Applications of In Vitro Tissue Culture Technologies in Breeding and Genetic Improvement of Wheat
Abstract
1. Introduction
2. Intraspecific Hybridization
3. Interspecific and Intergeneric Hybridization
3.1. Ph Locus
3.2. Synthetic Wheat
4. In Vitro Culture in Doubled Haploid Production
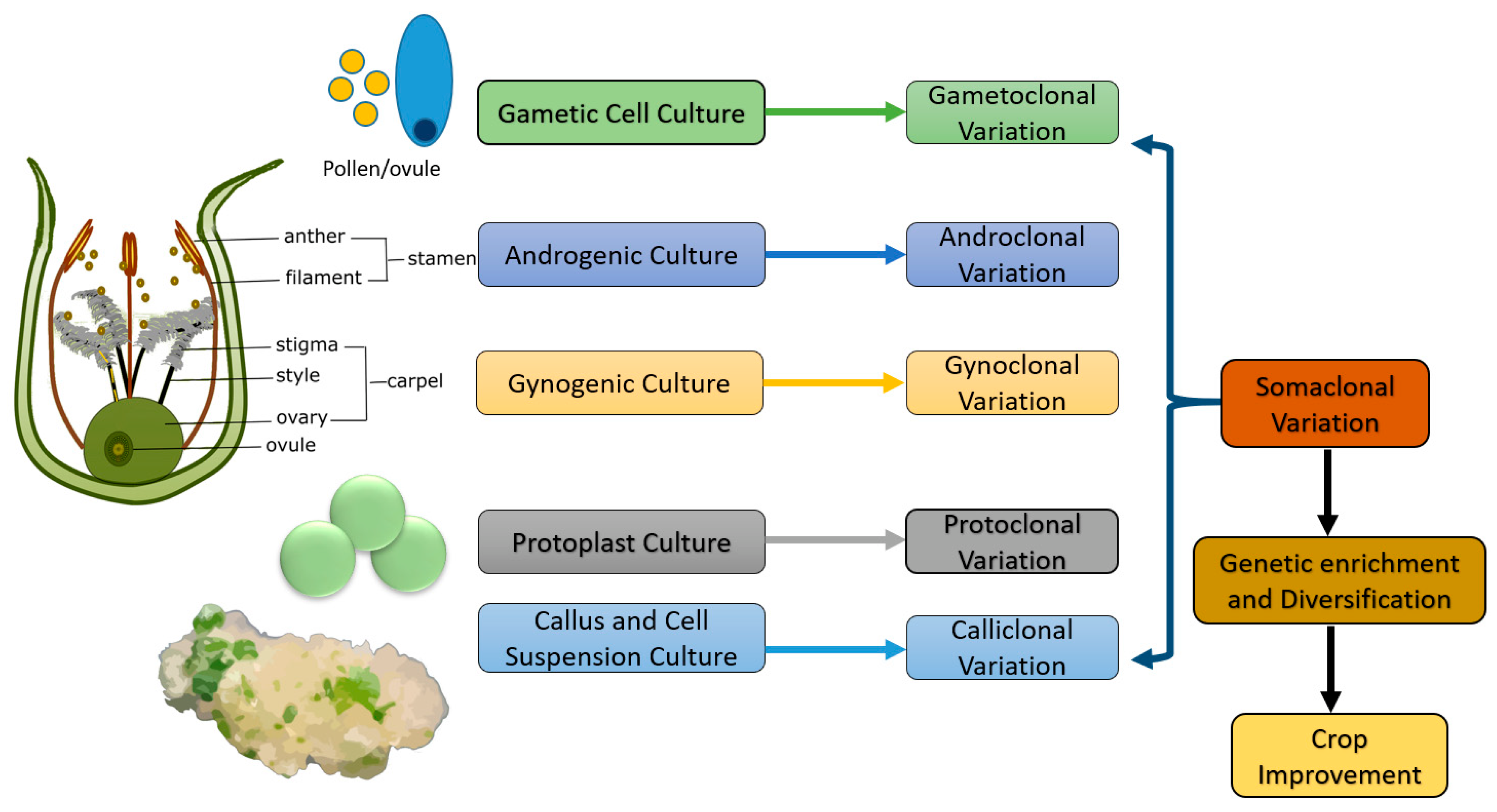
5. Somaclonal Variation
5.1. Gametoclonal Variation
5.2. Androclonal Variation
5.3. Gynoclonal Variation
5.4. Protoclonal Variation
5.5. Calliclonal Variation
6. Mutation Breeding
| Crop | In Vitro Techniques | Function/Traits | References |
|---|---|---|---|
| Interspecific hybridization | |||
| Pea (P. sativum × P. fulvum) | Immature embryo culture | Interspecific hybrid | [100] |
| Brassica (B. napus × B. rapa) | Embryo rescue via ovule culture | Interspecific hybrid | [101] |
| Rice (Oryza sativa × Oryza australiensis) | Young embryo rescue | Interspecific hybrid | [102] |
| Lentil (Lens culinaris × Lens tomentosus) | Ovule rescue technique | Interspecific hybrid between | [103] |
| Barley (diploid and tetraploid domestic barley × tetraploid wild barley) | Immature embryo culture | Induces genetic variation | [104] |
| Wheat (T. aestivum × Dasypyrum villosum) | Embryo rescue | Induces powdery mildew resistance | [105] |
| Wheat (T. aestivum × Dasypyrum villosum) | Embryo rescue | Induces stem rust resistance gene UG99 | [106] |
| Aegilops tauschii × Triticum aestivum Aegilops ovata × T. aestivum Aegilops cylindrica × T. aestivum Aegilops speltoides × T. aestivum | Embryo culture | Alien gene introgression | [107] |
| T. dicoccum × T. timopheevii | Immature embryo culture | Creation of genetic variation | [108] |
| Intergeneric hybridization | |||
| Rice (Oryza sativa × Leersia perrieri) | Embryo rescue | Intergeneric hybrid | [109] |
| Wheat × Barley (Triticum aestivum L. × Hordeum vulgare L.) | Embryo rescue | Intergeneric hybrid | [110] |
| Wheat (T. aestivum × Psathyrostachys huashanica) | Embryo culture | Resistance to powdery mildew | [111] |
| Wheat × rye (T. aestivum × Secale cereale) | Embryo rescue | Embryo lethality | [112] |
| Wheat × Rye (T. aestivum × Secale cereale) | -- | Resistance to powdery mildew by translocation of 4R chromosome | [113] |
| Brassica (Sinapis alba × Brassica juncea) | Somatic hybridization through protoplast fusion | Resistance to Alternaria brassicae and heat stress | [114] |
| Brassica (Sinapis alba × Brassica juncea) | Somatic hybridization through protoplast fusion | Broaden genetic variation along with resistance to Alternaria brassicae | [115] |
| Brassica (Brassica juncea × Sinapis alba) | Somatic hybridization through protoplast fusion | Development of a yellow-seeded stable allohexaploid | [116] |
| Doubled haploid | |||
| Durum wheat | Unpollinated ovary culture | Production of doubled haploid | [6] |
| Barley | Anther culture | Production transgenic homozygous DH lines | [117] |
| Barley | Anther culture | Haploid production | [118] |
| Lentil | Immature embryo culture | To shorten the breeding cycle | [119] |
| Wheat | Anther culture | Chromosome doubling | [120] |
| Wheat | Anther culture | Doubled haploid | [121] |
| Wheat | Microspore culture | Haploid production and resistance to Gibberella zeae | [122] |
| Lentil | Embryo rescue | Overcome reproductive barriers and hybrid recovery | [123] |
| Alloplasmic (H. vulgare × T. aestivum) and euplasmic line | Anther culture | Yield and quality traits, resistance to fungal diseases | [124] |
| Somaclonal variation | |||
| Barley | Endosperm-supported mature embryo | Somaclonal variation | [125] |
| Spelt wheat | Anther and isolated microspore culture | Induces genetic variation | [126] |
| Barley | Immature zygotic embryo culture | To modify tissue culture-induced variation | [87] |
| Egyptian barley | Mature embryo culture | Somaclonal variation | [127] |
| Maize | Immature embryo culture | Genetic variation | [128] |
| Wheat | Somatic hybridization | Stem rust | [129] |
| Maize | Cell culture from an immature embryo | Epigenomic variation | [130] |
| Mutation breeding | |||
| Wheat | Asymmetric somatic hybridization | Genome rearrangement, sequence elimination, and genetic variation via point mutations and indels | [131] |
| Wheat | Asymmetric somatic hybridization | Affects synonymous codon usage | [132] |
| Wheat | Asymmetric somatic hybridization | Induces genome-wide genetic variation | [133] |
| Wheat and maize | Immature embryo culture | Synthesis and study of a wheat/maize hybrid CENH3 gene | [134] |
7. Application of Biotechnology
7.1. Genome Editing
7.1.1. CRISPR/Cas9
7.1.2. CRISPR/Cpf1 System
7.1.3. Base Editing
7.1.4. Prime Editing
7.2. Genetic Transformation
7.2.1. Biological Systems
7.2.2. Non-Biological Systems
8. Discussion
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Grassini, P.; Eskridge, K.M.; Cassman, K.G. Distinguishing between yield advances and yield plateaus in historical crop production trends. Nat. Commun. 2013, 4, 2918. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Salina, E.A.; Adonina, I.G.; Badaeva, E.D.; Kroupin, P.Y.; Stasyuk, A.I.; Leonova, I.; Shishkina, A.A.; Divashuk, M.; Starikova, E.V.; Khuat, T.M.L.; et al. A Thinopyrum intermedium chromosome in bread wheat cultivars as a source of genes conferring resistance to fungal diseases. Euphytica 2015, 204, 91–101. [Google Scholar] [CrossRef]
- Kazi, A.M.; Ali, N.; Ibrahim, A.; Napar, A.A.; Jamil, M.; Hussain, S.; Mahmood, Z.; Delgado, R.; Rosas, V.; Cortes, A.; et al. Tissue Culture Mediated Allelic Diversification and Genomic Enrichment of Wheat to Combat Production Constraints and Address Food Security. Plant Tissue Cult. Biotechnol. 2017, 27, 89–140. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Britt, A.B.; Tripathi, L.; Sharma, S.; Upadhyaya, H.D.; Ortiz, R. Haploids: Constraints and opportunities in plant breeding. Biotechnol. Adv. 2015, 33, 812–829. [Google Scholar] [CrossRef]
- Slama, O.A.; Amara, H.S. Unpollinated Ovaries Used to Produce Doubled Haploid Lines in Durum Wheat. In Doubled Haploid Technology; Springer: Berlin/Heidelberg, Germany, 2021; Volume 2287, pp. 245–255. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. Plant tissue culture environment as a switch-key of (epi)genetic changes. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 140, 245–257. [Google Scholar] [CrossRef]
- Bridgen, M.P.; Van Houtven, W.; Eeckhaut, T. Plant Tissue Culture Techniques for Breeding. In Ornamental Crops; Van Huylenbroeck, J., Ed.; Handbook of Plant Breeding; Springer International Publishing: Cham, Switzerland, 2018; pp. 127–144. ISBN 978-3-319-90698-0. [Google Scholar]
- Niazian, M.; Shariatpanahi, M.E. In vitro-based doubled haploid production: Recent improvements. Euphytica 2020, 216, 1–21. [Google Scholar] [CrossRef]
- Borisjuk, N.; Kishchenko, O.; Eliby, S.; Schramm, C.; Anderson, P.; Jatayev, S.; Kurishbayev, A.; Shavrukov, Y. Genetic Modification for Wheat Improvement: From Transgenesis to Genome Editing. BioMed Res. Int. 2019, 2019, e6216304. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kaur, A.; Pandey, A.; Mamrutha, H.M.; Singh, G.P. CRISPR-based genome editing in wheat: A comprehensive review and future prospects. Mol. Biol. Rep. 2019, 46, 3557–3569. [Google Scholar] [CrossRef]
- Hu, D.; Jing, J.; Snowdon, R.J.; Mason, A.S.; Shen, J.; Meng, J.; Zou, J. Exploring the gene pool of Brassica napus by genomics-based approaches. Plant Biotechnol. J. 2021, 19, 1693–1712. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, C.; Wang, M.; Zhi, D.; Xia, G. Genomic changes at the early stage of somatic hybridization. Genet. Mol. Res. 2014, 13, 1938–1948. [Google Scholar] [CrossRef]
- Masood, S.; Naseem, Z.; Razzaq, H. Inheritance Study of Quantifiable Characters in Intraspecific Hybrids of Promising Triticum Aestivum (Bread Wheat) Lines under Normal Field Conditions. Int. J. Sci. Eng. Res. 2015, 6, 980–984. [Google Scholar]
- Antoshina, O.; Odnodushnova, J.; Fadkin, G.; Kondakova, I.; Fedosova, O. The study of the nature of the inheritance of quantitative traits in F1 hybrids of winter soft wheat. BIO Web Conf. 2020, 17, 00084. [Google Scholar] [CrossRef]
- Soomro, Z.A.; Khalid, M.; Abro, T.; Mangrio, G.; Channa, S.; Kasi, U.; Shar, P.; Hassni, M.; Shah, R. Response of Intraspecific Crosses in F1 and their Deterioration in F2 Generation of Bread Wheat (Triticum aestivum L.). Int. J. Sustain. Agric. Res. 2019, 6, 198–202. [Google Scholar] [CrossRef]
- Parfenova, E.; Utkina, E.; Nabatova, N.; Shamova, M.; Zhukova, M. Assessment of economic traits’ inheritance of winter rye at intraspecific hybridization. BIO Web Conf. 2021, 36, 01008. [Google Scholar] [CrossRef]
- Groszmann, M.; Gonzalez-Bayon, R.; Greaves, I.K.; Wang, L.; Huen, A.K.; Peacock, W.J.; Dennis, E.S. Intraspecific Arabidopsis Hybrids Show Different Patterns of Heterosis Despite the Close Relatedness of the Parental Genomes. Plant Physiol. 2014, 166, 265–280. [Google Scholar] [CrossRef]
- Alvarez, J.B.; Guzmán, C. Interspecific and intergeneric hybridization as a source of variation for wheat grain quality improvement. Theor. Appl. Genet. 2017, 131, 225–251. [Google Scholar] [CrossRef]
- Kaneko, Y.; Bang, S.W. Interspecific and intergeneric hybridization and chromosomal engineering of Brassicaceae crops. Breed. Sci. 2014, 64, 14–22. [Google Scholar] [CrossRef]
- Akmal, M. Embryo Culture and Embryo Rescue in Brassica. In Brassica Breeding and Biotechnology; IntechOpen: London, UK, 2021. [Google Scholar]
- Li, Q.; Chen, Y.; Yue, F.; Qian, W.; Song, H. Microspore culture reveals high fitness of B. napus-like gametes in an interspecific hybrid between Brassica napus and B. oleracea. PLoS ONE 2018, 13, e0193548. [Google Scholar] [CrossRef]
- Kiani, R.; Arzani, A.; Maibody, S.A.M.M.; Rahimmalek, M.; Ayers, T. Hybridization of wheat and Aegilops cylindrica: Development, karyomorphology, DNA barcoding and salt tolerance of the amphidiploids. J. Plant Biochem. Biotechnol. 2021, 30, 943–959. [Google Scholar] [CrossRef]
- Marcińska, I.; Czyczyło-Mysza, I.; Skrzypek, E.; Warchoł, M.; Zieliński, K.; Dubas, E. Obtaining of winter rye (Secale cereale L. ssp. cereale) haploid embryos through hybridization with maize (Zea mays L.). Cereal Res. Commun. 2018, 46, 521–532. [Google Scholar] [CrossRef]
- Maryenti, T.; Ishii, T.; Okamoto, T. Development and regeneration of wheat–rice hybrid zygotes produced by in vitro fertilization system. New Phytol. 2021, 232, 2369–2383. [Google Scholar] [CrossRef]
- Patial, M.; Chaudhary, H.K.; Sharma, N.; Gangwar, O.P.; Kishore, N.; Pal, D.; Pramanick, K.K.; Bhardwaj, S.C.; Chauhan, R. Developing genetic stock for yellow and brown rust resistance in Triticum aestivum L. via Imperata cylindrica-mediated doubled haploidy technique. Cereal Res. Commun. 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Xia, G.; Xiang, F.; Zhou, A.; Wang, H.; Chen, H. Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host) Nevishi. Theor. Appl. Genet. 2003, 107, 299–305. [Google Scholar] [CrossRef]
- Hao, M.; Zhang, L.; Ning, S.; Huang, L.; Yuan, Z.; Wu, B.; Yan, Z.; Dai, S.; Jiang, B.; Zheng, Y.; et al. The Resurgence of Introgression Breeding, as Exemplified in Wheat Improvement. Front. Plant Sci. 2020, 11, 252. [Google Scholar] [CrossRef]
- Fan, C.; Luo, J.; Zhang, S.; Liu, M.; Li, Q.; Li, Y.; Huang, L.; Chen, X.; Ning, S.; Yuan, Z.; et al. Genetic mapping of a major QTL promoting homoeologous chromosome pairing in a wheat landrace. Theor. Appl. Genet. 2019, 132, 2155–2166. [Google Scholar] [CrossRef]
- Roberts, M.A.; Reader, S.M.; Dalgliesh, C.; Miller, T.E.; Foote, T.N.; Fish, L.J.; Snape, J.; Moore, G. Induction and Characterization of Ph1 Wheat Mutants. Genetics 1999, 153, 1909–1918. [Google Scholar] [CrossRef]
- Svačina, R.; Sourdille, P.; Kopecký, D.; Bartoš, J. Chromosome Pairing in Polyploid Grasses. Front. Plant Sci. 2020, 11, 1056. [Google Scholar] [CrossRef]
- Koo, D.-H.; Friebe, B.; Gill, B.S. Homoeologous Recombination: A Novel and Efficient System for Broadening the Genetic Variability in Wheat. Agronomy 2020, 10, 1059. [Google Scholar] [CrossRef]
- Rey, M.-D.; Martín, A.C.; Higgins, J.; Swarbreck, D.; Uauy, C.; Shaw, P.; Moore, G. Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. 2017, 37, 1–11. [Google Scholar] [CrossRef]
- Kamboj, R.; Sharma, S.; Kumar, R.; Sharma, P.; Ravat, V.K.; Chhuneja, P.; Vyas, P.; Sheikh, I.; Dhaliwal, H.S. Introgression of powdery mildew resistance from Aegilops triuncialis into wheat through induced homeologous pairing. J. Plant Biochem. Biotechnol. 2020, 29, 418–426. [Google Scholar] [CrossRef]
- Serra, H.; Svačina, R.; Baumann, U.; Whitford, R.; Sutton, T.; Bartoš, J.; Sourdille, P. Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination. Nat. Commun. 2021, 12, 803. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Dundas, I.; Rasheed, A.; Ogbonnaya, F.; Kishii, M.; Bonnett, D.; Wang, R.R.-C.; Xu, S.; Chen, P.; Mahmood, T.; et al. Genetic Diversity for Wheat Improvement as a Conduit to Food Security. Adv. Agron. 2013, 122, 179–257. [Google Scholar] [CrossRef]
- Kihara, H.; Lilienfeld, F. A New Sinthesized 6x-Wheat. Hereditas 2010, 35, 307–319. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Gul, A.; Farooq, M.; Rizwan, S.; Ahmad, I. Rebirth of synthetic hexaploids with global implications for wheat improvement. Aust. J. Agric. Res. 2008, 59, 391–398. [Google Scholar] [CrossRef]
- Hao, M.; Li, A.; Shi, T.; Luo, J.; Zhang, L.; Zhang, X.; Ning, S.; Yuan, Z.; Zeng, D.; Kong, X.; et al. The abundance of homoeologue transcripts is disrupted by hybridization and is partially restored by genome doubling in synthetic hexaploid wheat. BMC Genom. 2017, 18, 149. [Google Scholar] [CrossRef]
- Gupta, P.K. Use of Alien Genetic Variation for Wheat Improvement. In Molecular Breeding for Sustainable Crop Improvement: Volume 2; Rajpal, V.R., Rao, S.R., Raina, S.N., Eds.; Sustainable Development and Biodiversity; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–30. ISBN 978-3-319-27090-6. [Google Scholar]
- Mujeeb-Kazi, A.; Miranda, J.L. Enhanced resolution of somatic chromosome constrictions as an aid to identifying intergeneric hybrids among some Triticeae. Cytologia 1985, 50, 701–709. [Google Scholar] [CrossRef]
- Kuzmanović, L.; Rossini, F.; Ruggeri, R.; Pagnotta, M.A.; Ceoloni, C. Engineered Durum Wheat Germplasm with Multiple Alien Introgressions: Agronomic and Quality Performance. Agronomy 2020, 10, 486. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, Y.; Zhao, X.; Lv, L.; Zhang, C.; Li, J.; Sun, G.; Li, S.; Song, C. Development and Utilization of Introgression Lines Using Synthetic Octaploid Wheat (Aegilops tauschii × Hexaploid Wheat) as Donor. Front. Plant Sci. 2018, 9, 1113. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Ma, P.; Zheng, Q.; Fu, S.; Li, L.; Han, F.; Han, G.; Wang, J.; Xu, Y.; Jin, Y.; et al. Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 2018, 132, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, J.; Liu, Q.; Liu, H.; Zhou, Y.; Yang, W.; Ma, W. Improvement and Re-Evolution of Tetraploid Wheat for Global Environmental Challenge and Diversity Consumption Demand. Int. J. Mol. Sci. 2022, 23, 2206. [Google Scholar] [CrossRef]
- Kasha, K.J.; Maluszynski, M. Production of Doubled Haploids in Crop Plants. An Introduction. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K.J., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 1–4. ISBN 978-94-017-1293-4. [Google Scholar]
- Ren, J.; Wu, P.; Trampe, B.; Tian, X.; Lübberstedt, T.; Chen, S. Novel technologies in doubled haploid line development. Plant Biotechnol. J. 2017, 15, 1361–1370. [Google Scholar] [CrossRef]
- Wędzony, M.; Forster, B.P.; Żur, I.; Golemiec, E.; Szechyńska-Hebda, M.; Dubas, E.; Gotębiowska, G.; Wędzony(✉), M. Progress in Doubled Haploid Technology in Higher Plants. In Advances in Haploid Production in Higher Plants; Touraev, A., Forster, B.P., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 1–33. ISBN 978-1-4020-8854-4. [Google Scholar]
- Kalinowska, K.; Chamas, S.; Unkel, K.; Demidov, D.; Lermontova, I.; Dresselhaus, T.; Kumlehn, J.; Dunemann, F.; Houben, A. State-of-the-art and novel developments of in vivo haploid technologies. Theor. Appl. Genet. 2018, 132, 593–605. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Corral-Martínez, P.; Parra-Vega, V.; González-García, B. Androgenesis in recalcitrant solanaceous crops. Plant Cell Rep. 2010, 30, 765–778. [Google Scholar] [CrossRef]
- Touraev, A.; Forster, B.P.; Jain, S.M. Advances in Haploid Production in Higher Plants; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Eliby, S.; Bekkuzhina, S.; Kishchenko, O.; Iskakova, G.; Kylyshbayeva, G.; Jatayev, S.; Soole, K.; Langridge, P.; Borisjuk, N.; Shavrukov, Y. Developments and prospects for doubled haploid wheat. Biotechnol. Adv. 2022, 60, 108007. [Google Scholar] [CrossRef]
- Esteves, P.; Belzile, F.J. TDZ in Cereal Gametic Embryogenesis. In Thidiazuron: From Urea Derivative to Plant Growth Regulator; Ahmad, N., Faisal, M., Eds.; Springer: Singapore, 2018; pp. 159–174. ISBN 978-981-10-8004-3. [Google Scholar]
- Chu, C. Establishment of an Efficient Medium for Another Culture of Rice through Comparative Experiments on the Nitrogen Sources. Sci. Sin. 1975, 18, 223–231. [Google Scholar]
- Ouyang, J.W.; Jia, S.E.; Zhang, C.; Chen, X.; Feng, G. A New Synthetic Medium (W14) for Wheat Anther Culture. Annu. Rep. 1989, 91–92. [Google Scholar]
- Gosal, S.S.; Wani, S.H. Cell and Tissue Culture Approaches in Relation to Crop Improvement. In Biotechnologies of Crop Improvement; Gosal, S.S., Wani, S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 1, pp. 1–55. ISBN 978-3-319-78283-6. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Eichten, S.R.; Hermanson, P.J.; Springer, N.M.; Kaeppler, S.M. Consistent and Heritable Alterations of DNA Methylation Are Induced by Tissue Culture in Maize. Genetics 2014, 198, 209–218. [Google Scholar] [CrossRef]
- Ranghoo-Sanmukhiya, V.M. Somaclonal Variation and Methods Used for Its Detection. In Propagation and Genetic Manipulation of Plants; Siddique, I., Ed.; Springer: Singapore, 2021; pp. 1–18. ISBN 9789811577369. [Google Scholar]
- Zhang, D.; Wang, Z.; Wang, N.; Gao, Y.; Liu, Y.; Wu, Y.; Bai, Y.; Zhang, Z.; Lin, X.; Dong, Y.; et al. Tissue Culture-Induced Heritable Genomic Variation in Rice, and Their Phenotypic Implications. PLoS ONE 2014, 9, e96879. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 1–18. [Google Scholar] [CrossRef]
- Machczyńska, J.; Orłowska, R.; Zimny, J.; Bednarek, P.T. Extended metAFLP approach in studies of tissue culture induced variation (TCIV) in triticale. Mol. Breed. 2014, 34, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Bhojwani, S.S.; Dantu, P.K. Plant Tissue Culture: An Introductory Text; Springer: New Delhi, India, 2013; ISBN 978-81-322-1025-2. [Google Scholar]
- Chadipiralla, K.; Gayathri, P.; Rajani, V.; Reddy, P.V.B. Plant Tissue Culture and Crop Improvement. In Sustainable Agriculture in the Era of Climate Change; Roychowdhury, R., Choudhury, S., Hasanuzzaman, M., Srivastava, S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 391–412. ISBN 978-3-030-45669-6. [Google Scholar]
- Shahzad, A.; Parveen, S.; Sharma, S.; Shaheen, A.; Saeed, T.; Yadav, V.; Akhtar, R.; Ahmad, Z.; Upadhyay, A. Plant Tissue Culture: Applications in Plant Improvement and Conservation. In Plant Biotechnology: Principles and Applications; Abdin, M.Z., Kiran, U., Kamaluddin, A.A., Eds.; Springer: Singapore, 2017; pp. 37–72. ISBN 978-981-10-2961-5. [Google Scholar] [CrossRef]
- Abu-Gammie, B.; Kasem, A.; Abdelrahem, A. Somaclonal Variation in Bread Wheat (Triticum aestivum L.). v. Meiotic Behavior of Some Gametoclones and Somaclones. Minia J. Agric. Res. Develop. 2016, 36, 91–110. [Google Scholar]
- Bashandy, T.; Hassan, M.S. Field Evaluation and Molecular Analysis of Some Bread Wheat Gametoclones and Somaclones under Natural Heat Stress. Minia J. Agric. Res. Develop. 2016, 36, 511–527. [Google Scholar]
- Savenko, E.G.; Mukhina, Z.M.; Glazyrina, V.A.; Korotenko, T.L.; Garkusha, S.V. Phenotypic evaluation of DH-lines developed on the base of rice samples with blast resistance genes. E3S Web Conf. 2021, 285, 02021. [Google Scholar] [CrossRef]
- Grauda, D.; Miķelsone, A.; Ļisina, N.; Žagata, K.; Ornicāns, R.; Fokina, O.; Lapiņa, L.; Rashal, I. Anther Culture Effectiveness in Producing Doubled Haploids of Cereals/Putekđòu Kultûras Efektivitâte Graudaugu Dubultoto Haploîdu Izveidođanâ. Proc. Latv. Acad. Sci. Sect. B. Nat. Exact Appl. Sci. 2014, 68, 142–147. [Google Scholar] [CrossRef]
- Grauda, D.; Žagata, K.; Lanka, G.; Strazdina, V.; Fetere, V.; Lisina, N.; Krasnevska, N.; Fokina, O.; Mikelsone, A.; Ornicans, R.; et al. Genetic diversity of wheat (Triticum aestivum L.) plants-regenerants produced by anther culture. Vavilov J. Genet. Breed. 2016, 20, 537–544. [Google Scholar] [CrossRef]
- El-Hennawy, M.A.; Al-Ashkar, I.; Abdelfattah, E. Genetic Analysis of Anther Culture Response in Some Bread Wheat Crosses under Drought Stress Conditions. Environ. Sci. 2018, 13, 555–581. [Google Scholar]
- Tripathy, S.K.; Swain, D.; Mohapatra, P.M.; Prusti, A.M.; Sahoo, B.; Panda, S.; Dash, M.; Chakma, B.; Behera, S.K. Exploring factors affecting anther culture in rice (Oryza sativa L.). J. Appl. Biol. Biotechnol. 2019, 7, 87–92. [Google Scholar] [CrossRef]
- Watts, A.; Kumar, V.; Raipuria, R.K.; Bhattacharya, R.C. In Vivo Haploid Production in Crop Plants: Methods and Challenges. Plant Mol. Biol. Rep. 2018, 36, 685–694. [Google Scholar] [CrossRef]
- Bohanec, B. Doubled Haploids via Gynogenesis. In Advances in Haploid Production in Higher Plants; Touraev, A., Forster, B.P., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 35–46. ISBN 978-1-4020-8854-4. [Google Scholar]
- Weyen, J. Applications of Doubled Haploids in Plant Breeding and Applied Research. Methods Mol. Biol. 2021, 2287, 23–39. [Google Scholar] [CrossRef]
- Ayed-Slama, O.; Ayed, S.; Slim-Amara, H. Selection of Tolerant Lines to Salinity Derived from Durum Wheat (Triticum durum Desf.) in Vitro Culture. Agric. Sci. 2015, 06, 699–706. [Google Scholar] [CrossRef]
- Slama-Ayed, O.; Bouhaouel, I.; Ayed, S.; De Buyser, J.; Picard, E.; Amara, H.S. Efficiency of three haplomethods in durum wheat (Triticum turgidum subsp. durum Desf.): Isolated microspore culture, gynogenesis and wheat × maize crosses. Czech J. Genet. Plant Breed. 2019, 55, 101–109. [Google Scholar] [CrossRef]
- El-Esawi, M.A. Micropropagation Technology and Its Applications for Crop Improvement. In Plant Tissue Culture: Propagation, Conservation and Crop Improvement; Anis, M., Ahmad, N., Eds.; Springer: Singapore, 2016; pp. 523–545. ISBN 978-981-10-1917-3. [Google Scholar]
- Compton, M.E.; Saunders, J.A.; Veilleux, R.E. Use of Protoplasts for Plant Improvement. In Plant Tissue Culture Concepts and Laboratory Exercises; Routledge: London, UK, 2000; ISBN 978-0-203-74313-3. [Google Scholar]
- Mwangangi, I.M.; Muli, J.; Neondo, J.O. Plant Hybridization as an Alternative Technique in Plant Breeding Improvement. Asian J. Res. Crop Sci. 2019, 4, 1–11. [Google Scholar] [CrossRef]
- YuJi, L.; GuangZhe, L.; Qin, Z. Tri-Parental Protoplast Fusion of Brassica Species to Produce Somatic Hybrids with High Genetic and Phenotypic Variability. Indian J. Genet. Plant Breed. 2015, 75, 497–505. [Google Scholar]
- Chakraborty, S. Protoplast Culture and Somatic Hybridization, a Promising Frontier of Plant Biotechnology. J. Arts Sci. Teach. 2016, 2, 9. [Google Scholar]
- Reed, K.M.; Bargmann, B.O.R. Protoplast Regeneration and Its Use in New Plant Breeding Technologies. Front. Genome Ed. 2021, 3, 20. [Google Scholar] [CrossRef]
- Thorat, A.S.; Sonone, N.A.; Choudhari, V.V.; Devarumath, R.M.; Babu, K.H. Plant regeneration from cell suspension culture in Saccharum officinarum L. and ascertaining of genetic fidelity through RAPD and ISSR markers. 3 Biotech 2017, 7, 16. [Google Scholar] [CrossRef]
- Ahmadpour, R.; Zare, N.; Asghari-Zakarta, R.; Sheikhzadeh, P. Efficient In Vitro Somatic Embryogenesis and Plant Regeneration from Mature and Immature Embryos of Wheat (Triticum aestivum L.). Braz. Arch. Biol. Technol. 2016, 59, 1–12. [Google Scholar] [CrossRef][Green Version]
- Dey, T.; Saha, S.; Ghosh, P. Somaclonal variation among somatic embryo derived plants—Evaluation of agronomically important somaclones and detection of genetic changes by RAPD in Cymbopogon winterianus. S. Afr. J. Bot. 2015, 96, 112–121. [Google Scholar] [CrossRef]
- Kacem, N.S.; Delporte, F.; Muhovski, Y.; Djekoun, A.; Watillon, B. In vitro screening of durum wheat against water-stress mediated through polyethylene glycol. J. Genet. Eng. Biotechnol. 2017, 15, 239–247. [Google Scholar] [CrossRef]
- Orłowska, R. Barley somatic embryogenesis-an attempt to modify variation induced in tissue culture. J. Biol. Res. 2021, 28, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Delporte, F.; Pretova, A.; du Jardin, P.; Watillon, B. Morpho-histology and genotype dependence of in vitro morphogenesis in mature embryo cultures of wheat. Protoplasma 2014, 251, 1455–1470. [Google Scholar] [CrossRef] [PubMed]
- Machczyńska, J.; Zimny, J.; Bednarek, P.T. Tissue culture-induced genetic and epigenetic variation in triticale (× Triticosecale spp. Wittmack ex A. Camus 1927) regenerants. Plant Mol. Biol. 2015, 89, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Bado, S.; Forster, B.P.; Nielen, S.; Ali, A.M.; Lagoda, P.J.; Till, B.J.; Laimer, M. Plant Mutation Breeding: Current Progress and Future Assessment. Plant Breed. Rev. 2015, 39, 23–88. [Google Scholar]
- EFSA Panel on Genetically Modified Organisms (GMO); Mullins, E.; Bresson, J.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Moreno, F.J.; et al. In vivo and in vitro random mutagenesis techniques in plants. EFSA J. 2021, 19, e06611. [Google Scholar] [CrossRef]
- Abaza, G.M.; Awaad, H.A.; Attia, Z.M.; Abdel-Lateif, K.S.; Gomaa, M.A.; Abaza, S.M.; Mansour, E. Inducing Potential Mutants in Bread Wheat Using Different Doses of Certain Physical and Chemical Mutagens. Plant Breed. Biotechnol. 2020, 8, 252–264. [Google Scholar] [CrossRef]
- Thamodharan, M.; Pillai, A. In Vitro Mutagenesis to Improve Salt Tolerance in Rice (Oryza sativa L.). Electron. J. Plant Breed. 2020, 11, 638–644. [Google Scholar]
- Raghavi, G.; Kumari, S.M.P.; ArumugamPillai, M.; Saravanan, S.; Pushpam, A.K. Improvement of Rice Cultivar Kavuni for Semi Dwarf Character by In Vitro Chemical Mutation Using Ethyl Methyl Sulphonate. Pharma Innov. J. 2021, 10, 1483–1488. [Google Scholar]
- Nikam, A.A.; Devarumath, R.M.; Ahuja, A.; Babu, H.; Shitole, M.G.; Suprasanna, P. Radiation-induced in vitro mutagenesis system for salt tolerance and other agronomic characters in sugarcane (Saccharum officinarum L.). Crop J. 2015, 3, 46–56. [Google Scholar] [CrossRef]
- Kona, P.; Hemanth Kumar, M.; Balaji, A. Effect of 2,4D & EMS on In Vitro Regeneration in Sugarcane Cultivar. Int. J. Microbiol. Res. 2018, 10, 1090–1093. [Google Scholar]
- Gil, J.; Andrade-Martínez, J.S.; Duitama, J. Accurate, Efficient and User-Friendly Mutation Calling and Sample Identification for TILLING Experiments. Front. Genet. 2021, 12, 624513. [Google Scholar] [CrossRef] [PubMed]
- Serrat, X.; Esteban, R.; Guibourt, N.; Moysset, L.; Nogués, S.; Lalanne, E. EMS mutagenesis in mature seed-derived rice calli as a new method for rapidly obtaining TILLING mutant populations. Plant Methods 2014, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Holme, I.; Gregersen, P.; Brinch-Pedersen, H. Induced Genetic Variation in Crop Plants by Random or Targeted Mutagenesis: Convergence and Differences. Front. Plant Sci. 2019, 10, 1468. [Google Scholar] [CrossRef]
- Bobkov, S.V.; Selikhova, T.N. Obtaining interspecific hybrids for introgressive pea breeding. Russ. J. Genet. Appl. Res. 2017, 7, 145–152. [Google Scholar] [CrossRef]
- Niemann, J.; Olender, M.; Wojciechowski, A. Interspecific Hybridization between Selected Brassica Napus and Brassica Rapa Ssp. Chinensis Genotypes through Embryo Rescue and Their Evaluation for Crossability. BioTechnol. J. Biotechnol. Comput. Biol. Bionanotechnol. 2015, 96, 1. [Google Scholar]
- Yi, C.; Hu, D.; Zhang, J.; Jin, W.; Li, W.; Zhou, Y.; Liang, G.; Gu, M. Development and identification of synthetic interspecific hybrids between Oryza sativa and Oryza australiensis. Indian J. Genet. Plant Breed. 2018, 78, 174–179. [Google Scholar] [CrossRef]
- Suvorova, G. Hybridization of cultivated lentil Lens culinaris Medik. and wild species Lens tomentosus Ladizinsky. Czech J. Genet. Plant Breed. 2014, 50, 130–134. [Google Scholar] [CrossRef]
- Mohammed, A.H.; Morrison, J.I.; Baldwin, B.S. Interspecific crosses between domestic and wild barley and embryo rescue to overcome sexual incompatibilities. Agrosyst. Geosci. Environ. 2020, 3, e20130. [Google Scholar] [CrossRef]
- Zhang, R.; Xiong, C.; Mu, H.; Yao, R.; Meng, X.; Kong, L.; Xing, L.; Wu, J.; Feng, Y.; Cao, A. Pm67, a new powdery mildew resistance gene transferred from Dasypyrum villosum chromosome 1V to common wheat (Triticum aestivum L.). Crop J. 2020, 9, 882–888. [Google Scholar] [CrossRef]
- Ando, K.; Krishnan, V.; Rynearson, S.; Rouse, M.N.; Danilova, T.; Friebe, B.; See, D.; Pumphrey, M.O. Introgression of a Novel Ug99-Effective Stem Rust Resistance Gene into Wheat and Development of Dasypyrum villosum Chromosome-Specific Markers via Genotyping-by-Sequencing (GBS). Plant Dis. 2019, 103, 1068–1074. [Google Scholar] [CrossRef]
- Yuan, B.; Cao, X.; Lv, A. Gene introgression from common wheat into Aegilops L. Saudi J. Biol. Sci. 2016, 24, 813–816. [Google Scholar] [CrossRef][Green Version]
- Miroshnichenko, D.; Klementyeva, A.; Pushin, A.; Dolgov, S. A competence of embryo-derived tissues of tetraploid cultivated wheat species Triticum dicoccum and Triticum timopheevii for efficient and stable transgenesis mediated by particle inflow gun. BMC Plant Biol. 2020, 20, 442. [Google Scholar] [CrossRef] [PubMed]
- Ballesfin, M.L.E.; Vinarao, R.B.; Sapin, J.; Kim, S.-R.; Jena, K.K. Development of an intergeneric hybrid between Oryza sativa L. and Leersia perrieri (A. Camus) Launert. Breed. Sci. 2018, 68, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Polgári, D.; Mihók, E.; Sági, L. Composition and random elimination of paternal chromosomes in a large population of wheat × barley (Triticum aestivum L. × Hordeum vulgare L.) hybrids. Plant Cell Rep. 2019, 38, 767–775. [Google Scholar] [CrossRef]
- Han, J.; Liu, Y.; Hou, C.; Li, J.; Wang, J.; Zhang, Q.; Yang, Q.; Chen, X.; Wu, J. A 1Ns Disomic Addition from Psathyrostachys Huashanica Keng Confers Resistance to Powdery Mildew in Wheat. Agronomy 2020, 10, 312. [Google Scholar] [CrossRef]
- Tikhenko, N.; Rutten, T.; Senula, A.; Rubtsova, M.; Keller, E.R.J.; Börner, A. The changes in the reproductive barrier between hexaploid wheat (Triticum aestivum L.) and rye (Secale cereale L.): Different states lead to different fates. Planta 2017, 246, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Han, G.; Zheng, Q.; Liu, S.; Han, F.; Wang, J.; Luo, Q.; An, D. Development of Novel Wheat-Rye Chromosome 4R Translocations and Assignment of Their Powdery Mildew Resistance. Plant Dis. 2020, 104, 260–268. [Google Scholar] [CrossRef]
- Kumari, P.; Bisht, S.; Bhat, R. Stable, Fertile Somatic Hybrids between Sinapis Alba and Brassica Juncea Show Resistance to Alternaria Brassicae and Heat Stress. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 133, 77–86. [Google Scholar] [CrossRef]
- Kumari, P.; Singh, K.P.; Bisht, D.; Kumar, S. Somatic Hybrids of Sinapis Alba+ Brassica Juncea: Study of Backcross Progenies for Morphological Variations, Chromosome Constitution and Reaction to Alternaria Brassicae. Euphytica 2020, 216, 1–14. [Google Scholar] [CrossRef]
- Kumari, P.; Singh, K.P.; Kumar, S.; Yadava, D.K. Development of a Yellow-Seeded Stable Allohexaploid Brassica Through Inter-Generic Somatic Hybridization With a High Degree of Fertility and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2020, 11, 1830. [Google Scholar] [CrossRef]
- Ohnoutková, L.; Vlčko, T. Homozygous Transgenic Barley (Hordeum vulgare L.) Plants by Anther Culture. Plants 2020, 9, 918. [Google Scholar] [CrossRef]
- Bilynska, O.; Dulnyev, P. Natural and Chemically Modified Starches as Gelling Agents of Nutrient Media for Barley Haploid Production in Anther Culture in Vitro. In Proceedings of the Life Sciences in the Dialogue of Generations: Connections between Universities, Academia and Business Community, Chişinău, Moldova, 21–22 October 2019; pp. 13–14. [Google Scholar]
- Bermejo, C.; Gatti, I.; Cointry, E. In vitro embryo culture to shorten the breeding cycle in lentil (Lens culinaris Medik). Plant Cell Tissue Organ Cult. (PCTOC) 2016, 127, 585–590. [Google Scholar] [CrossRef]
- Broughton, S.; Castello, M.; Liu, L.; Killen, J.; Hepworth, A.; O’Leary, R. The Effect of Caffeine and Trifluralin on Chromosome Doubling in Wheat Anther Culture. Plants 2020, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.M.; Allue, S.; Costar, A.; Alvaro, F.; Valles, M.P. Doubled Haploid Production from Spanish and Central European Spelt by Anther Culture. J. Agric. Sci. Technol. 2019, 21, 1313–1324. [Google Scholar]
- Coelho, M.B.; Scagliusi, S.M.M.; Lima, M.I.P.M.; Consoli, L.; Grando, M.F. Androgenic response of wheat genotypes resistant to fusariosis. Pesqui. Agropecuária Bras. 2018, 53, 575–582. [Google Scholar] [CrossRef]
- Saha, S.; Tullu, A.; Yuan, H.Y.; Lulsdorf, M.M.; Vandenberg, A. Improvement of embryo rescue technique using 4-chloroindole-3 acetic acid in combination with in vivo grafting to overcome barriers in lentil interspecific crosses. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 120, 109–116. [Google Scholar] [CrossRef]
- Pershina, L.; Trubacheeva, N.; Badaeva, E.; Belan, I.; Rosseeva, L. Study of Androgenic Plant Families of Alloplasmic Introgression Lines (H. vulgare) –T. aestivum and the Use of Sister DH Lines in Breeding. Plants 2020, 9, 764. [Google Scholar] [CrossRef]
- Aydin, M.; Arslan, E.; Taspinar, M.S.; Karadayi, G.; Agar, G. Analyses of somaclonal variation in endosperm-supported mature embryo culture of rye (Secale cereale L.). Biotechnol. Biotechnol. Equip. 2016, 30, 1082–1089. [Google Scholar] [CrossRef]
- Lantos, C.; Bóna, L.; Nagy, É.; Békés, F.; Pauk, J. Induction of in vitro androgenesis in anther and isolated microspore culture of different spelt wheat (Triticum spelta L.) genotypes. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 133, 385–393. [Google Scholar] [CrossRef]
- Rashad, S.E.; Abdel-Tawab, F.M.; Fahmy, E.M.; Sakr, M.M. Somaclonal Variation from Mature Embryo Ex-Plants of Some Egyptian Barley Genotypes. Egypt. J. Genet. Cytol. 2020, 49, 103–121. [Google Scholar]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Upadhyaya, N.M.; Sperschneider, J.; Matny, O.; Nguyen-Phuc, H.; Mago, R.; Raley, C.; Miller, M.E.; Silverstein, K.A.T.; Henningsen, E.; et al. Emergence of the Ug99 lineage of the wheat stem rust pathogen through somatic hybridisation. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Crisp, P.; Stelpflug, S.; Kaeppler, S.M.; Li, Q.; Springer, N.M. Heritable Epigenomic Changes to the Maize Methylome Resulting from Tissue Culture. Genetics 2018, 209, 983–995. [Google Scholar] [CrossRef]
- Wang, M.; Liu, C.; Xing, T.; Wang, Y.; Xia, G. Asymmetric somatic hybridization induces point mutations and indels in wheat. BMC Genom. 2015, 16, 807. [Google Scholar] [CrossRef]
- Xu, W.; Li, Y.; Li, Y.; Liu, C.; Wang, Y.; Xia, G.; Wang, M. Asymmetric Somatic Hybridization Affects Synonymous Codon Usage Bias in Wheat. Front. Genet. 2021, 12, 896. [Google Scholar] [CrossRef]
- Wang, M.; Ji, Y.; Feng, S.; Liu, C.; Xiao, Z.; Wang, X.; Wang, Y.; Xia, G. The non-random patterns of genetic variation induced by asymmetric somatic hybridization in wheat. BMC Plant Biol. 2018, 18, 244. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, J.; Yu, W. Synthesis of a Wheat/Maize Hybrid CENH3 Gene, the Genetic Transformation of Wheat, Its Chromosomal Localization and Effects on Chromosome Behaviors in Wheat/Maize Somatic Hybrids. Agric. Sci. 2019, 10, 985–1014. [Google Scholar] [CrossRef][Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Ahmar, S.; Saeed, S.; Khan, M.; Khan, S.U.; Mora-Poblete, F.; Kamran, M.; Faheem, A.; Maqsood, A.; Rauf, M.; Saleem, S.; et al. A Revolution toward Gene-Editing Technology and Its Application to Crop Improvement. Int. J. Mol. Sci. 2020, 21, 5665. [Google Scholar] [CrossRef]
- Ansari, W.A.; Chandanshive, S.U.; Bhatt, V.; Nadaf, A.B.; Vats, S.; Katara, J.L.; Sonah, H.; Deshmukh, R. Genome Editing in Cereals: Approaches, Applications and Challenges. Int. J. Mol. Sci. 2020, 21, 4040. [Google Scholar] [CrossRef]
- Iqbal, Z.; Iqbal, M.S.; Ahmad, A.; Memon, A.G.; Ansari, M.I. New prospects on the horizon: Genome editing to engineer plants for desirable traits. Curr. Plant Biol. 2020, 24, 100171. [Google Scholar] [CrossRef]
- Matres, J.M.; Hilscher, J.; Datta, A.; Armario-Nájera, V.; Baysal, C.; He, W.; Huang, X.; Zhu, C.; Valizadeh-Kamran, R.; Trijatmiko, K.R.; et al. Genome editing in cereal crops: An overview. Transgenic Res. 2021, 30, 461–498. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, A.; Ezura, H. Genome Editing for Improving Crop Nutrition. Front. Genome Ed. 2022, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Patron, N.; Kay, P.; Wong, D.; Buchanan, M.; Cao, Y.-Y.; Sawbridge, T.; Davies, J.P.; Mason, J.; Webb, S.R.; et al. Zinc finger nuclease-mediated precision genome editing of an endogenous gene in hexaploid bread wheat (Triticum aestivum) using a DNA repair template. Plant Biotechnol. J. 2018, 16, 2088–2101. [Google Scholar] [CrossRef]
- Bilichak, A.; Sastry-Dent, L.; Sriram, S.; Simpson, M.; Samuel, P.; Webb, S.; Jiang, F.; Eudes, F. Genome editing in wheat microspores and haploid embryos mediated by delivery of ZFN proteins and cell-penetrating peptide complexes. Plant Biotechnol. J. 2019, 18, 1307–1316. [Google Scholar] [CrossRef]
- Ibrahim, S.; Saleem, B.; Rehman, N.; Zafar, S.A.; Naeem, M.K.; Khan, M.R. CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. Adv. Res. 2021, 37, 33–41. [Google Scholar] [CrossRef]
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 2019, 51, 1099–1105. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Zhang, D.; Zhao, X.; Cao, X.; Dong, L.; Liu, J.; Chen, K.; Zhang, H.; Gao, C.; et al. Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 2018, 94, 857–866. [Google Scholar] [CrossRef]
- Jung, Y.-J.; Nogoy, F.M.; Lee, S.-K.; Cho, Y.-G.; Kang, K.-K. Application of ZFN for Site Directed Mutagenesis of Rice SSIVa Gene. Biotechnol. Bioprocess Eng. 2018, 23, 108–115. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR–Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.-L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef]
- Char, S.N.; Unger-Wallace, E.; Frame, B.; Briggs, S.A.; Main, M.; Spalding, M.H.; Vollbrecht, E.; Wang, K.; Yang, B. Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol. J. 2015, 13, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.-Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2017, 217, 1161–1176. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Mai, H.T.T.; Moukouanga, D.; Lebrun, M.; Bellafiore, S.; Champion, A. CRISPR/Cas9-Mediated Gene Editing of the Jasmonate Biosynthesis OsAOC Gene in Rice. In Jasmonate in Plant Biology: Methods and Protocols; Champion, A., Laplaze, L., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 199–209. ISBN 978-1-07-160142-6. [Google Scholar] [CrossRef]
- Zeng, Z.; Han, N.; Liu, C.; Buerte, B.; Zhou, C.; Chen, J.; Wang, M.; Zhang, Y.; Tang, Y.; Zhu, M.; et al. Functional dissection of HGGT and HPT in barley vitamin E biosynthesis via CRISPR/Cas9-enabled genome editing. Ann. Bot. 2020, 126, 929–942. [Google Scholar] [CrossRef]
- Karunarathne, S.D.; Han, Y.; Zhang, X.; Li, C. CRISPR/Cas9 gene editing and natural variation analysis demonstrate the potential for HvARE1 in improvement of nitrogen use efficiency in barley. J. Integr. Plant Biol. 2022, 64, 756–770. [Google Scholar] [CrossRef]
- Kapusi, E.; Corcuera-Gómez, M.; Melnik, S.; Stoger, E. Heritable Genomic Fragment Deletions and Small Indels in the Putative ENGase Gene Induced by CRISPR/Cas9 in Barley. Front. Plant Sci. 2017, 8, 540. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-León, S.; Gil Humanes, J.; Ozuna, C.V.; Gimenez, M.J.; Sousa, C.; Voytas, D.; Barro, F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 Activity Facilitates Multiplex Gene Editing in Allopolyploid Wheat. CRISPR J. 2018, 1, 65–74. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Ma, L. Recent advances in CRISPR/Cas9 and applications for wheat functional genomics and breeding. aBIOTECH 2021, 2, 375–385. [Google Scholar] [CrossRef]
- Kim, D.; Hager, M.; Brant, E.; Budak, H. Efficient genome editing in wheat using Cas9 and Cpf1 (AsCpf1 and LbCpf1) nucleases. Funct. Integr. Genom. 2021, 21, 355–366. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2019, 71, 1337–1349. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Marx, V. Base editing a CRISPR way. Nat. Chem. Biol. 2018, 15, 767–770. [Google Scholar] [CrossRef]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R.; Gao, C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Soto, A.; Echeverría-Beirute, F.; Abdelnour-Esquivel, A.; Valdez-Melara, M.; Boch, J.; Gatica-Arias, A. Rice breeding in the new era: Comparison of useful agronomic traits. Curr. Plant Biol. 2021, 27, 100211. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Keshavareddy, G.; Kumar, A.; Ramu, V.S. Methods of Plant Transformation—A Review. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 2656–2668. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Nishimura, A.; Ashikari, M.; Lin, S.; Takashi, T.; Angeles, E.R.; Yamamoto, T.; Matsuoka, M. Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proc. Natl. Acad. Sci. USA 2005, 102, 11940–11944. [Google Scholar] [CrossRef]
- Ishida, Y.; Tsunashima, M.; Hiei, Y.; Komari, T. Wheat (Triticum aestivum L.) Transformation Using Immature Embryos. In Agrobacterium Protocols; Wang, K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; Volume 1, pp. 189–198. ISBN 978-1-4939-1695-5. [Google Scholar]
- Herrera-Estrella, L.R.; Depicker, A.; Van Montagu, M.; Schell, J. Expression of chimaeric genes transferred into plant cells using a Ti-plasmid-derived vector. Nature 1983, 303, 209–213. [Google Scholar] [CrossRef]
- Ramkumar, T.R.; Lenka, S.K.; Arya, S.S.; Bansal, K.C. A Short History and Perspectives on Plant Genetic Transformation. In Biolistic DNA Delivery in Plants: Methods and Protocols; Rustgi, S., Luo, H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 39–68. ISBN 978-1-07-160356-7. [Google Scholar]
- Bhatt, R.; Asopa, P.; Jain, R.; Kothari-Chajer, A.; Kothari, S.; Kachhwaha, S. Optimization of Agrobacterium Mediated Genetic Transformation in Paspalum scrobiculatum L. (Kodo Millet). Agronomy 2021, 11, 1104. [Google Scholar] [CrossRef]
- Che, P.; Anand, A.; Wu, E.; Sander, J.D.; Simon, M.K.; Zhu, W.; Sigmund, A.L.; Zastrow-Hayes, G.; Miller, M.; Liu, D.; et al. Developing a flexible, high-efficiency Agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnol. J. 2018, 16, 1388–1395. [Google Scholar] [CrossRef]
- Ehiei, Y.; Eishida, Y.; Ekomari, T. Progress of cereal transformation technology mediated by Agrobacterium tumefaciens. Front. Plant Sci. 2014, 5, 628. [Google Scholar] [CrossRef]
- Shrawat, A.K.; Good, A.G. Agrobacterium Tumefaciens-Mediated Genetic Transformation of Cereals Using Immature Embryos. In Plant Embryo Culture: Methods and Protocols; Thorpe, T.A., Yeung, E.C., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; pp. 355–372. ISBN 978-1-61737-988-8. [Google Scholar]
- Shrawat, A.K.; Lörz, H. Agrobacterium-mediated transformation of cereals: A promising approach crossing barriers. Plant Biotechnol. J. 2006, 4, 575–603. [Google Scholar] [CrossRef] [PubMed]
- Levée, V.; Garin, E.; Klimaszewska, K.; Seguin, A. Stable genetic transformation of white pine (Pinus strobus L.) after cocultivation of embryogenic tissues with Agrobacterium tumefaciens. Mol. Breed. 1999, 5, 429–440. [Google Scholar] [CrossRef]
- Liu, S.; Ma, J.; Liu, H.; Guo, Y.; Li, W.; Niu, S. An efficient system for Agrobacterium-mediated transient transformation in Pinus tabuliformis. Plant Methods 2020, 16, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.S.; Mohammadi, K.; Ji, K.S. Study on factors influencing transformation efficiency in Pinus massoniana using Agrobacterium tumefaciens. Plant Cell, Tissue Organ Cult. (PCTOC) 2018, 133, 437–445. [Google Scholar] [CrossRef]
- Morris, J.W.; Castle, L.A.; Morris, R.O. Efficacy of different Agrobacterium tumefaciens strains in transformation of pinaceous gymnosperms. Physiol. Mol. Plant Pathol. 1989, 34, 451–461. [Google Scholar] [CrossRef]
- Kumar, S.V.; Misquitta, R.W.; Reddy, V.S.; Rao, B.J.; Rajam, M.V. Genetic transformation of the green alga—Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004, 166, 731–738. [Google Scholar] [CrossRef]
- Broothaerts, W.; Mitchell, H.J.; Weir, B.; Kaines, S.; Smith, L.M.A.; Yang, W.; Mayer, J.E.; Roa-Rodríguez, C.; Jefferson, R. Gene transfer to plants by diverse species of bacteria. Nature 2005, 433, 629–633. [Google Scholar] [CrossRef]
- Chung, S.-M.; Vaidya, M.; Tzfira, T.; Chung, S.-M.; Vaidya, M.; Tzfira, T.; Chung, S.-M.; Vaidya, M.; Tzfira, T.; Chung, S.-M.; et al. Agrobacterium is not alone: Gene transfer to plants by viruses and other bacteria. Trends Plant Sci. 2006, 11, 1–4. [Google Scholar] [CrossRef]
- Abid, N.; Maqbool, A.; Malik, K.A. Screening Commercial Wheat (Triticum aestivum L.) Varieties for Agrobacterium Mediated Transformation Ability. Pak. J. Agric. Sci. 2014, 51, 83–89. [Google Scholar]
- Richardson, T.; Thistleton, J.; Higgins, T.J.; Howitt, C.; Ayliffe, M. Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 647–659. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Du, L.; Ye, X. Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 2016, 15, 614–623. [Google Scholar] [CrossRef]
- Bouchabké-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; Van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic Expression of BABY BOOM Triggers a Conversion from Vegetative to Embryonic Growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Leafy cotyledon2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef]
- Luo, G.; Palmgren, M. GRF-GIF Chimeras Boost Plant Regeneration. Trends Plant Sci. 2020, 26, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef]
- Sparks, C.A.; Jones, H.D. Transformation of Wheat by Biolistics. In Transgenic Crops of the World; Springer: Dordrecht, The Netherlands, 2004; pp. 19–34. [Google Scholar]
- Sparks, C.A. Biolistics Transformation of Wheat. In Transgenic Wheat, Barley and Oats; Springer: Berlin/Heidelberg, Germany, 2009; pp. 71–92. [Google Scholar]
- Ismagul, A.; Yang, N.; Maltseva, E.; Iskakova, G.; Mazonka, I.; Skiba, Y.; Bi, H.; Eliby, S.; Jatayev, S.; Shavrukov, Y.; et al. A biolistic method for high-throughput production of transgenic wheat plants with single gene insertions. BMC Plant Biol. 2018, 18, 135. [Google Scholar] [CrossRef]
- Kohli, A.; Gahakwa, D.; Vain, P.; Laurie, D.A.; Christou, P. Transgene expression in rice engineered through particle bombardment: Molecular factors controlling stable expression and transgene silencing. Planta 1999, 208, 88–97. [Google Scholar] [CrossRef]
- Agrawal, P.K.; Kohli, A.; Twyman, R.M.; Christou, P. Transformation of Plants with Multiple Cassettes Generates Simple Transgene Integration Patterns and High Expression Levels. Mol. Breed. 2005, 16, 247–260. [Google Scholar] [CrossRef]
- Srivastava, V.; Vasil, V.; Vasil, I.K. Molecular Characterization of the Fate of Transgenes in Transformed Wheat (Triticum aestivum L.). Theor. Appl. Genet. 1996, 92, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Stoeger, E.; Williams, S.; Keen, D.; Christou, P. Molecular Characteristics of Transgenic Wheat and the Effect on Transgene Expression. Transgenic Res. 1998, 7, 463–471. [Google Scholar] [CrossRef]
- Abdul, R.; Ma, Z.; Wang, H. Genetic Transformation of Wheat (Triticum aestivum L): A Review. Triticeae Genom. Genet. 2009, 1, 1–7. [Google Scholar] [CrossRef]
- Tassy, C.; Partier, A.; Beckert, M.; Feuillet, C.; Barret, P. Biolistic transformation of wheat: Increased production of plants with simple insertions and heritable transgene expression. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 171–181. [Google Scholar] [CrossRef]
- Bechtold, N.; Jaudeau, B.; Jolivet, S.; Maba, B.; Vezon, D.; Voisin, R.; Pelletier, G. The Maternal Chromosome Set Is the Target of the T-DNA in the in Planta Transformation of Arabidopsis thaliana. Genetics 2000, 155, 1875–1887. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Linghu, Q.; Nagira, Y.; Miki, R.; Taoka, N.; Imai, R. An in planta biolistic method for stable wheat transformation. Sci. Rep. 2017, 7, 11443. [Google Scholar] [CrossRef]
- Rod-in, W.; Sujipuli, K.; Ratanasut, K. The Floral-Dip Method for Rice (Oryza sativa) Transformation. J. Agric. Technol. 2014, 10, 467–474. [Google Scholar]
- Trieu, A.T.; Burleigh, S.H.; Kardailsky, I.V.; Maldonado-Mendoza, I.; Versaw, W.K.; Blaylock, L.A.; Shin, H.; Chiou, T.-J.; Katagi, H.; Dewbre, G.R.; et al. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 2000, 22, 531–541. [Google Scholar] [CrossRef]
- Yasmeen, A.; Mirza, B.; Inayatullah, S.; Safdar, N.; Jamil, M.; Ali, S.; Choudhry, M.F. In Planta Transformation of Tomato. Plant Mol. Biol. Report. 2009, 27, 20–28. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Cigan, A.M. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]

| Crop | Genome Editing Technique | Targeted Gene | Molecular Function | Type of Editing | Effect | Reference |
|---|---|---|---|---|---|---|
| Wheat | ZFN | AHAS | Role in branched amino acid formation | Insertion and replacement | Resistance against herbicide | [141] |
| ZFN | IPK1 | Phytate formation | Deletion | Removal of antinutritional phytate, mineral accumulation against abiotic stress (Fe, Zn) | [142] | |
| CRISPR/Cas9 | IPK1 | Phytic acid biosynthesis | Deletion and insertion | Reduced phytic acid and enhanced Fe and Zn in wheat grains | [143] | |
| CRISPR/Cas9 | HRC | Encodes a putative histidine-rich calcium-binding protein | Deletion and insertion | Reduced fusarium head blight severity | [144] | |
| CRISPR/Cas9 | GW2 | Genetic determinant of grain weight | Deletion | Increase in thousand-grain weight and grain protein content | [145] | |
| Rice | ZFN | SSIVa | Involved in the starch biosynthesis pathway | Deletion | Improve eating quality | [146] |
| TALENs | 11N3 | Rice bacterial blight susceptibility gene | Deletion and insertion | Increase resistance to rice bacterial blight | [147] | |
| CRISPR/Cas9 | SAPK2 | Regulate drought response | Deletion | Improved drought tolerance | [148] | |
| CRISPR/Cas9 | OsPIN5b GS3 OsMYB30 | Regulate panicle length, grain size, and cold tolerance | Mutation deletion | Increased panicle length, enlarged grain size, and increased cold tolerance | [149] | |
| CRISPR/Cas9 | NRT1.1B | Control yield and early maturation | Base editing | Increased yield | [150] | |
| Maize | ZFN | IPK1 | Catalyses the final step in phytate biosynthesis in maize seeds | Deletion and insertion | Removal of antinutritional phytate herbicide tolerance | [151] |
| TALENs | gl2 | Cuticular lipid functions | Deletion | Reduce epicuticular wax | [152] | |
| CRISPR/Cas9 | RR22 | Salinity tolerance | Deletion, insertion, substitution | Regulation of salt tolerance | [153] | |
| CRISPR/Cas9 | NC1 QTL and, HKT1 | Encodes an HKT-type transporter | Deletion | Reduce salt tolerance | [154] | |
| CRISPR/Cas9 | AOC | Jasmonic acid biosynthesis pathway | Deletion, insertion, substitution | Efficient coordination with the environment | [155] | |
| Barley | CRISPR/Cas9 | HPT HGT | Biosynthesis of tocotrienols and tocopherol | Deletion and insertion | Decreased grain size and weight Shrunken phenotype Lower total starch content in grains | [156] |
| ARE1 | Involved in nitrogen use efficiency | Missense and/or frameshift mutations | Increase in plant height, tiller number, grain protein content, yield, and chlorophyll content | [157] | ||
| CRISPR/Cas9 | ENGase | Production of GN1 type FNGs (Free N Glycans | Indels and deletions | Increased abiotic tolerance | [158] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wijerathna-Yapa, A.; Ramtekey, V.; Ranawaka, B.; Basnet, B.R. Applications of In Vitro Tissue Culture Technologies in Breeding and Genetic Improvement of Wheat. Plants 2022, 11, 2273. https://doi.org/10.3390/plants11172273
Wijerathna-Yapa A, Ramtekey V, Ranawaka B, Basnet BR. Applications of In Vitro Tissue Culture Technologies in Breeding and Genetic Improvement of Wheat. Plants. 2022; 11(17):2273. https://doi.org/10.3390/plants11172273
Chicago/Turabian StyleWijerathna-Yapa, Akila, Vinita Ramtekey, Buddhini Ranawaka, and Bhoja Raj Basnet. 2022. "Applications of In Vitro Tissue Culture Technologies in Breeding and Genetic Improvement of Wheat" Plants 11, no. 17: 2273. https://doi.org/10.3390/plants11172273
APA StyleWijerathna-Yapa, A., Ramtekey, V., Ranawaka, B., & Basnet, B. R. (2022). Applications of In Vitro Tissue Culture Technologies in Breeding and Genetic Improvement of Wheat. Plants, 11(17), 2273. https://doi.org/10.3390/plants11172273







