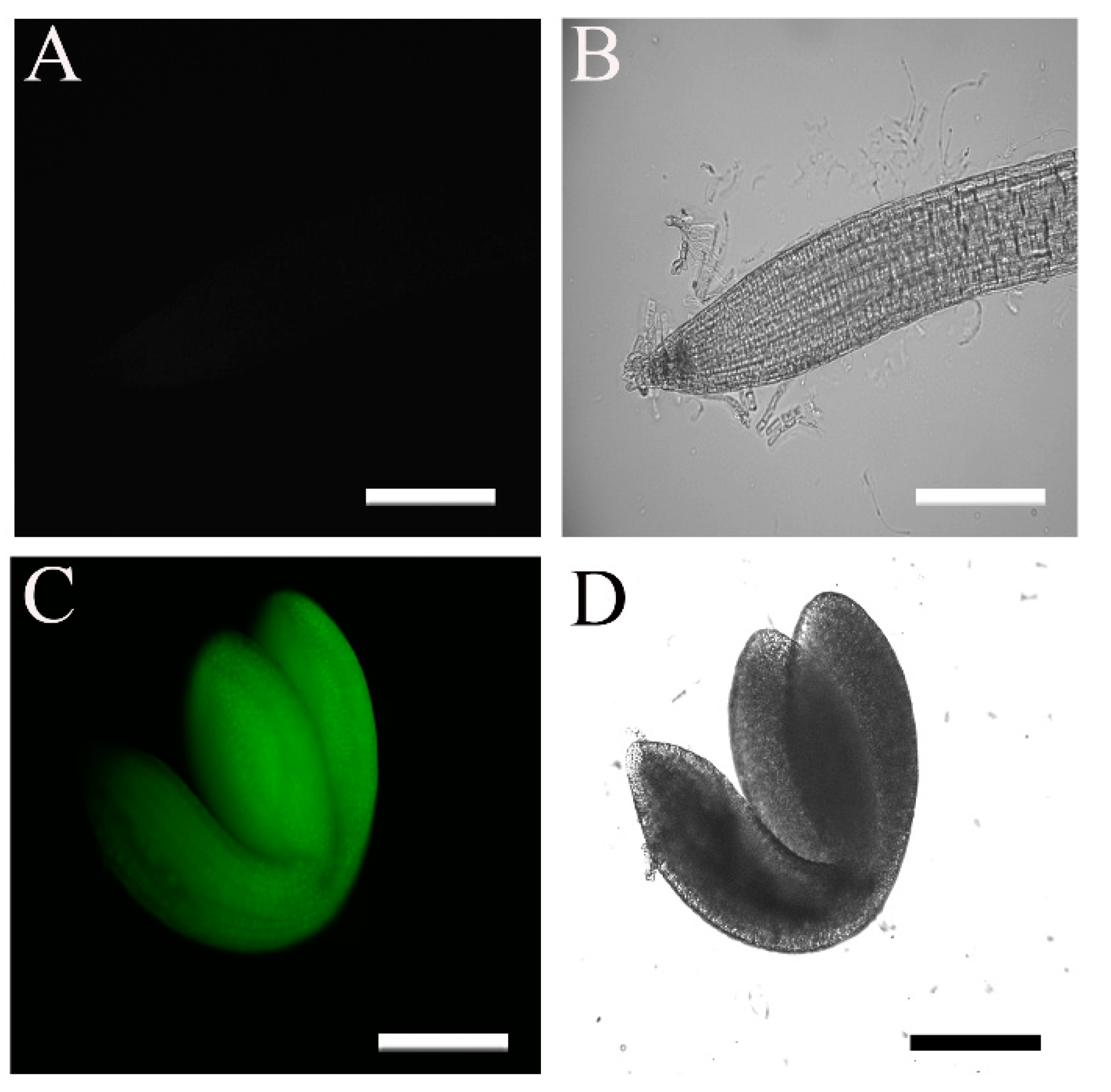
2.1. The 35S::HA-FLAG-ABI4-eGFP Construct Encodes a Biologically Active Protein
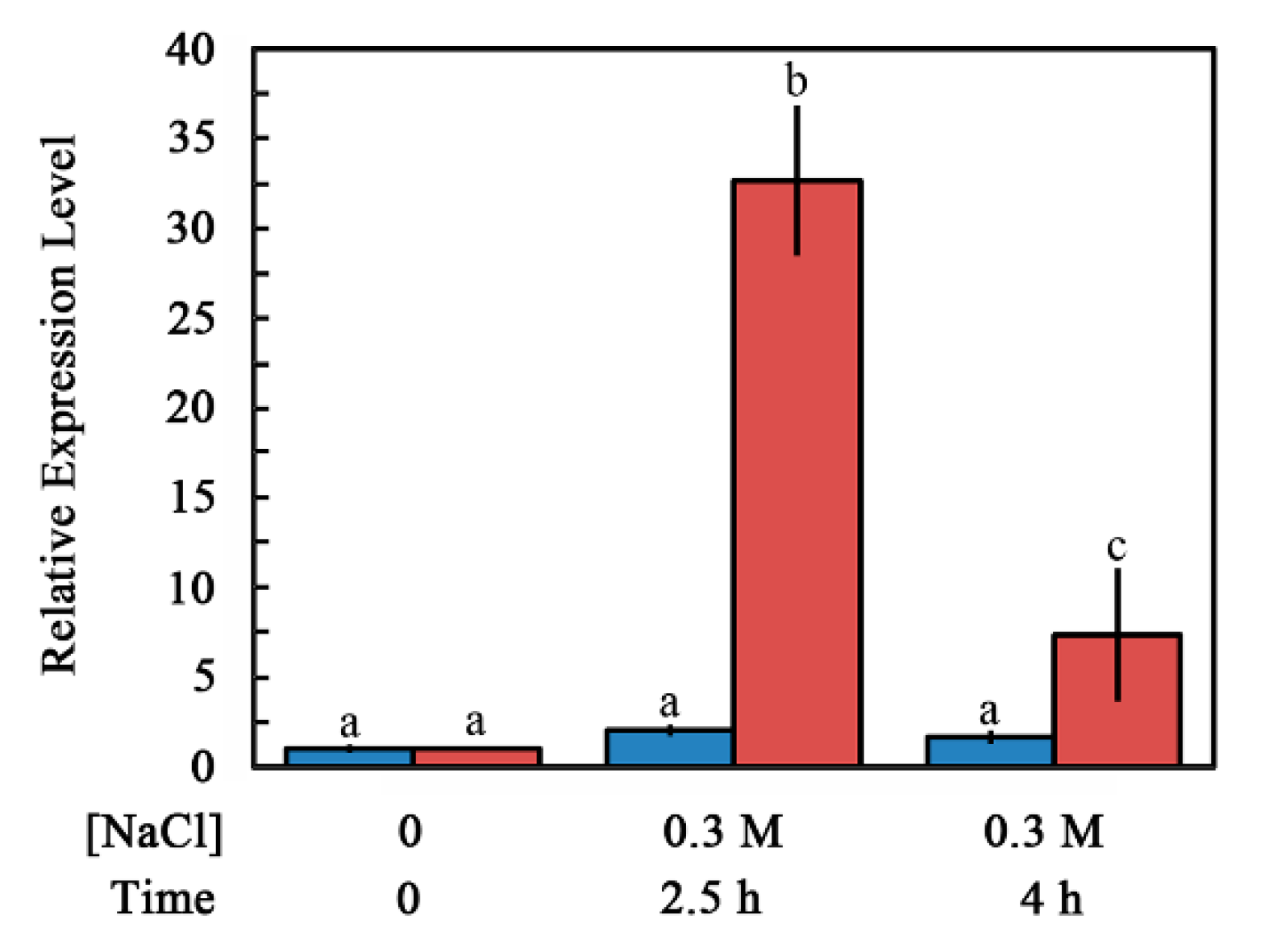
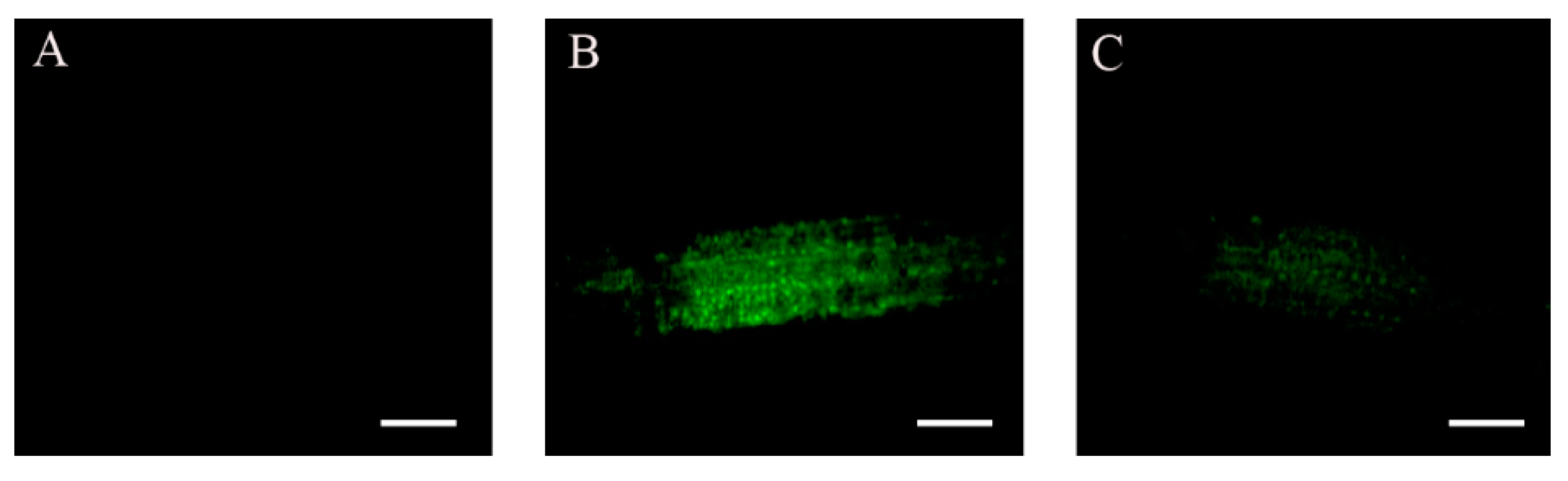
2.2. Accumulation of ABI4-eGFP Is NaCl-Dependent
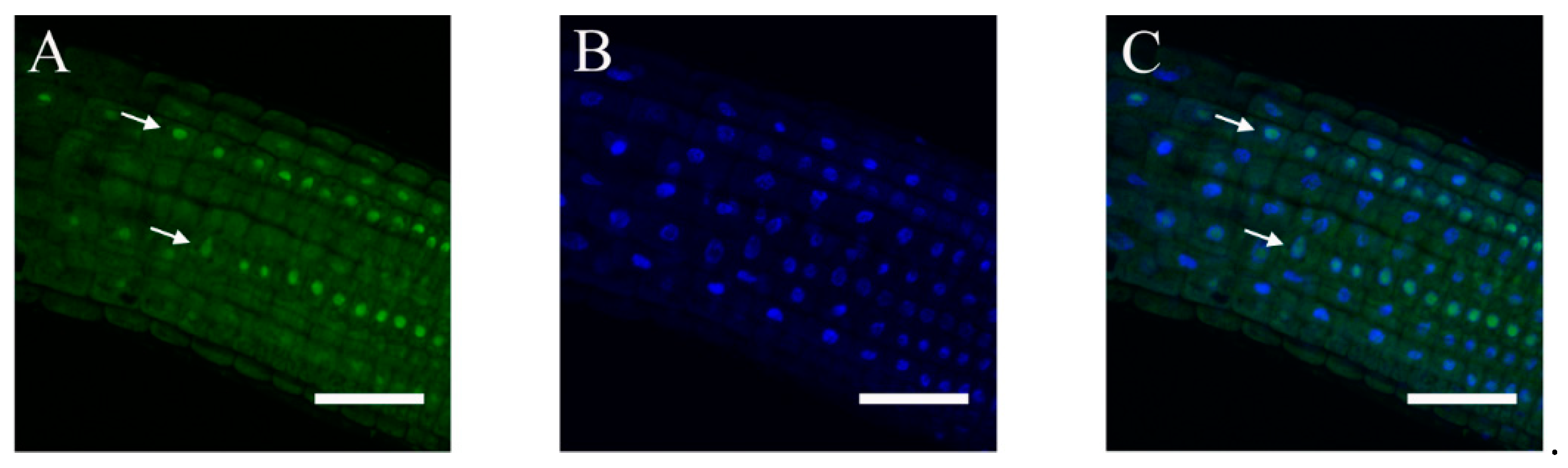
2.3. Subcellular Localization of ABI4-GFP following NaCl Treatment Is Cell-Type-Specific
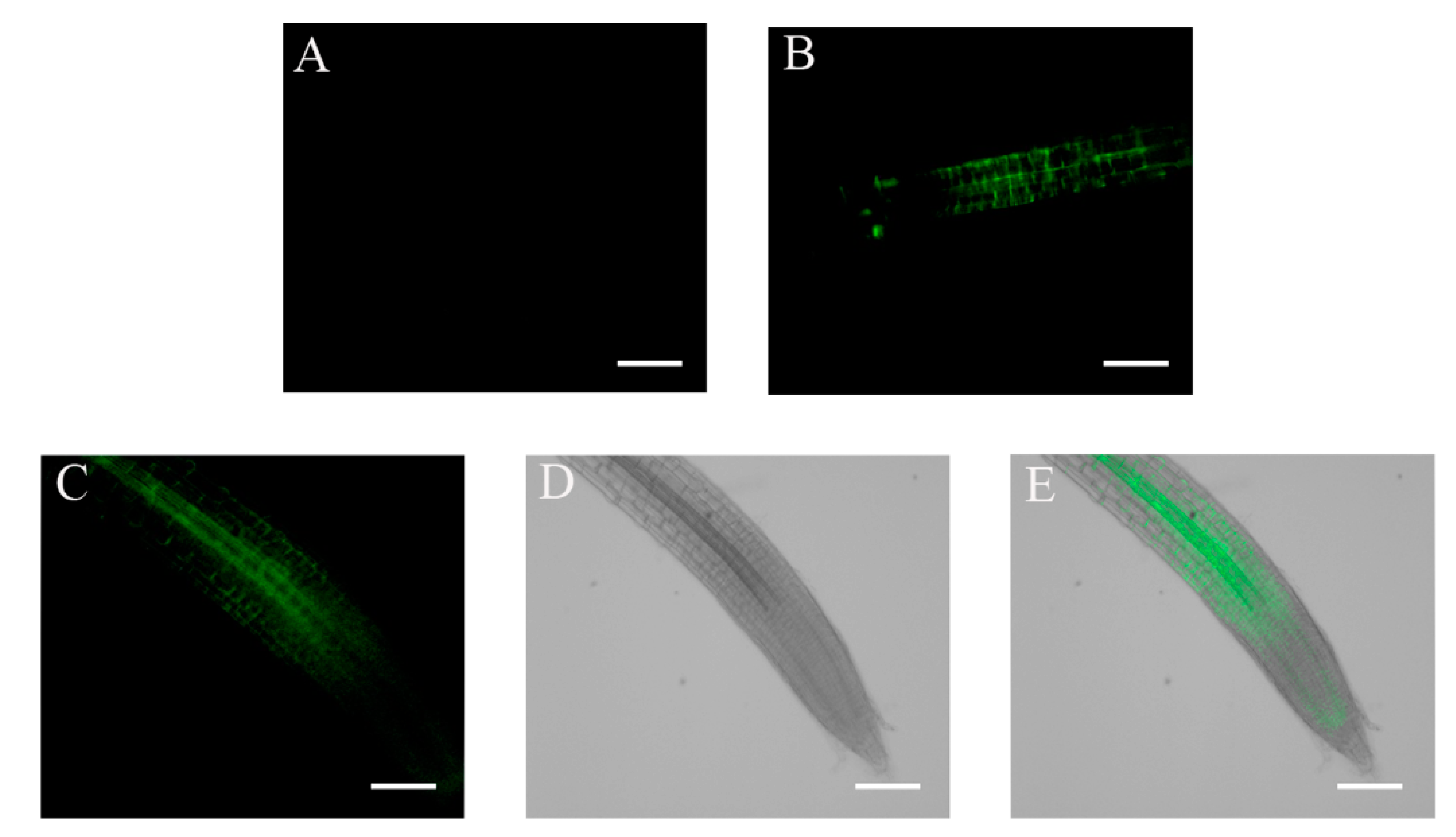
2.4. ABA and Glucose Treatment Enhance ABI4-eGFP Protein Levels
2.5. Auxin Counteracts the NaCl-Induced Increase in ABI4-eGFP Levels
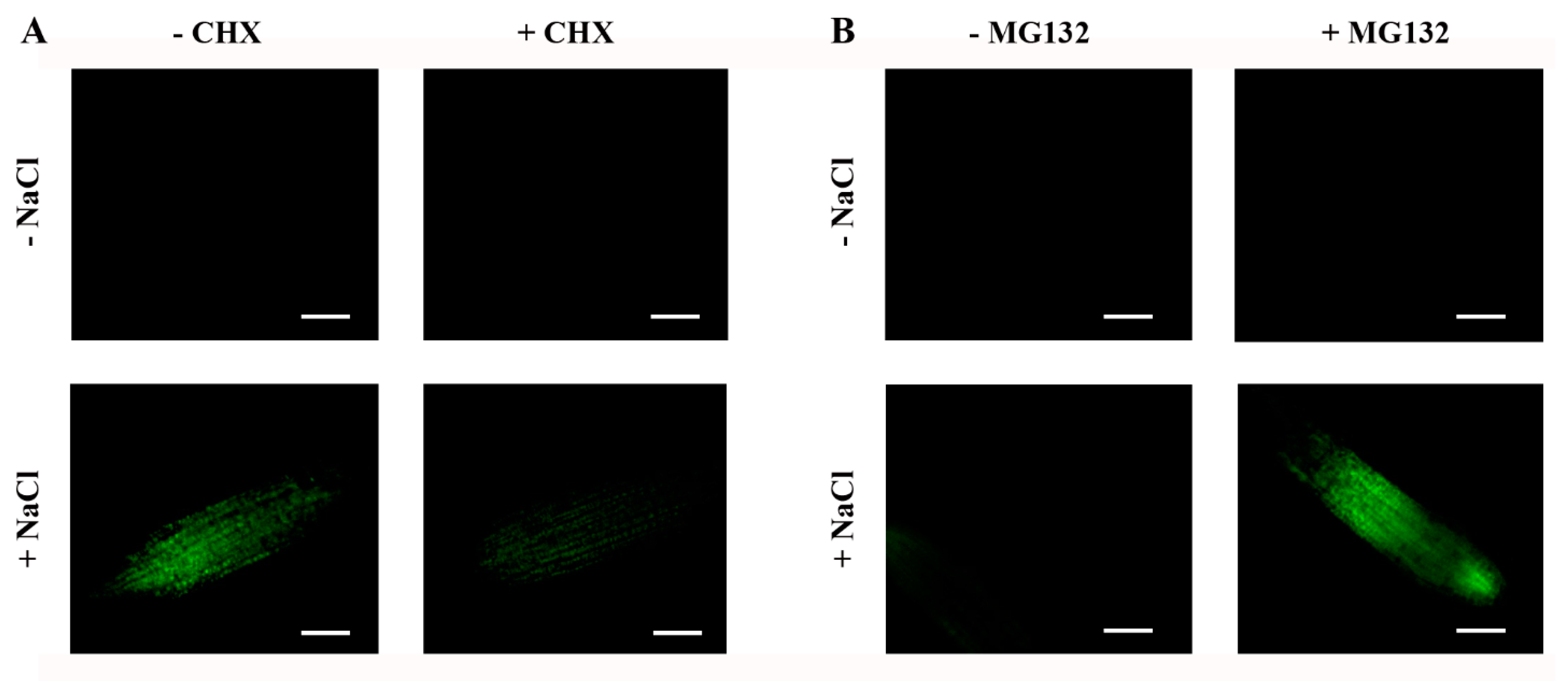
2.6. Steady-State Levels of ABI4-eGFP Are Controlled by De Novo Translation and Degradation by the 26S Proteasome
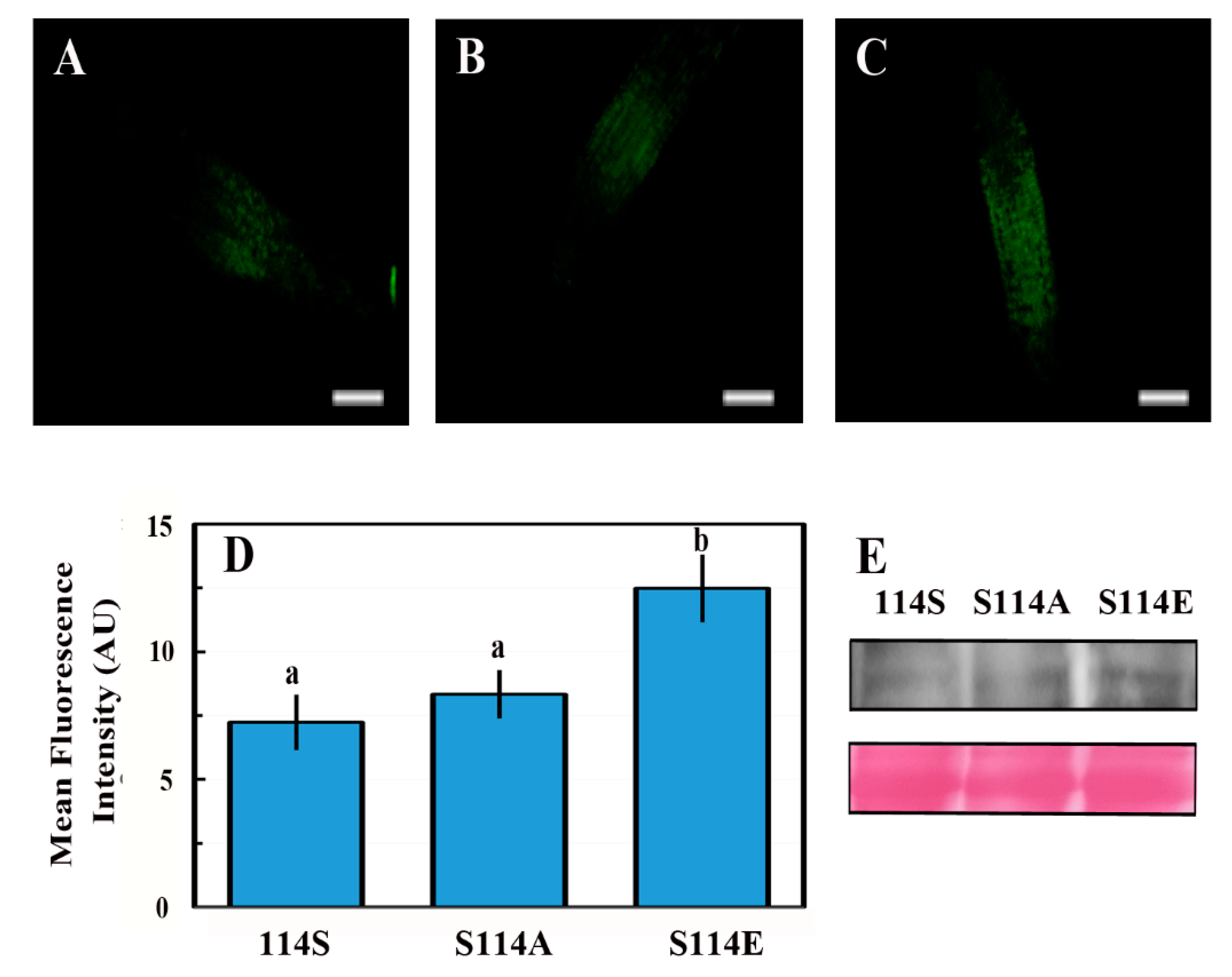
2.7. The Phosphorylation State of Serine 114 Affects the Stability of the ABI4 Protein
3. Discussion
3.1. ABI4 Is a Lowly Expressed and Highly Regulated Gene
3.2. ABI4 Is a Post-Transcriptionally Regulated Low-Level Protein
3.3. ABI4 Is Stabilized by External Signals
3.4. Phosphorylation of S114 Stabilizes ABI4
3.5. MAPK Regulates ABI4 Both Transcriptionally and Post-Transcriptionally
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Constructs and Plant Transformation
4.3. Germination Assay
4.4. Plant Treatment
4.5. Microscopy
4.6. Embryo Excision
4.7. Protein Extraction, SDS-PAGE, and Western Blot Analysis
4.8. Quantitative RT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Wang, M.L.; Lynch, T.J.; Rao, S.; Goodman, H.M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 1998, 10, 1043–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jofuku, K.D.; Den Boer, B.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed] [Green Version]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994, 6, 765–771. [Google Scholar] [CrossRef]
- Quesada, V.; Ponce, M.R.; Micol, J.L. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 2000, 154, 421–436. [Google Scholar] [CrossRef]
- Arenas-Huertero, F.; Arroyo, A.; Zhou, L.; Sheen, J.; León, P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000, 14, 2085–2096. [Google Scholar] [CrossRef]
- Laby, R.J.; Kincaid, M.S.; Kim, D.; Gibson, S.I. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000, 23, 587–596. [Google Scholar] [CrossRef]
- Huijser, C.; Kortstee, A.; Pego, J.; Weisbeek, P.; Wisman, E.; Smeekens, S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 2000, 23, 577–585. [Google Scholar] [CrossRef]
- Rook, F.; Corke, F.; Card, R.; Munz, G.; Smith, C.; Bevan, M.W. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001, 26, 421–433. [Google Scholar] [CrossRef]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S.C. ABI4: Versatile activator and repressor. Trends Plant Sci. 2013, 18, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Kacprzak, S.M.; Mochizuki, N.; Naranjo, B.; Xu, D.; Leister, D.; Kleine, T.; Okamoto, H.; Terry, M.J. Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol. 2019, 179, 18–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Söderman, E.M.; Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000, 124, 1752–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef]
- Penfield, S.; Li, Y.; Gilday, A.D.; Graham, S.; Graham, I.A. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 2006, 18, 1887–1899. [Google Scholar] [CrossRef] [Green Version]
- Shkolnik-Inbar, D.; Adler, G.; Bar-Zvi, D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 2013, 73, 993–1005. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef] [Green Version]
- Shkolnik-Inbar, D.; Bar-Zvi, D. Expression of ABSCISIC ACID INSENSITIVE 4 (ABI4) in developing Arabidopsis seedlings. Plant Signal. Behav. 2011, 6, 694–696. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, A.; Bossi, F.; Finkelstein, R.R.; León, P. Three genes that affect sugar sensing (Abscisic Acid Insensitive 4, Abscisic acid Insensitive 5, and Constitutive Triple Response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 2003, 133, 231–242. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.; Lynch, T.; Reeves, W.; Petitfils, M.; Mostachetti, M. Accumulation of the transcription factor ABA-insensitive (ABI)4 is tightly regulated post-transcriptionally. J. Exp. Bot. 2011, 62, 3971–3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregorio, J.; Hernández-Bernal, A.F.; Cordoba, E.; León, P. Characterization of evolutionarily conserved motifs involved in activity and regulation of the ABA-INSENSITIVE (ABI) 4 transcription factor. Mol. Plant 2014, 7, 422–436. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chi, W.; Sun, X.; Feng, P.; Guo, H.; Li, J.; Lin, R.; Lu, C.; Wang, H.; Leister, D.; et al. Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat. Plants 2016, 2, 16066. [Google Scholar] [CrossRef] [PubMed]
- Popescu, S.C.; Popescu, G.V.; Bachan, S.; Zhang, Z.; Gerstein, M.; Snyder, M.; Dinesh-Kumar, S.P. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009, 23, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Feng, P.; Chi, W.; Sun, X.; Xu, X.; Li, Y.; Ren, D.; Lu, C.; Rochaix, J.D.; Leister, D. Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat. Commun. 2016, 7, 12173. [Google Scholar] [CrossRef]
- Bai, Z.; Zhang, J.; Ning, X.; Guo, H.; Xu, X.; Huang, X.; Wang, Y.; Hu, Z.; Lu, C.; Zhang, L. A kinase–phosphatase–transcription factor module regulates adventitious root emergence in Arabidopsis root–hypocotyl junctions. Mol. Plant. 2020, 13, 1162–1177. [Google Scholar] [CrossRef]
- Eisner, N.; Maymon, T.; Sanchez, E.C.; Bar-Zvi, D.; Brodsky, S.; Finkelstein, R.; Bar-Zvi, D. Phosphorylation of serine 114 of the transcription factor ABSCISIC ACID INSENSITIVE 4 is essential for activity. Plant Sci. 2021, 305, 110847. [Google Scholar] [CrossRef]
- Cinelli, R.A.; Ferrari, A.; Pellegrini, V.; Tyagi, M.; Giacca, M.; Beltram, F. The enhanced green fluorescent protein as a tool for the analysis of protein dynamics and localization: Local fluorescence study at the single-molecule level. Photochem. Photobiol. 2000, 71, 771–776. [Google Scholar] [CrossRef]
- Odell, J.T.; Nagy, F.; Chua, N.H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812. [Google Scholar] [CrossRef]
- Finkelstein, R.R. Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant (lea) gene. Mol. Gen. Genet. 1993, 238, 401–408. [Google Scholar] [CrossRef]
- Amack, S.C.; Antunes, M.S. CaMV35S promoter—A plant biology and biotechnology workhorse in the era of synthetic biology. Curr. Plant Biol. 2020, 24, 100179. [Google Scholar] [CrossRef]
- Chandrasekaran, U.; Luo, X.F.; Zhou, W.G.; Shu, K. Multifaceted signaling networks mediated by Abscisic Acid Insensitive 4. Plant Commun. 2020, 1, 100040. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Zhang, X.; Gong, Z.; Yang, S.; Shi, Y. ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 2017, 89, 354–365. [Google Scholar] [CrossRef] [Green Version]
- Aviña-Padilla, K.; Ramírez-Rafael, J.A.; Herrera-Oropeza, G.E.; Muley, V.Y.; Valdivia, D.I.; Díaz-Valenzuela, E.; García-García, A.; Varela-Echavarría, A.; Hernández-Rosales, M. Evolutionary perspective and expression analysis of intronless genes highlight the conservation of their regulatory role. Front. Genet. 2021, 12, 654256. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, H.-M.; Zhu, K.; Van de Peer, Y.; Cheng, Z.-M. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef]
- Zhang, G.; Gurtu, V.; Kain, S.R. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem. Biophys. Res. Commun. 1996, 227, 707–711. [Google Scholar] [CrossRef]
- Birnbaum, K.; Shasha, D.E.; Wang, J.Y.; Jung, J.W.; Lambert, G.M.; Galbraith, D.W.; Benfey, P.N. A gene expression map of the Arabidopsis root. Science 2003, 302, 1956–1960. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Ba, Q.; Lu, D.; Li, W.; Salovska, B.; Hou, P.; Mueller, T.; Rosenberger, G.; Gao, E.; Di, Y.; et al. Global and site-specific effect of phosphorylation on protein turnover. Dev. Cell 2021, 56, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The Antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 2004, 16, 3386–3399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Bonilla, L.D.; Eschen-Lippold, L.; Gago-Zachert, S.; Tabassum, N.; Bauer, N.; Scheel, D.; Lee, J. The Arabidopsis tandem zinc finger 9 protein binds RNA and mediates pathogen-associated molecular pattern-triggered immune responses. Plant Cell Physiol. 2014, 55, 412–425. [Google Scholar] [CrossRef] [Green Version]
- Bigeard, J.; Hirt, H. Nuclear Signaling of Plant MAPKs. Front. Plant Sci. 2018, 9, 469. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Chen, W.H.; Jia, W.; Zhang, J. Mitogen-activated protein kinase kinase 5 (MKK5)-mediated signalling cascade regulates expression of iron superoxide dismutase gene in Arabidopsis under salinity stress. J. Exp. Bot. 2015, 66, 5971–5981. [Google Scholar] [CrossRef] [Green Version]
- Ichimura, K.; Mizoguchi, T.; Yoshida, R.; Yuasa, T.; Shinozaki, K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000, 24, 655–665. [Google Scholar] [CrossRef]
- Menges, M.; Dóczi, R.; Ökrész, L.; Morandini, P.; Mizzi, L.; Soloviev, M.; Murray, J.A.; Bögre, L. Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol. 2008, 179, 643–662. [Google Scholar] [CrossRef]
- Sheikh, A.H.; Eschen-Lippold, L.; Pecher, P.; Hoehenwarter, W.; Sinha, A.K.; Scheel, D.; Lee, J. Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 61. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-S.; Chao, Y.-C.; Tseng, T.-W.; Huang, C.-K.; Lo, P.-C.; Lu, C.-A. Two MYB-related transcription factors play opposite roles in sugar signaling in Arabidopsis. Plant Mol. Biol. 2017, 93, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Feng, C.Z.; Ye, Q.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 2016, 12, e1005833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhong, M.; Qu, L.; Yang, J.; Liu, X.; Zhao, Q.; Liu, X.; Zhao, X. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021, 310, 110983. [Google Scholar] [CrossRef]
- Guo, Z.; Xu, H.; Lei, Q.; Du, J.; Li, C.; Wang, C.; Yang, Y.; Yang, Y.; Sun, X. The Arabidopsis transcription factor LBD15 mediates ABA signaling and tolerance of water-deficit stress by regulating ABI4 expression. Plant J. 2020, 104, 510–521. [Google Scholar] [CrossRef]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef]
- Shang, Y.; Yan, L.; Liu, Z.-Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.-Q.; Wang, X.-F.; Du, S.-Y.; Jiang, T. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Seo, P.J. Coordination of seed dormancy and germination processes by MYB96. Plant Signal. Behav. 2015, 10, e1056423. [Google Scholar] [CrossRef] [Green Version]
- Reeves, W.M.; Lynch, T.J.; Mobin, R.; Finkelstein, R.R. Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 2011, 75, 347–363. [Google Scholar] [CrossRef] [Green Version]
- Bossi, F.; Cordoba, E.; Dupré, P.; Mendoza, M.S.; Román, C.S.; León, P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 2009, 59, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maymon, T.; Eisner, N.; Bar-Zvi, D. The ABCISIC ACID INSENSITIVE (ABI) 4 Transcription Factor Is Stabilized by Stress, ABA and Phosphorylation. Plants 2022, 11, 2179. https://doi.org/10.3390/plants11162179
Maymon T, Eisner N, Bar-Zvi D. The ABCISIC ACID INSENSITIVE (ABI) 4 Transcription Factor Is Stabilized by Stress, ABA and Phosphorylation. Plants. 2022; 11(16):2179. https://doi.org/10.3390/plants11162179
Chicago/Turabian StyleMaymon, Tzofia, Nadav Eisner, and Dudy Bar-Zvi. 2022. "The ABCISIC ACID INSENSITIVE (ABI) 4 Transcription Factor Is Stabilized by Stress, ABA and Phosphorylation" Plants 11, no. 16: 2179. https://doi.org/10.3390/plants11162179
APA StyleMaymon, T., Eisner, N., & Bar-Zvi, D. (2022). The ABCISIC ACID INSENSITIVE (ABI) 4 Transcription Factor Is Stabilized by Stress, ABA and Phosphorylation. Plants, 11(16), 2179. https://doi.org/10.3390/plants11162179




