In Vitro and In Silico Study of the α-Glucosidase and Lipase Inhibitory Activities of Chemical Constituents from Piper cumanense (Piperaceae) and Synthetic Analogs
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phytochemistry
2.2. Synthesis of Analogs and Derivatives
2.3. Inhibition of Digestive Enzymes
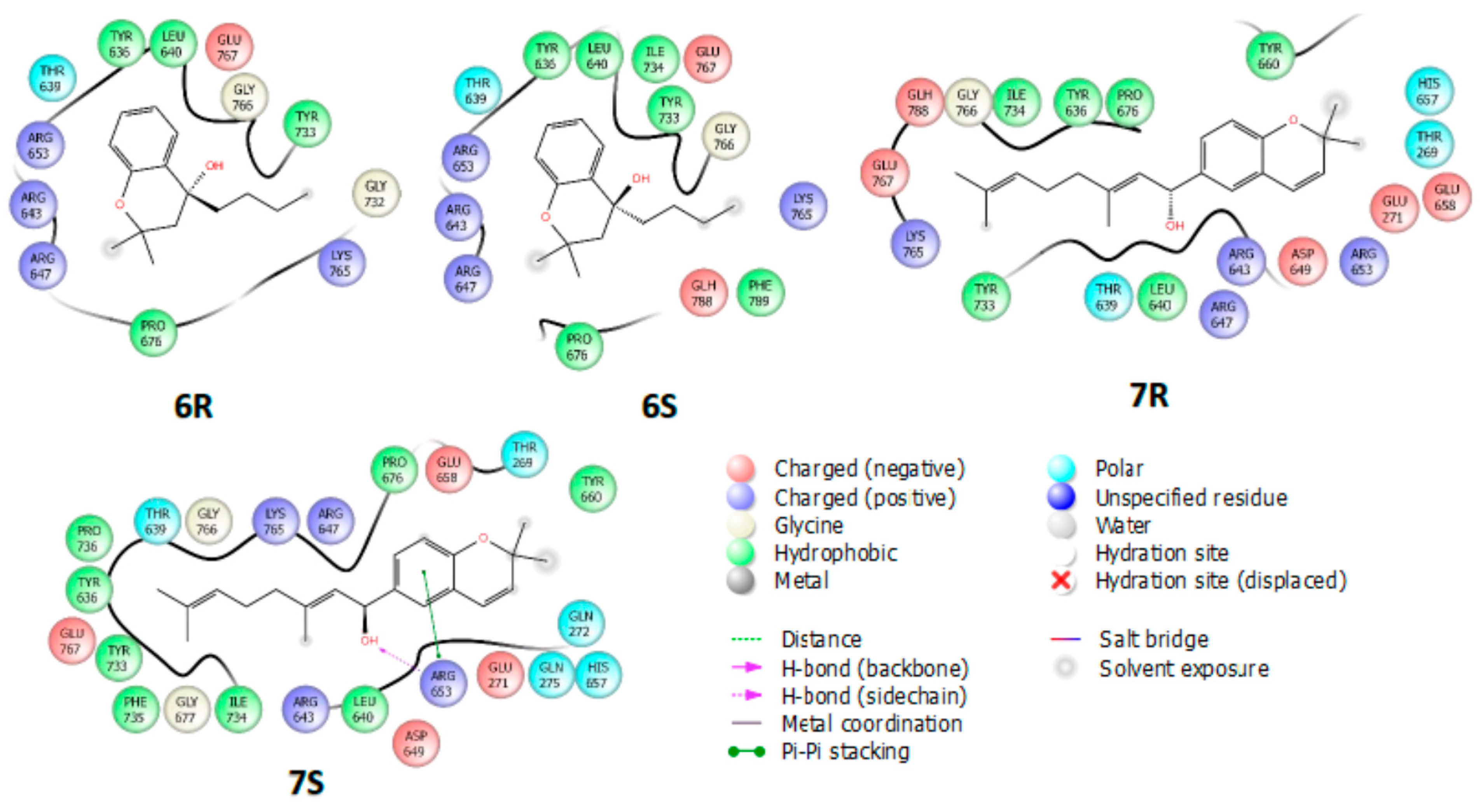
2.4. Molecular Docking
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Preparation of Analogs and Derivatives
3.5. Biological Activity–Determination of Enzymatic Inhibition against Pancreatic Lipase (PL) and α-Glucosidase (AG)
3.5.1. PL Inhibition Assay
3.5.2. AG Inhibition Assay
3.5.3. Kinetic Study
3.6. Molecular Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardozo-Muñoz, J.; Cuca-Suárez, L.E.; Prieto-Rodríguez, J.A.; Lopez-Vallejo, F.; Patiño-Ladino, O.J. Multitarget Action of Xanthones from Garcinia mangostana against α-Amylase, α-Glucosidase and Pancreatic Lipase. Molecules 2022, 27, 3283. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Prasad, A.; Haque, F.; Ray, S.; De, B.; Ray, S.S. In silico investigation of black tea components on α-amylase, α-glucosidase and lipase. J. Appl. Pharm. Sci. 2015, 5, 42–47. [Google Scholar] [CrossRef]
- Macabeo, A.P.G.; Pilapil, L.A.E.; Garcia, K.Y.M.; Quimque, M.T.J.; Phukhamsakda, C.; Cruz, A.J.C.; Hyde, K.D.; Stadler, M. Alpha-Glucosidase- and Lipase-Inhibitory Phenalenones from a New Species of Pseudolophiostoma Originating from Thailand. Molecules 2020, 25, 965. [Google Scholar] [CrossRef] [PubMed]
- Foddai, M.; Kasabri, V.; Afifi, F.U.; Azara, E.; Petretto, G.L.; Pintore, G. In Vitro Inhibitory Effects of Sardinian Pistacia lentiscus L. and Pistacia terebinthus L. on Metabolic Enzymes: Pancreatic Lipase, α-Amylase, and α-Glucosidase. Starch-Stärke 2015, 67, 204–212. [Google Scholar] [CrossRef]
- Shi, Y.; Burn, P. Lipid Metabolic Enzymes: Emerging Drug Targets for the Treatment of Obesity. Nat. Rev. Drug Discov. 2004, 3, 695–710. [Google Scholar] [CrossRef]
- Liu, T.-T.; Liu, X.-T.; Chen, Q.-X.; Shi, Y. Lipase Inhibitors for Obesity: A Review. Biomed. Pharmacother. 2020, 128, 110314. [Google Scholar] [CrossRef]
- Klein, S.; Gastaldelli, A.; Yki-Järvinen, H.; Scherer, P.E. Why Does Obesity Cause Diabetes? Cell Metab. 2022, 34, 11–20. [Google Scholar] [CrossRef]
- Reed, J.; Bain, S.; Kanamarlapudi, V. A Review of Current Trends with Type 2 Diabetes Epidemiology, Aetiology, Pathogenesis, Treatments and Future Perspectives. Diabetes Metab. Syndr. Obes. Targets Ther. 2021, 14, 3567–3602. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Abdullah, N.; Zain, W.; Hamid, H.; Ramli, N. Essential Oil from Piperaceae as a Potential for Biopesticide Agents: A Review. Food Res. 2020, 4, 1–10. [Google Scholar] [CrossRef]
- Salehi, B.; Zakaria, Z.A.; Gyawali, R.; Ibrahim, S.A.; Rajkovic, J.; Shinwari, Z.K.; Khan, T.; Sharifi-Rad, J.; Ozleyen, A.; Turkdonmez, E.; et al. Piper Species: A Comprehensive Review on Their Phytochemistry, Biological Activities and Applications. Molecules 2019, 24, 1364. [Google Scholar] [CrossRef] [PubMed]
- Mgbeahuruike, E.E.; Yrjönen, T.; Vuorela, H.; Holm, Y. Bioactive Compounds from Medicinal Plants: Focus on Piper Species. S. Afr. J. Bot. 2017, 112, 54–69. [Google Scholar] [CrossRef]
- Ekeuku, S.O.; Nur Azlina, M.F.; Chin, K.-Y. Effects of Piper Sarmentosum on Metabolic Syndrome and Its Related Complications: A Review of Preclinical Evidence. Appl. Sci. 2021, 11, 9860. [Google Scholar] [CrossRef]
- Njeri, L.K.; Eliud, N.M.N. Anti-Diabetic Activity in Mice of Piper Capence Used Traditionally in the Management of Diabetes Mellitus in Kenya. J. Diabetes Metab. 2017, 8, 737–743. [Google Scholar] [CrossRef]
- Yadav, V.; Krishnan, A.; Vohora, D. A Systematic Review on Piper longum L.: Bridging Traditional Knowledge and Pharmacological Evidence for Future Translational Research. J. Ethnopharmacol. 2020, 247, 112255. [Google Scholar] [CrossRef]
- Sun, X.; Chen, W.; Dai, W.; Xin, H.; Rahmand, K.; Wang, Y.; Zhang, J.; Zhang, S.; Xu, L.; Han, T. Piper Sarmentosum Roxb.: A Review on Its Botany, Traditional Uses, Phytochemistry, and Pharmacological Activities. J. Ethnopharmacol. 2020, 263, 112897. [Google Scholar] [CrossRef] [PubMed]
- Takooree, H.; Aumeeruddy, M.Z.; Rengasamy, K.R.R.; Venugopala, K.N.; Jeewon, R.; Zengin, G.; Mahomoodally, M.F. A Systematic Review on Black Pepper (Piper nigrum L.): From Folk Uses to Pharmacological Applications. Crit. Rev. Food Sci. Nutr. 2019, 59, S210–S243. [Google Scholar] [CrossRef]
- Nabi, S.A.; Kasetti, R.B.; Sirasanagandla, S.; Tilak, T.K.; Kumar, M.V.J.; Rao, C.A. Antidiabetic and Antihyperlipidemic Activity of Piper longum Root Aqueous Extract in STZ Induced Diabetic Rats. BMC Complement. Altern. Med. 2013, 13, 37. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, S.; Vasudeva, N. Screening of Antidiabetic and Antihyperlipidemic Potential of Oil from Piper longum and Piperine with Their Possible Mechanism. Expert Opin. Pharmacother. 2013, 14, 1723–1736. [Google Scholar] [CrossRef]
- Thent, Z.C.; Seong Lin, T.; Das, S.; Zakaria, Z. Effect of Piper sarmentosum Extract on the Cardiovascular System of Diabetic Sprague-Dawley Rats: Electron Microscopic Study. Evid.-Based Complement. Altern. Med. 2012, 2012, 628750. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.R.; Mohd Ramli, E.S.; Abdul Nasir, N.A.; Mohd Ismail, N.; Mohd Fahami, N.A. Methanolic Extract of Piper Sarmentosum Attenuates Obesity and Hyperlipidemia in Fructose-Induced Metabolic Syndrome Rats. Molecules 2021, 26, 3985. [Google Scholar] [CrossRef] [PubMed]
- Damsud, T.; Adisakwattana, S.; Phuwapraisirisan, P. Three New Phenylpropanoyl Amides from the Leaves of Piper sarmentosum and Their α-Glucosidase Inhibitory Activities. Phytochem. Lett. 2013, 6, 350–354. [Google Scholar] [CrossRef]
- Daud, D.; Jalil, U.J.S.A.; Sahrol, N.A.A.; Zulkefli, S.Z.; Adam, N.A.; Ishak, N.K.; Norzehan, E.; Mahbob, M.; Tawang, A. Comparative Anti-Obesity and Anti-Diabetic Properties of Piper sarmentosum and Piper Betle Aqueous Extracts via in-Vitro System. J. Pharm. Adv. Res. 2021, 4, 1421–1427. [Google Scholar]
- Yogeswari, S.; Bindu, K.H.; Kamalraj, S.; Ashokkumar, V.; Jayabaskaran, C. Antidiabetic, Antithrombin and Cytotoxic Bioactive Compounds in Five Cultivars of Piper betle L. Environ. Technol. Innov. 2020, 20, 101140. [Google Scholar] [CrossRef]
- Kaleem, M.; Sheema; Sarmad, H.; Bano, B. Protective Effects of Piper nigrum and Vinca rosea in Alloxan Induced Diabetic Rats. Indian J. Physiol. Pharmacol. 2005, 49, 65–71. [Google Scholar]
- Magaña-Barajas, E.; Buitimea-Cantúa, G.V.; Hernández-Morales, A.; Torres-Pelayo, V.D.R.; Vázquez-Martínez, J.; Buitimea-Cantúa, N.E. In Vitro α-Amylase and α-Glucosidase Enzyme Inhibition and Antioxidant Activity by Capsaicin and Piperine from Capsicum Chinense and Piper nigrum Fruits. J. Environ. Sci. Health Part B 2021, 56, 282–291. [Google Scholar] [CrossRef]
- Parra Amin, J.E.; Cuca, L.E.; González-Coloma, A. Antifungal and Phytotoxic Activity of Benzoic Acid Derivatives from Inflorescences of Piper cumanense. Nat. Prod. Res. 2021, 35, 2763–2771. [Google Scholar] [CrossRef]
- Parra, J.E.; Patiño, O.J.; Prieto, J.A.; Delgado, W.A.; Cuca, L.E. A New Benzoic Acid Derivative Isolated from Piper Cf. Cumanense Kunth (Piperaceae). Phytochem. Lett. 2013, 6, 590–592. [Google Scholar] [CrossRef]
- Parra, J.E.; Delgado, W.A.; Cuca, L.E. Cumanensic Acid, a New Chromene Isolated from Piper Cf. Cumanense Kunth. (Piperaceae). Phytochem. Lett. 2011, 4, 280–282. [Google Scholar] [CrossRef]
- Ruiz, P.G.; Garavito, G.; Acebey, C.L.; Arteaga, L.; Pinzon, R.; Gimenez, T.A. Actividad Leishmanicida y Tripanocida de Algunas Plantas Reportadas Como Medicinales En Colombia. Biofarbo 2004, 13, 27–30. [Google Scholar]
- Sánchez-Suárez, J.; Albarracín, D.; Rojas, M.; Rincón, J.; Robledo, S.; Muñoz, D.L.; Oviedo, J.J.; Calderón, M.N.; Fernández, N.; Delgado, G. Evaluación de la actividad citotóxica y leishmanicida de extractos y fracciones de Piper cumanense y Piper holtonii Evaluation of the cytotoxic and leishmanicidal activity of extracts and fractions of Piper cumanense and Piper holtonii. Rev. Colomb. Cienc. Quim. Farm. 2010, 39, 21–29. [Google Scholar]
- Rapado, L.N.; Freitas, G.C.; Polpo, A.; Rojas-Cardozo, M.; Rincón, J.V.; Scotti, M.T.; Kato, M.J.; Nakano, E.; Yamaguchi, L.F. A Benzoic Acid Derivative and Flavokawains from Piper Species as Schistosomiasis Vector Controls. Molecules 2014, 19, 5205–5218. [Google Scholar] [CrossRef] [PubMed]
- Velandia, S.A.; Quintero, E.; Stashenko, E.E.; Ocazionez, R.E. Actividad Antiproliferativa de Aceites Esenciales de Plantas Cultivadas En Colombia. Acta Biológica Colomb. 2018, 23, 189–198. [Google Scholar] [CrossRef]
- Muñoz, D.; Sandoval-Hernandez, A.; Delgado, W.; Arboleda, G.; Cuca, L. In Vitro Anticancer Screening of Colombian Plants from Piper genus (Piperaceae). J. Pharmacogn. Phytother. 2018, 10, 174–181. [Google Scholar] [CrossRef]
- Gaia, A.M.; Yamaguchi, L.F.; Jeffrey, C.S.; Kato, M.J. Age-Dependent Changes from Allylphenol to Prenylated Benzoic Acid Production in Piper gaudichaudianum Kunth. Phytochemistry 2014, 106, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Lago, J.H.G.; Ito, A.T.; Fernandes, C.M.; Young, M.C.M.; Kato, M.J. Secondary Metabolites Isolated from Piper chimonantifolium and Their Antifungal Activity. Nat. Prod. Res. 2012, 26, 770–773. [Google Scholar] [CrossRef]
- Lago, J.H.G.; Ramos, C.S.; Casanova, D.C.C.; Morandim, A.D.A.; Bergamo, D.C.B.; Cavalheiro, A.J.; Bolzani, V.D.S.; Furlan, M.; Guimarães, E.F.; Young, M.C.M.; et al. Benzoic Acid Derivatives from Piper Species and Their Fungitoxic Activity against Cladosporium Cladosporioides and C. Sphaerospermum. J. Nat. Prod. 2004, 67, 1783–1788. [Google Scholar] [CrossRef]
- Cao, S.; Rossant, C.; Ng, S.; Buss, A.D.; Butler, M.S. Phenolic Derivatives from Wigandia Urens with Weak Activity against the Chemokine Receptor CCR5. Phytochemistry 2003, 64, 987–990. [Google Scholar] [CrossRef]
- Faraone, I.; Russo, D.; Genovese, S.; Milella, L.; Monné, M.; Epifano, F.; Fiorito, S. Screening of in Vitro and in Silico α-Amylase, α-Glucosidase, and Lipase Inhibitory Activity of Oxyprenylated Natural Compounds and Semisynthetic Derivatives. Phytochemistry 2021, 187, 112781. [Google Scholar] [CrossRef]
- Kwiecień, H.; Smist, M.; Wrzesniewska, A. Synthesis of Aryl-Fused 1,4-Oxazepines and Their Oxo Derivatives: A Review. Curr. Org. Synth. 2012, 9, 828–850. [Google Scholar] [CrossRef]
- Osipov, D.; Osyanin, V.; Klimochkin, Y. Reactions of 5-Formyl-and 5-Acyl-3,4-Dihydro-2H-Pyrans and Their Annelated Analogs with Nucleophiles. Targets Heterocycl. Syst. 2018, 22, 436–467. [Google Scholar] [CrossRef]
- Iguchi, D.; Erra-Balsells, R.; Bonesi, S.M. Expeditious Photochemical Reaction toward the Preparation of Substituted Chroman-4-Ones. Tetrahedron Lett. 2014, 55, 4653–4656. [Google Scholar] [CrossRef]
- López, C.S.; Erra-Balsells, R.; Bonesi, S.M. A Mild and Convenient One-Pot Photochemical Synthesis of Chroman-4-One Derivatives. The Photo-Fries Rearrangement of (Hetero)Aryl 3-Methyl-2-Butenoate Esters under Basic Catalysis. Tetrahedron Lett. 2010, 51, 4387–4390. [Google Scholar] [CrossRef]
- Souza, G.H.B.; da Silva Filho, A.A.; de Souza, V.A.; Pereira, A.C.; Royo, V.A.; e Silva, M.L.A.; da Silva, R.; Donate, P.M.; Carvalho, J.C.T.; Bastos, J.K. Analgesic and anti-inflammatory activities evaluation of (-)-O-acetyl,(-)-O-methyl,(-)-O-dimethylethylamine cubebin and their preparation from (-)-cubebin. Il Farm. 2004, 59, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Martinez-Gonzalez, A.I.; Alvarez-Parrilla, E.; Díaz-Sánchez, Á.G.; de la Rosa, L.A.; Núñez-Gastélum, J.A.; Vazquez-Flores, A.A.; Gonzalez-Aguilar, G.A. In Vitro Inhibition of Pancreatic Lipase by Polyphenols: A Kinetic, Fluorescence Spectroscopy and Molecular Docking Study. Food Technol. Biotechnol. 2017, 55, 519–530. [Google Scholar] [CrossRef]
- Li, Y.-Q.; Yang, P.; Gao, F.; Zhang, Z.-W.; Wu, B. Probing the Interaction between 3 Flavonoids and Pancreatic Lipase by Methods of Fluorescence Spectroscopy and Enzymatic Kinetics. Eur. Food Res. Technol. 2011, 233, 63–69. [Google Scholar] [CrossRef]
- Wu, X.; He, W.; Zhang, H.; Li, Y.; Liu, Z.; He, Z. Acteoside: A Lipase Inhibitor from the Chinese Tea Ligustrum Purpurascens Kudingcha. Food Chem. 2014, 142, 306–310. [Google Scholar] [CrossRef]
- You, Q.; Chen, F.; Wang, X.; Jiang, Y.; Lin, S. Anti-Diabetic Activities of Phenolic Compounds in Muscadine against Alpha-Glucosidase and Pancreatic Lipase. LWT Food Sci. Technol. 2012, 46, 164–168. [Google Scholar] [CrossRef]
- Ghani, U.; Ashraf, S.; Ul-Haq, Z.; Mujamammi, A.H.; Özkay, Y.; Demirci, F.; Kaplancikli, Z.A. Dithiocarbamate Derivatives Inhibit α-Glucosidase through an Apparent Allosteric Site on the Enzyme. Chem. Biol. Drug Des. 2021, 98, 283–294. [Google Scholar] [CrossRef]
- Liu, S.-K.; Hao, H.; Bian, Y.; Ge, Y.-X.; Lu, S.; Xie, H.-X.; Wang, K.-M.; Tao, H.; Yuan, C.; Zhang, J.; et al. Discovery of New α-Glucosidase Inhibitors: Structure-Based Virtual Screening and Biological Evaluation. Front. Chem. 2021, 9, 639279. [Google Scholar] [CrossRef] [PubMed]
- Lopéz, D.; Cherigo, L.; Mejia, L.C.; Loza-Mejía, M.A.; Martínez-Luis, S. α-Glucosidase Inhibitors from a Mangrove Associated Fungus, Zasmidium sp. Strain EM5-10. BMC Chem. 2019, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, C.; Pettus, T.R.R. Strategies for the Preparation of Differentially Protected Ortho-Prenylated Phenols. Synlett 2003, 2003, 127–137. [Google Scholar] [CrossRef]
- Walunj, R.M.; Natu, A.D.; Paradkar, M.V.; Rojatkar, S.R. Efficient Total Synthesis of Naturally Occurring Anti-TMV Compound Gramniphenol G. Synth. Commun. 2016, 46, 1425–1431. [Google Scholar] [CrossRef]
- Morelli, C.F.; Biagiotti, M.; Pappalardo, V.M.; Rabuffetti, M.; Speranza, G. Chemistry of α-Mangostin. Studies on the Semisynthesis of Minor Xanthones from Garcinia mangostana. Nat. Prod. Res. 2015, 29, 750–755. [Google Scholar] [CrossRef]
- Bustanji, Y.; Al-Masri, I.M.; Mohammad, M.; Hudaib, M.; Tawaha, K.; Tarazi, H.; Alkhatib, H.S. Pancreatic Lipase Inhibition Activity of Trilactone Terpenes of Ginkgo Biloba. J. Enzyme Inhib. Med. Chem. 2011, 26, 453–459. [Google Scholar] [CrossRef]
- Whitcomb, D.C.; Lowe, M.E. Human Pancreatic Digestive Enzymes. Dig. Dis. Sci. 2007, 52, 1–17. [Google Scholar] [CrossRef]
- Ryu, H.W.; Cho, J.K.; Curtis-Long, M.J.; Yuk, H.J.; Kim, Y.S.; Jung, S.; Kim, Y.S.; Lee, B.W.; Park, K.H. α-Glucosidase Inhibition and Antihyperglycemic Activity of Prenylated Xanthones from Garcinia mangostana. Phytochemistry 2011, 72, 2148–2154. [Google Scholar] [CrossRef]
- Iwai, K.; Kim, M.Y.; Onodera, A.; Matsue, H. α-Glucosidase Inhibitory and Antihyperglycemic Effects of Polyphenols in the Fruit of Viburnum dilatatum Thunb. J. Agric. Food Chem. 2006, 54, 4588–4592. [Google Scholar] [CrossRef]
- Suzuki, Y.; Hayashi, K.; Sakane, I.; Kakuda, T. Effect and Mode of Action of Banaba (Lagerstroemia speciosa L.) Leaf Extracts on Postprandial Blood Glucose in Rats. J. Jpn. Soc. Nutr. Food Sci. 2001, 54, 131–137. [Google Scholar] [CrossRef]
- Cheng, Y.-C.; Prusoff, W.H. Relationship between the Inhibition Constant (KI) and the Concentration of Inhibitor Which Causes 50 per Cent Inhibition (I50) of an Enzymatic Reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef] [PubMed]
- Luyen, N.T.; Tram, L.H.; Hanh, T.T.H.; Binh, P.T.; Dang, N.H.; Van Minh, C.; Dat, N.T. Inhibitors of A-Glucosidase, a-Amylase and Lipase from Chrysanthemum morifolium. Phytochem. Lett. 2013, 6, 322–325. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Preto, J.; Gentile, F. Assessing and Improving the Performance of Consensus Docking Strategies Using the DockBox Package. J. Comput. Aided Mol. Des. 2019, 33, 817–829. [Google Scholar] [CrossRef]
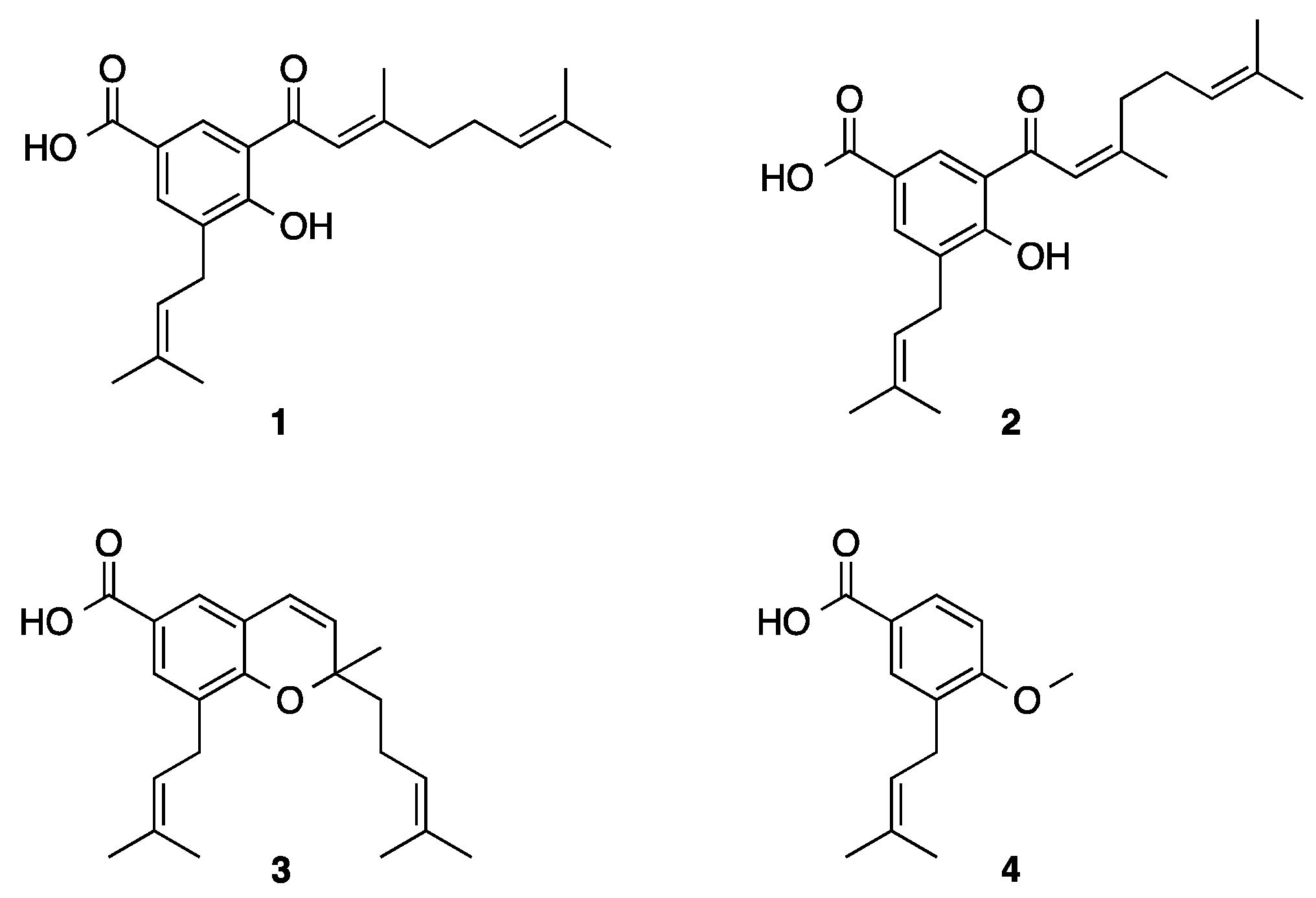
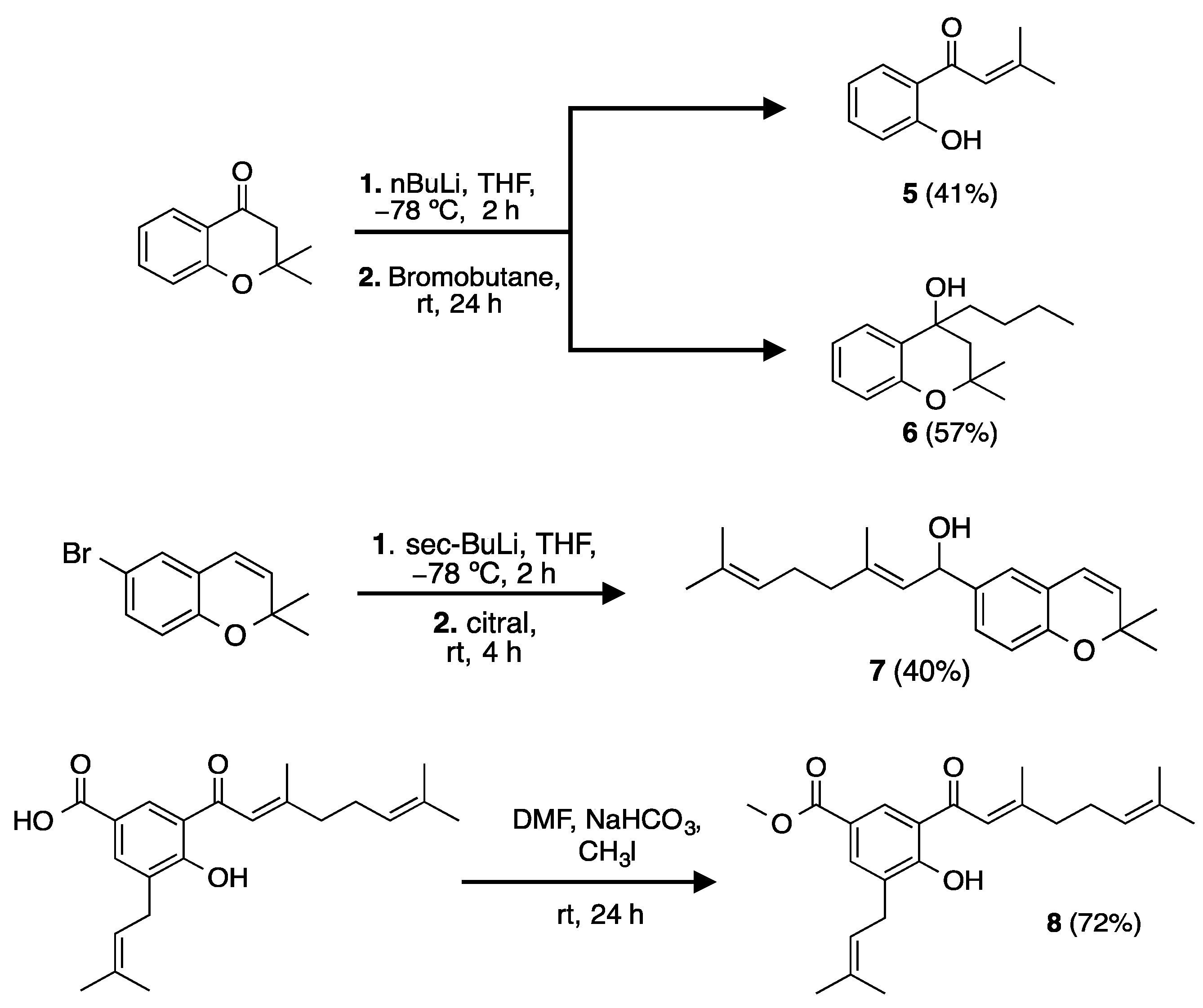
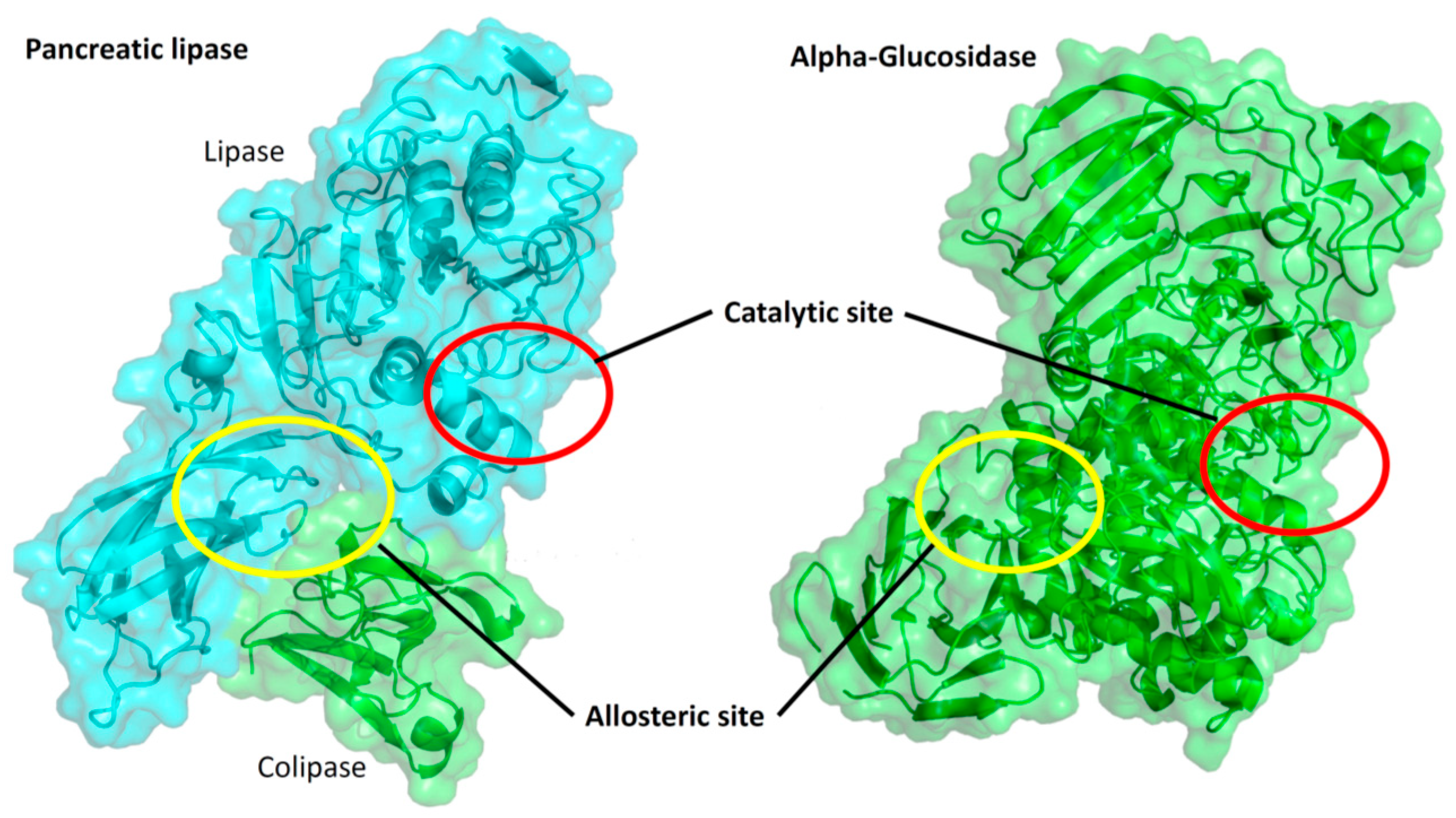
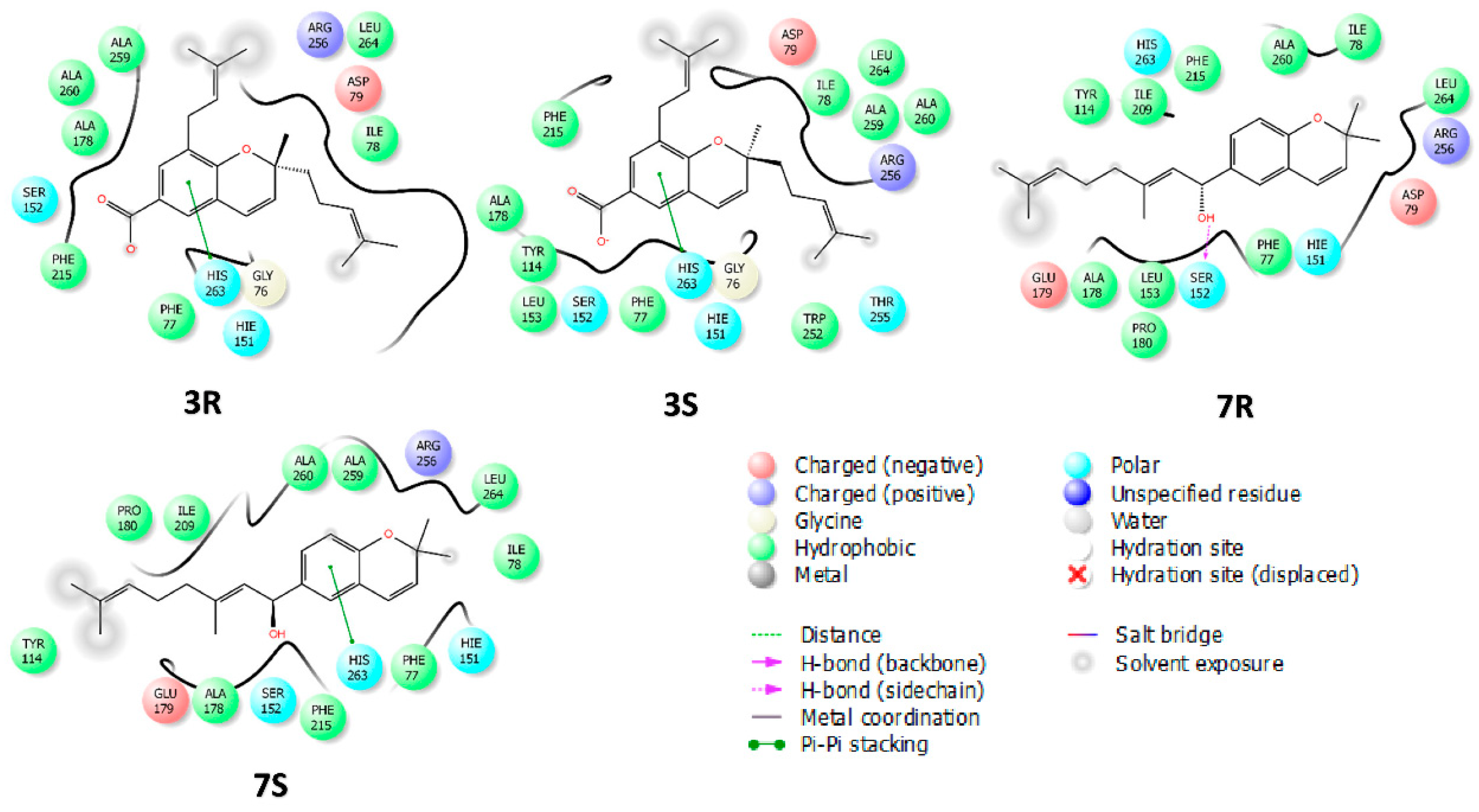

| Compound/Positive Control | Pancreatic Lipase IC50 µM | Ki (µM) | Inhibition Type | α-Glucosidase IC50 µM | Ki (µM) | Inhibition Type |
|---|---|---|---|---|---|---|
| 1 | 33.78 ± 5.95 * | 7.88 | Uncompetitive | NA | − | − |
| 2 | 36.24 ± 5.05 * | 8.46 | Uncompetitive | NA | − | − |
| 3 | 36.30 ± 6.50 * | 18.01 | Competitive | NA | − | − |
| 4 | NA | − | − | NA | − | − |
| 5 | 48.34 ± 2.57 | 48.34 | Noncompetitive | NA | − | − |
| 6 | 28.32 ± 2.90 * | 28.32 | Noncompetitive | 153.90 ± 4.65 ** | 51.30 | Mixed |
| 7 | 33.08 ± 5.28 * | 25.35 | Competitive | 134.20 ± 5.60 ** | 135.81 | Noncompetitive |
| 8 | 55.78 ± 2.98 | 42.75 | Mixed | NA | − | − |
| Orlistat | 0.73 ± 0.02 *** | 0.24 | Irreversible | − | − | − |
| Acarbose | − | − | − | 345.90 ± 3.21 * | 130.07 | Competitive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prieto-Rodríguez, J.A.; Lévuok-Mena, K.P.; Cardozo-Muñoz, J.C.; Parra-Amin, J.E.; Lopez-Vallejo, F.; Cuca-Suárez, L.E.; Patiño-Ladino, O.J. In Vitro and In Silico Study of the α-Glucosidase and Lipase Inhibitory Activities of Chemical Constituents from Piper cumanense (Piperaceae) and Synthetic Analogs. Plants 2022, 11, 2188. https://doi.org/10.3390/plants11172188
Prieto-Rodríguez JA, Lévuok-Mena KP, Cardozo-Muñoz JC, Parra-Amin JE, Lopez-Vallejo F, Cuca-Suárez LE, Patiño-Ladino OJ. In Vitro and In Silico Study of the α-Glucosidase and Lipase Inhibitory Activities of Chemical Constituents from Piper cumanense (Piperaceae) and Synthetic Analogs. Plants. 2022; 11(17):2188. https://doi.org/10.3390/plants11172188
Chicago/Turabian StylePrieto-Rodríguez, Juliet A., Kevin P. Lévuok-Mena, Juan C. Cardozo-Muñoz, Jorge E. Parra-Amin, Fabián Lopez-Vallejo, Luis E. Cuca-Suárez, and Oscar J. Patiño-Ladino. 2022. "In Vitro and In Silico Study of the α-Glucosidase and Lipase Inhibitory Activities of Chemical Constituents from Piper cumanense (Piperaceae) and Synthetic Analogs" Plants 11, no. 17: 2188. https://doi.org/10.3390/plants11172188
APA StylePrieto-Rodríguez, J. A., Lévuok-Mena, K. P., Cardozo-Muñoz, J. C., Parra-Amin, J. E., Lopez-Vallejo, F., Cuca-Suárez, L. E., & Patiño-Ladino, O. J. (2022). In Vitro and In Silico Study of the α-Glucosidase and Lipase Inhibitory Activities of Chemical Constituents from Piper cumanense (Piperaceae) and Synthetic Analogs. Plants, 11(17), 2188. https://doi.org/10.3390/plants11172188







