An Overview of Pentatricopeptide Repeat (PPR) Proteins in the Moss Physcomitrium patens and Their Role in Organellar Gene Expression
Abstract
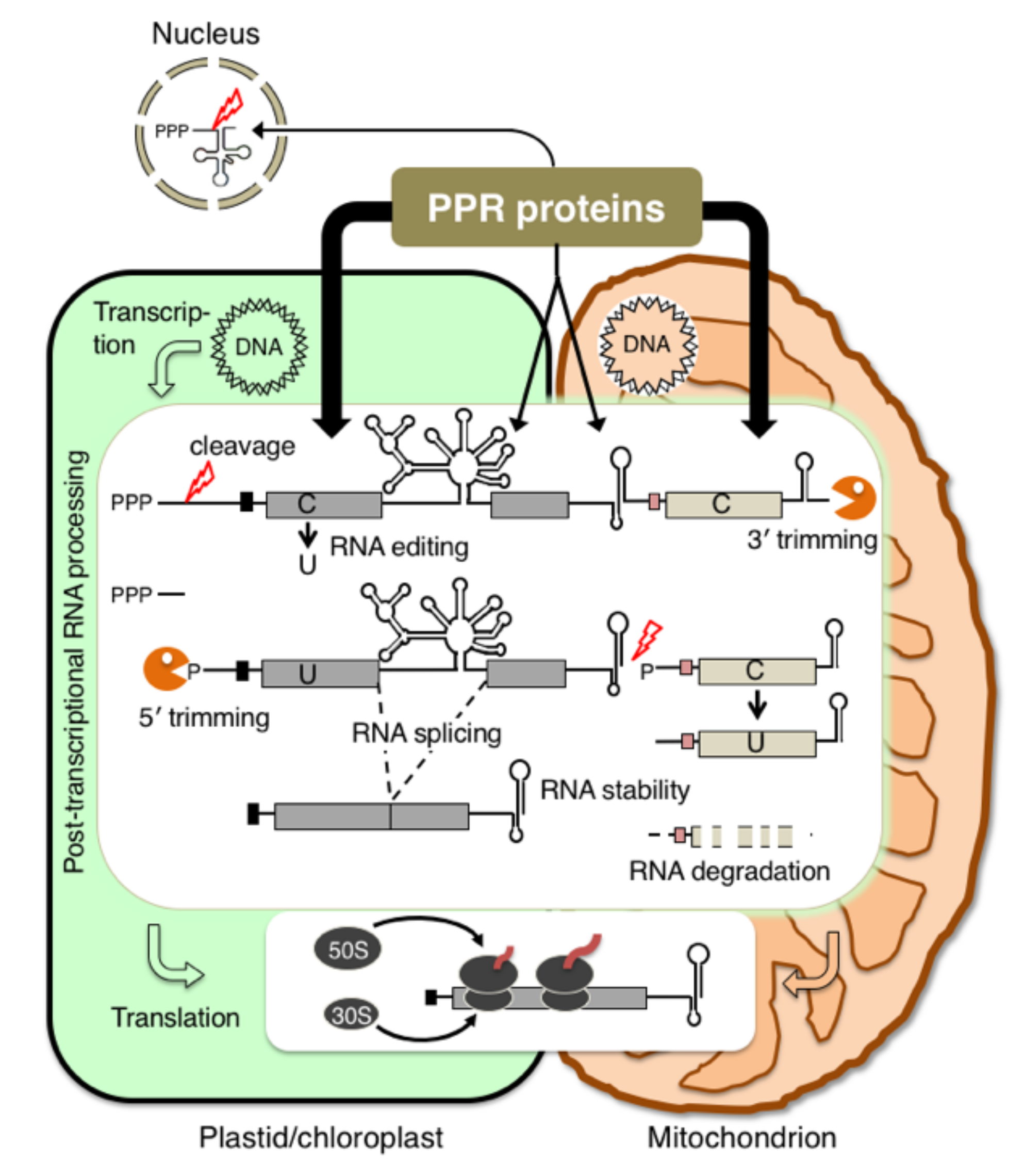
:1. Introduction
2. P. patens Is a Model Plant for Studying the Molecular Function of PPR Genes
3. The Compact PPR Gene Family in Physcomitrium
| Protein Name | Gene Locus ID 1 | Type 2 | PPR Motif Bead Patterns and Additional Domain/Motif in Parenthesis 3 | Subcellular Localization 4 | Phenotype of Moss Colony of Gene Knockout Lines | Function Identified in P. patens | Arabidopsis Homologs | Function Identified in Arabidopsis | Refs. | |
|---|---|---|---|---|---|---|---|---|---|---|
| Pred | Exp | |||||||||
| PpPPR_1 | Pp3c3_19290 | P | P-P-P-P-P-P-P-P-P-P-P-P-P | m | At5g50280 (EMB1006) | required for embryo development | [14] | |||
| PpPPR_2 | Pp3c16_9240 | P | P-P-P-P-P-P-P-P-P-P-P | c | C | Smaller than WT | ||||
| PpPPR_3 | Pp3c5_10110 | P | (RRM)-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P | other | C | WT-like | At5g04810 (AtPPR4) | rps12-intron 1 trans-splicing | [37,38] | |
| PpPPR_4 | Pp3c17_11510 | P | P-P-Pi-P-P-P-P-P-P-P-P | c | C | Very small colony | RNA splicing of group II intron in pre-tRNAIle | [39] | ||
| PpPPR_5 | Pp3c21_11730 | P | P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_6 | Pp3c5_21760 | P | P-P-P-P | c | C | |||||
| PpPPR_7 | Pp3c25_10050 | P | P-P-P-P-P-P-Pi-P-P-P-(LAGLIDADG) | c | At2g15820 (OTP51) | ycf3-intron 2 splicing | [40] | |||
| PpPPR_8 | Pp3c14_19310 | P | P-P-P-Pi-P-P-P-P-P-P-P | m | Smaller than WT | |||||
| PpPPR_9 | Pp3c24_14870 | PLS | P1-L1-S1-P1-L1-S1-P1-L1-S1-P2 | m | M | Smaller than WT | RNA splicing of cox1 intron 3 | [41] | ||
| PpPPR_10 | Pp3c21_550 | P | P-P | c | At4g21190 (EMB1417) | chloroplast localized, required for embryo development | [16] | |||
| PpPPR_11 | Pp3c3_2440 | P | P-P-P-P-P | other | M | Smaller than WT | stabilization of nad7 mRNA | Unpublished | ||
| PpPPR_12 | Pp3c7_22430 | P | P-Pi-P-P-P-P-P-P-P-P-P | c | ||||||
| PpPPR_13 | Pp3c7_17210 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P | c | ||||||
| PpPPR_14 | Pp3c26_10760 | P | P-P-P-P-P-P-P-P-P-P-P | c | At3g46610 (LPE1) | binds to the 5′ UTR of psbA mRNA | [42] | |||
| PpPPR_15 | Pp3c17_6450 | P | P1-SS-P1-P1-P1 | c | C | |||||
| PpPPR_16 | Pp3c14_20030 | P | P-P-P-P-P-P | m | ||||||
| PpPPR_17 | Pp3c8_4580 | P | P-P-P-P-P-P-P-P-P-P-P | c | C | Smaller than WT | At4g39620 (AtPPR5) | trnG splicing and intron stabilization | [43] | |
| PpPPR_18 | Pp3c10_3690 | P | P-P-P-P-P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_19 | Pp3c10_19570 | P | P-Pi-P-P-P-P-P-P-P-P-P-P | c | C | WT-like | At4g34830 (MRL1) | stabilizes rbcL 5’ end | [44] | |
| PpPPR_20 | Pp3c10_14800 | P | P-P-P-P-P-P-P-P | c | C | At1g01970 | chloroplast localized, function unknown | [16] | ||
| PpPPR_21 | Pp3c22_3230 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P | c | C | Smaller than WT | stablilization of psbI-ycf12 mRNA | At5g02860 (AtPPR21L) | chloroplast localized, probably required for embryo development | [45] |
| PpPPR_22 | Pp3c16_23700 | P | P-P-P-P-P-P-P-P-P-P-(LAGLIDADG) | m | At2g15820 (OTP51) | ycf3 -intron 2 splicing | [40] | |||
| PpPPR_23 | Pp3c26_11100 | P | P-P-P-P-P-P-P-P-P-P | c | Smaller than WT | At3g59040 (PPR287) | crucial for chloroplast function and plant development | [46] | ||
| PpPPR_24 | Pp3c2_3210 | P | P-P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_25 | Pp3c16_5160 | PLS | P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1 | m | M | |||||
| PpPPR_26 | Pp3c10_24680 | P | P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_27 | Pp3c20_15110 | P | P-P-P-P-P-P-P-P-P-P | other | C | WT-like | At3g53170 | chloroplast nucleoid | [47] | |
| PpPPR_28 | Pp3c6_24160 | P | P-P-P-P-P-P-P-P-P-P-P-P | c | C | Smaller than WT | ||||
| PpPPR_29 | Pp3c5_2770 | P | P-P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_30 | Pp3c7_10060 | P | P-P-P-P-P-P-P-P-(SMR) | m | At1g18900 | mitochondrial localized, function unknown | [16] | |||
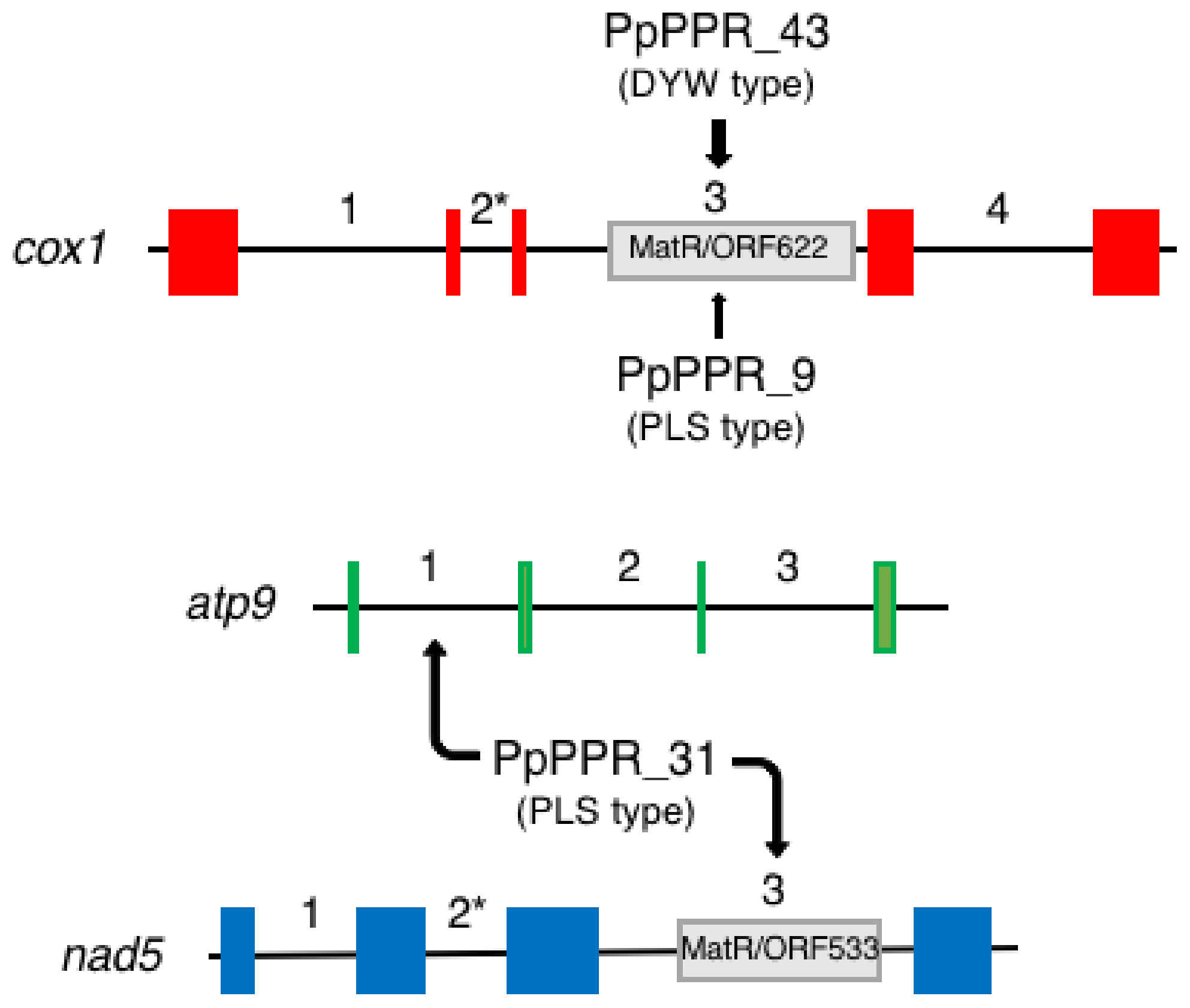
| PpPPR_31 | Pp3c6_23550 | PLS | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1 | m | M | Smaller than WT | RNA splicing of atp9 intron 1 and nad5 intron 3 | [41] | ||
| PpPPR_32 | Pp3c8_15500 | P | P-P-P-P-P-P-P-P-P-P-P-P-P | c | C | Smaller than WT | psaC mRNA accumulation | [48] | ||
| PpPPR_33 | Pp3c1_37670 | P | P-P-P-P-P-P-P-Pi-P-P | m | ||||||
| PpPPR_34 | Pp3c22_1710 | PLS | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1 | other | C | |||||
| PpPPR_35 | Pp3c8_13830 | P | P-P-P-P-P-P-P-P-P-P | other | ||||||
| PpPPR_36 | Pp3c4_14490 | P | P-P-P-P-P | m | ||||||
| PpPPR_37 | Pp3c4_14140 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P | m | M | Bigger than WT | ||||
| PpPPR_38 | Pp3c6_26920 | P | P-P-P-P-P-P-P-P-P-P-P | c | C | Very small colony | RNA processing of clpP-5′-rps12 mRNA | [49,50] | ||
| PpPPR_39 | Pp3c4_3090 | P | P-P-P-P-P-Pi-P-P | c | At3g42630 (PBF2) | RNA splicing of ycf3 | [51] | |||
| PpPPR_40 | Pp3c23_13490 | P | P-P-P-P-P-P-P-P-P | c | C | At5g67570 (DG1) | involved in the regulation of early chloroplast development | [52] | ||
| PpPPR_41 | Pp3c13_18720 | P | P-Pi-P-P-P-P | other | C | |||||
| PpPPR_42 | Pp3c21_12360 | P | P-P-P-P-P-P-P-P-P-P-(SMR) | m | At5g02830 | chloroplast nucleoid | [47] | |||
| PpPPR_43 | Pp3c3_24770 | DYW | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | m | M | Very small colony | RNA splicing of cox1 intron 3 | [53] | ||
| PpPPR_44 | Pp3c17_24090 | P | P-P-P-P-P-P-Pi-P-P-P | m | ||||||
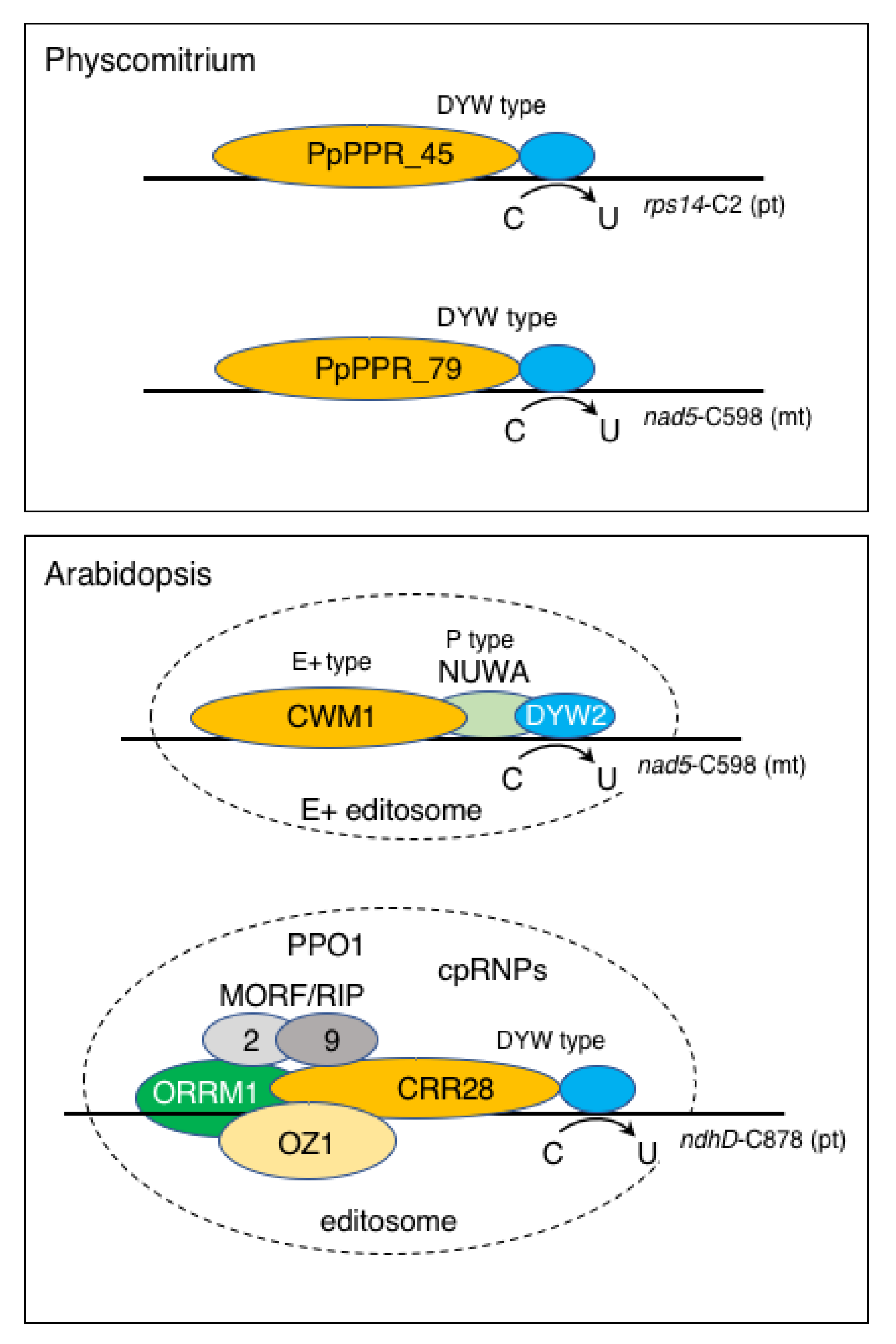
| PpPPR_45 | Pp3c11_7720 | DYW | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | c | C | RNA editing at rps14-C2 | [54] | |||
| PpPPR_46 | Pp3c3_5760 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P | c | At5g39980 (PDM3) | essential for chloroplast development | [55] | |||
| PpPPR_47 | Pp3c4_17980 | P | P-Pi-P-P-P-P-P-P-P-P | other | ||||||
| PpPPR_48 | Pp3c24_4430 | P | P-P-P-P-P-P-P-P-P-P-P-P-Pi-P-P | c | M | WT-like | At1g10910 (PDM2) | involved in the regulation of chloroplast development | [56] | |
| PpPPR_49 | Pp3c1_6490 | P | P-P-P-P-P-P-P-P-P-Pi-P-P-P-P-P-P-P-P-P-P-P-P-P-P | m | M | WT-like | ||||
| PpPPR_50 | Pp3c11_2930 | P | P-P-P-Pi-P-Pi-Pi-P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_51 | Pp3c12_14320 | P | P-P-P-P-P-P-P-P-P-P | c | C | At4g34830 (MRL1) | stabilizes rbcL 5′ end | [44] | ||
| PpPPR_52 | Pp3c25_7340 | P | P-P-P-P-P | c | C | |||||
| PpPPR_53 | Pp3c13_17120 | P | P-P-P-P-P-P-P-P-P-P | c | C | WT-like | At5g02860 | chloroplast nucleoid | [47] | |
| PpPPR_54 | Pp3c12_26140 | P | P-P | m | C/M | Bigger than WT | ||||
| PpPPR_55 | Pp3c6_14920 | P | P-P-P | c | At3g46870 (THA8-like) | ycf3-intron 2 and trnA splicing | [57] | |||
| PpPPR_56 | Pp3c19_930 | DYW | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | m | M | WT-like | RNA editing at nad3-C230, nad4-C272 | [58] | ||
| PpPPR_57 | Pp3c12_4690 | P | P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_58 | Pp3c1_21850 | P | P-P-P-P-Pi-Pi-P-P-P-P | m | At4g35850 | present in mitochondrial complexome | [59] | |||
| PpPPR_59 | Pp3c22_3070 | P | P-P-P-P-P-P-P-P-P-P-P-P-P (SMR) | c | C | WT-like | At5g02830 | chloroplast nucleoid | [47] | |
| PpPPR_60 | Pp3c16_9420 | P | P-P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_61 | Pp3c14_7210 | P | P-P-P-P-Pi-Pi-P-P-P-P | m | At4g35850 | present in mitochondrial complexome | [59] | |||
| PpPPR_62 | Pp3c16_9880 | P | P-P-P-P-(SMR) | m | At2g17033 | chloroplast localized, function unknown | [16] | |||
| PpPPR_63 | Pp3c7_17100 | P | P-Pi-P-(NYN) | other | NUC | Very small colony | 5’-end processing of pre-tRNA | At2g16650 (PRORP2)At4g21900 (PRORP3) | 5’-end processing of pre-tRNA | [60,61] |
| PpPPR_64 | Pp3c11_11830 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-Pi-P-(SMR) | other | C | Smaller than WT | Expression of psaA-psaB-rps14 | At1g74850 (pTAC2) | involved in transcription by PEP | [62,63] |
| PpPPR_65 | Pp3c4_16600 | DYW | S2-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | m | M | Very small colony | RNA editing at ccmFc-C103 | [64,65] | ||
| PpPPR_66 | Pp3c16_5890 | P | P-P-P-P-P-P-P-P-P-P-P | c | C | WT-like | RNA splicing of ndhA | At2g35130 (AtPPR66L) | RNA splicing of ndhA | [66] |
| PpPPR_67 | Pp3c2_30390 | P | P-P-P-(NYN) | other | C/M | WT-like | 5′-end processing of pre-tRNA | At2g32230 (RPORP1) | 5′-end processing of pre-tRNA | [60,61] |
| PpPPR_68 | Pp3c2_27580 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_69 | Pp3c17_5040 | PLS | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2 | sp | C | WT-like | At4g18520 (PDM1/SEL1) | RNA editing of accD, trnK-intron splicing, RNA processing of rpoA | [67] | |
| PpPPR_70 | Pp3c8_4280 | P | P-P-P-P-P-P-P-P-P-(CBS) | c | C/NUC | At5g10690 (CBSPPR1) | chloroplast nucleoid, possibly play a role in regulating transcription or replication | [47,68] | ||
| PpPPR_71 | Pp3c14_16110 | DYW | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | m | M | Very small colony | RNA editing at ccmFc-C122 | [69] | ||
| PpPPR_72 | Pp3c6_26210 | P | P-P-P-P-P-P-P-P-P-P-P | c | C/M | function unknown | At2g35130 (AtPPR66L) | RNA splicing of ndhA | [66] | |
| PpPPR_73 | Pp3c3_31860 | P | P-P-P-P-P-Pi-Pi-P | m | C/M | |||||
| PpPPR_74 | Pp3c17_13520 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P | c | C | Smaller than WT | ||||
| PpPPR_75 | Pp3c5_16950 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-(SMR) | c | C | At2g31400 (GUN1) | involved in retrograde signaling to the nucleus. | [70] | ||
| PpPPR_76 | Pp3c16_280 | P | (RRM)-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P | other | C | At5g04810 (AtPPR4) | rps12-intron 1 trans-splicing | [37,38] | ||
| PpPPR_77 | Pp3c5_15090 | DYW | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | other | M | Very small colony | RNA editing at cox2-C370, cox3-C733 | [71] | ||
| PpPPR_78 | Pp3c2_12230 | DYW | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | other | M | WT-like | RNA editing at rps14-C137, cox1-C755 | [71,72] | ||
| PpPPR_79 | Pp3c5_7610 | DYW | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | m | M | Smaller than WT | RNA editing at nad5-C598 | [58] | ||
| PpPPR_80 | Pp3c4_9690 | P | P-P-P-P-P-P-P-P-P-P | c | C | At4g39620 (AtPPR5) | trnG splicing and intron stabilization | [43] | ||
| PpPPR_81 | Pp3c14_15490 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-(SMR) | other | C | Bigger than WT | ||||
| PpPPR_82 | Pp3c9_6880 | P | P-P-P-P-P-P-P-P-P | other | C | |||||
| PpPPR_83 | Pp3c2_1940 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-Pi-P-P-P | other | At2g41720 (EMB2654) | rps12-intron 1 trans-splicing | [73,74] | |||
| PpPPR_84 | Pp3c15_12010 | P | P-P-P-P-P-P-P-P-P-P | c | ||||||
| PpPPR_85 | Pp3c6_15900 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-(SMR) | c | C | WT-like | At2g31400 (GUN1) | involved in retrograde signaling to the nucleus | [70] | |
| PpPPR_86 | Pp3c17_2130 | P | P-P-Pi-P-Pi-P-P-P-P-P-P-P-P-P-P-P-P-Pi-P | c | ||||||
| PpPPR_87 | Pp3c8_15040 | P | P-P-P-P-P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_88 | Pp3c18_8600 | P | P-P-P-P-P-P-P-P-P | m | ||||||
| PpPPR_89 | Pp3c1_28760 | P | P-P-P-P-P-P-P | m | M | |||||
| PpPPR_90 | Pp3c11_21930 | 15P | P-P-Pi-P-P-P-P-P-P-P-P-P-P-P-P | other | At5g42310 (AtCRP1) | stabilizes 5′ and 3′ ends in petB-petD intergenic region; activates petA, psaC, and petD translation | [17,18,75,76] | |||
| PpPPR_91 | Pp3c17_23250 | DYW | P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-P1-L1-S1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | m | M | Very small colony | RNA editing at nad5-C730 | [58] | ||
| PpPPR_92 | Pp3c5_2530 | P | P-P-P-P-P-P-P-Pi-P-P-P-P-P-P-P-P-P-P-P-P | c | C | Smaller than WT | At4g30825 (BFA2) | accumulation of the atpH/F transcript | [77] | |
| PpPPR_93 | Pp3c6_3910 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P | other | ||||||
| PpPPR_94 | Pp3c16_4140 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P-P | c | C | At4g30825 (BFA2) | accumulation of the atpH/F transcript | [77] | ||
| PpPPR_95 | Pp3c5_26320 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P | c | ||||||
| PpPPR_96 | Pp3c4_4900 | P | P-P-P-P-P-P-P-P-P-P-Pi-P-(SMR) | c | C | |||||
| PpPPR_97 | Pp3c3_19780 | P | P-P-P-P-P-P-P-P-P-P-P-P-P-P | m | M | |||||
| PpPPR_98 | Pp3c27_5540 | DYW | P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-L2-S2-E1-E2-DYW | m | M | WT-like | RNA editing at atp9-C92 | [64,65] | ||
| PpPPR_99 | Pp3c12_2390 | P | P-P-P-P-P-P-P-P-P-P-P-P-P | c | C | At3g09650 (HCF152) | stabilizes 5′ and 3′ ends in psbH-petB intergennic region, also stimulates petB splicing | [20,78] | ||
| PpPPR_100 | Pp3c12_8330 | P | P-P-P-P-Pi-Pi-P-P-P | c | At2g30100 | chloroplast localized, function unknown | [16] | |||
| PpPPR_101 | Pp3c2_36070 | P | P1-P1-P2-L2-P1-P1-P2-P1-P1-P1-P1-L2-P1-P1 | m | ||||||
| PpPPR_102 | Pp3c6_11500 | P | P-P-P-P | c | C | WT-like | ||||
| PpPPR_103 | Pp3c17_4890 | P | P1-L1-P2 | other | ||||||
| PpPPR_104 | Pp3c10_16850 | P | P-P-Pi-P-(NYN) | m | C/M | Smaller than WT | 5’-end processing of pre-tRNA | At2g32230 (RPORP1) | 5’-end processing of pre-tRNA | [60,61] |
| PpPPR_105 | Pp3c24_8560 | PLS | L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P1-L1-S1-P2-S1-P1-L1-S2 | m | C | WT-like | ||||
| PpPPR_106 | Pp3c22_3080 | P | P-P-(SMR) | sp | ||||||
| PpPPR_107 | Pp3c1_8170 | P | P-P-P-P-P-P-(SAP)-P | m | C | At3g04260 (pTAC3) | light-dependent transcription | [79] | ||
4. Subcellular Localization of PPR Proteins in Physcomitrium
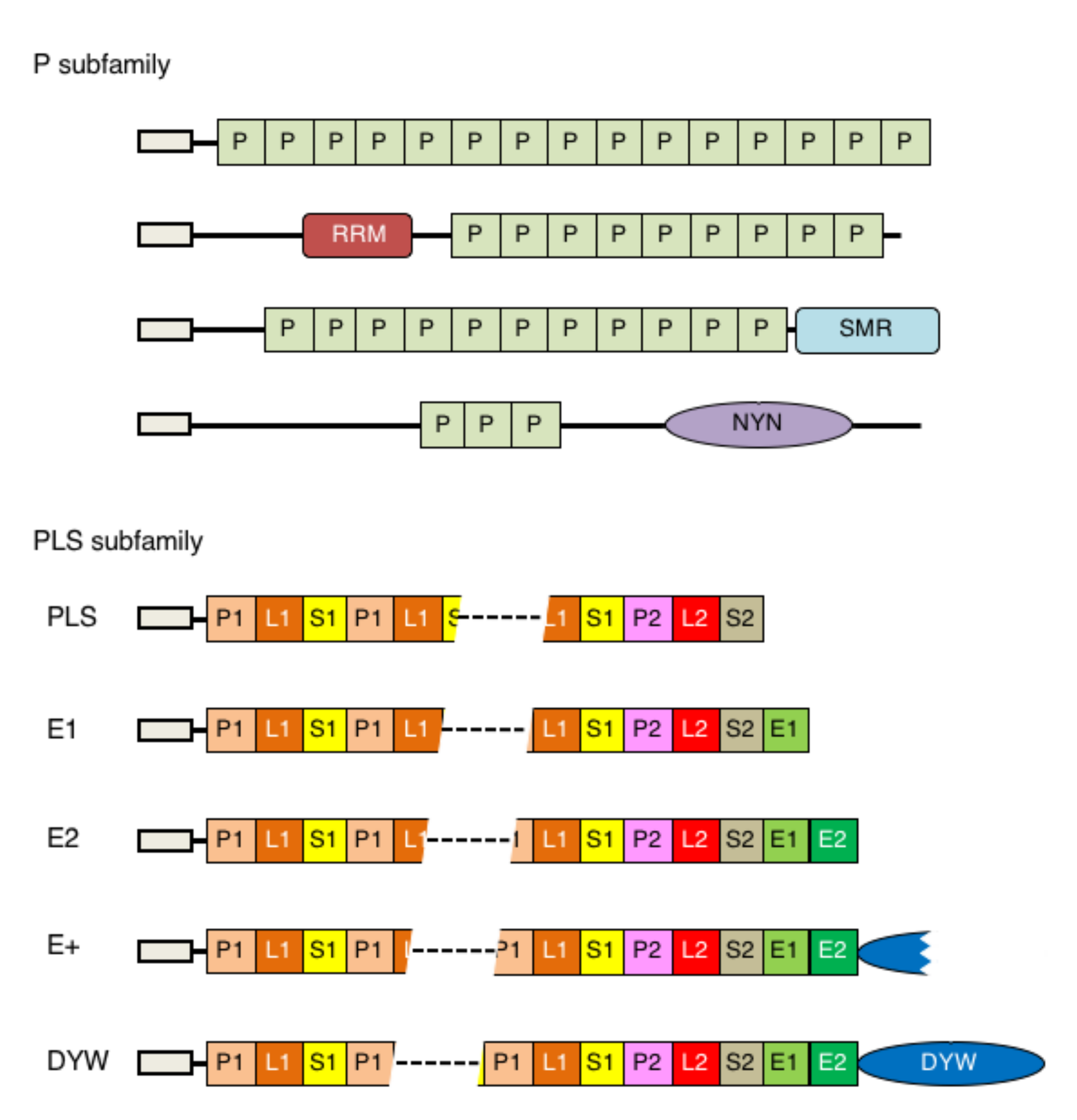
5. P-Type of PPR Proteins in Physcomitrium
5.1. P-Type Proteins Involved in RNA Processing/Stabilization
5.1.1. PpPPR_38
5.1.2. PpPPR_21
5.1.3. PpPPR_32
5.2. P-Type Proteins Involved in RNA Splicing
5.2.1. PpPPR_4
5.2.2. PpPPR_66
5.3. P-Type Proteins with an NYN Domain
5.4. P-Type Proteins with an SMR Domain
6. PLS-Type PPR Proteins in Physcomitrium
7. DYW-Type PPR Proteins in Physcomitrium
8. Molecular Basis of RNA Editing in Physcomitrium
9. Additional Function of the DYW Domain
10. Conclusions and Future Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gray, M.W. The evolutionary origins of organelles. Trends Genet. 1989, 5, 294–299. [Google Scholar] [CrossRef]
- Herrmann, R.G.; Westhoff, P.; Link, G. Biogenesis of plastids in higher plants. In Cell Organelles. Plant Gene Research; Herrmann, R.G., Ed.; Springer: Vienna, Austria, 1992; Volume 6, pp. 275–349. [Google Scholar] [CrossRef]
- Binder, S.; Brennicke, A. Gene expression in plant mitochondria: Transcriptional and post–transcriptional control. Phil. Trans. R. So. Lond. B 2003, 358, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988, 7, 2637–2644. [Google Scholar] [CrossRef] [PubMed]
- Sugita, M.; Sugiura, M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 1996, 32, 315–326. [Google Scholar] [CrossRef]
- Stern, D.B.; Goldschmidt-Clermont, M.; Hanson, M.R. Chloroplast RNA metabolism. Ann. Rev. Plant Biol. 2010, 61, 125–155. [Google Scholar] [CrossRef]
- Hammani, K.; Giegé, P. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014, 19, 380–389. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008, 13, 663–670. [Google Scholar] [CrossRef]
- Gutmann, B.; Gobert, A.; Giegé, P. Mitochondrial genome evolution and the emergence of PPR proteins. In Advances in Botanical Research 63, Mitochondrial Genome Evolution. Edition: Mitochondria, RNA Binding Proteins, Post-Transcriptional Regulation; Drouard, L., Ed.; Elsevier: Amsterdam, The Netherlands, 2012; Chapter 10; pp. 253–313. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Small, I.D.; Peeters, N. The PPR-motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 46–47. [Google Scholar] [CrossRef]
- Aubourg, S.; Boudet, N.; Kreis, M.; Lecharny, A. In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol. Biol. 2000, 42, 603–613. [Google Scholar] [CrossRef]
- Groves, M.R.; Barford, D. Topological characteristics of helical repeat protein. Curr. Opin. Struct. Biol. 1999, 9, 383–389. [Google Scholar] [CrossRef]
- Lurin, C.; Andrés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Cheng, S.; Gutmann, B.; Zhong, X.; Ye, Y.; Fisher, M.F.; Bai, F.; Castleden, I.; Song, Y.; Song, B.; Huang, J.; et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016, 85, 532–547. [Google Scholar] [CrossRef]
- Colcombet, J.; Lopez-Obando, M.; Heurtevin, L.; Bernard, C.; Martin, K.; Berthomé, R.; Lurin, C. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 2013, 10, 1557–1575. [Google Scholar] [CrossRef]
- Barkan, A.; Walker, M.; Nolasco, M.; Johnson, D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994, 13, 3170–3181. [Google Scholar] [CrossRef]
- Fisk, D.G.; Walker, M.B.; Barkan, A. Molecular cloning of the maize gene crp1reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999, 18, 2621–2630. [Google Scholar] [CrossRef]
- Bentolila, S.; Alfonso, A.A.; Hanson, M.R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 2002, 99, 10887–10892. [Google Scholar] [CrossRef]
- Meierhoff, K.; Felder, S.; Nakamura, T.; Bechtold, N.; Schuster, G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 2003, 15, 1480–1495. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Endo, T.; Peltier, G.; Tasaka, M.; Shikanai, T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 2003, 36, 541–549. [Google Scholar] [CrossRef]
- Kazama, T.; Toriyama, K. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 2003, 544, 1873–3468. [Google Scholar] [CrossRef]
- Williams, P.M.; Barkan, A. A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 2003, 36, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Tasaka, M.; Shikanai, T. PPR motif of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004, 38, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Hattori, M.; Hasebe, M.; Sugita, M. Identification and characterization of cDNAs encoding pentatricopeptide repeat proteins in the basal land plant, the moss Physcomitrella patens. Gene 2004, 343, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Cove, D. The moss Physcomitrella patens. Annu. Rev. Genet. 2005, 39, 339–358. [Google Scholar] [CrossRef]
- Cove, D.; Bezanilla, M.; Harries, P.; Quatrano, R. Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 2006, 57, 497–520. [Google Scholar] [CrossRef]
- Sugiura, C.; Kobayashi, Y.; Aoki, S.; Sugita, C.; Sugita, M. Complete chloroplast DNA sequence of the moss Physcomitrella patens: Evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res. 2003, 31, 5324–5331. [Google Scholar] [CrossRef]
- Terasawa, K.; Odahara, M.; Kabeya, Y.; Kikugawa, T.; Sekine, Y.; Fugiwara, M.; Sato, N. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol. Biol. Evol. 2007, 24, 699–709. [Google Scholar] [CrossRef]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.-F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, D.G.; Zrÿd, J.-P. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997, 11, 1195–1206. [Google Scholar] [CrossRef]
- Sugiura, C.; Sugita, M. Plastid transformation reveals that moss tRNAArg-CCG is not essential for plastid function. Plant J. 2004, 40, 314–321. [Google Scholar] [CrossRef]
- Nishiyama, T.; Hiwatashi, Y.; Sakakibara, K.; Kato, M.; Hasebe, M. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 2000, 7, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Sakayama, H.; de Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell 2018, 174, 448–464.e24. [Google Scholar] [CrossRef]
- O’Toole, N.; Hattori, M.; Andres, C.; Iida, K.; Lurin, C.; Schmitz-Linneweber, C.; Sugita, M.; Small, I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008, 25, 1120–1128. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Williams-Carrier, R.E.; Williams-Voelker, P.M.; Kroeger, T.S.; Vichas, A.; Barkan, A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 2006, 18, 2650–2663. [Google Scholar] [CrossRef]
- Tahini, L.; Ferrari, R.; Lehninger, M.-K.; Mizzotti, C.; Moratti, F.; Resentini, F.; Colombo, M.; Costa, A.; Masiero, S.; Pesaresi, P. Trans-splicing of plastid rps12 transcripts, mediated by AtPPR4, is essential for embryo patterning in Arabidopsis thaliana. Planta 2018, 248, 257–265. [Google Scholar] [CrossRef]
- Goto, S.; Kawaguchi, Y.; Sugita, C.; Ichinose, M.; Sugita, M. P-class pentatricopeptide repeat protein PTSF1 is required for splicing of the plastid pre-tRNAIle in Physcomitrella patens. Plant J. 2016, 86, 493–503. [Google Scholar] [CrossRef]
- de Longevialle, A.F.; Hendrickson, L.; Taylor, N.L.; Delannoy, E.; Lurin, C.; Badger, M.; Millar, A.H.; Small, I. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008, 56, 157–168. [Google Scholar] [CrossRef]
- Ichinose, M.; Ishimaru, A.; Nakajima, K.; Kawaguchi, Y.; Sugita, M. Two novel PLS-class pentatricopeptide repeat proteins are involved in the group II intron splicing of mitochondrial transcripts in the moss Physcomitrella patens. Plant Cell Physiol. 2020, 61, 1687–1698. [Google Scholar] [CrossRef]
- Jin, H.; Fu, M.; Duan, Z.; Duan, S.; Li, M.; Dong, X.; Liu, B.; Feng, D.; Wang, J.; Peng, L.; et al. Low photosynthetic efficiency 1 is required for light-regulated photosystem II biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E6075–E6084. [Google Scholar] [CrossRef]
- Beick, S.; Schmitz-Linneweber, C.; Williams-Carrier, R.; Jensen, B.; Barkan, A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 2008, 28, 5337–5347. [Google Scholar] [CrossRef] [PubMed]
- Johnson, X.; Wostrikoff, K.; Finazzi, G.; Kuras, R.; Schwarz, C.; Bujaldon, S.; Nickelsen, J.; Stern, D.B.; Wollman, F.-A.; Vallon, O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 2010, 22, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, T.; Matsuda, Y.; Sugita, C.; Ichinose, M.; Yamamoto, H.; Shikanai, T.; Sugita, M. The P-class pentatricopeptide repeat protein PpPPR_21 is needed for accumulation of the psbI-ycf12 dicistronic mRNA in Physcomitrella chloroplasts. Plant J. 2019, 97, 1120–1131. [Google Scholar] [CrossRef]
- Lee, K.; Park, S.J.; Han, J.H.; Jeon, Y.; Pai, H.-S.; Kang, H. A chloroplast-targeted pentatricopeptide repeat protein PPR287 is crucial for chloroplast function and Arabidopsis development. BMC Plant Biol. 2019, 19, 244. [Google Scholar] [CrossRef]
- Majeran, W.; Friso, G.; Asakura, Y.; Qu, X.; Hung, M.; Ponnala, L.; Watkins, K.P.; Barkan, A.; van Wijk, K.J. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 2012, 158, 156–189. [Google Scholar] [CrossRef]
- Suzuki, R.; Sugita, C.; Aoki, S.; Sugita, M. Physcomitrium patens pentatricopeptide repeat protein PpPPR_32 is involved in the accumulation of psaC mRNA encoding the iron sulfur protein of photosystem I. Genes Cells 2022, 27, 293–304. [Google Scholar] [CrossRef]
- Hattori, M.; Miyake, H.; Sugita, M. A pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. J. Biol. Chem. 2007, 282, 10773–10782. [Google Scholar] [CrossRef]
- Hattori, M.; Sugita, M. A moss pentatricopeptide repeat protein binds to the 3’ end of plastid clpP pre-mRNA and assists with the mRNA maturation. FEBS J. 2009, 276, 5860–5869. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Z.; Zhang, Y.; Zhou, W.; Zhang, A.; Lu, C. Pentatricopeptide repeat protein PHOTOSYSTEM I BIOGENESIS FACTOR2 is required for splicing of ycf3. J. Integr. Plant Biol. 2020, 62, 1741–1761. [Google Scholar] [CrossRef]
- Chi, W.; Ma, J.; Zhang, D.; Guo, J.; Chen, F.; Lu, C.; Zhang, L. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 2008, 147, 573–584. [Google Scholar] [CrossRef]
- Ichinose, M.; Tasaki, E.; Sugita, C.; Sugita, M. A PPR-DYW protein is required for splicing of a group II intron of cox1 pre-mRNA in Physcomitrella patens. Plant J. 2012, 70, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Uchida, M.; Sugita, M. Identification of a pentatricopeptide repeat RNA editing factor in Physcomitrella patens chloroplasts. FEBS Lett. 2014, 588, 4060–4064, Erratum in FEBS Lett. 2015, 589, 1171. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, J.; Li, Y.; Su, B.; Xu, H.; Shan, X.; Song, C.; Xie, J.; Li, R. PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 2017, 68, 5615–5627. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhang., J.; Qu, S.; Zhao, Y.; Su, B.; Lv, X.; Li, R.; Wan, Y.; Xiao, J. The pentratricopeptide repeat protein pigment-defective mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant Cell Physiol. 2017, 58, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Khrouchtchova, A.; Monde, R.A.; Barkan, A. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 2012, 18, 1197–1209. [Google Scholar] [CrossRef]
- Ohtani, S.; Ichinose, M.; Tasaki, E.; Aoki, Y.; Komura, Y.; Sugita, M. Targeted gene disruption identifies three PPR-DYW proteins involved in RNA editing for five editing sites of the moss mitochondrial transcripts. Plant Cell Physiol. 2010, 51, 1942–1949. [Google Scholar] [CrossRef]
- Senkler, J.; Senkler, M.; Eubel, H.; Hildebrandt, T.; Lengwenus, C.; Schertl, P.; Schwarzländer, M.; Wagner, S.; Wittig, I.; Braun, H.-P. The mitochondrial complexome of Arabidopsis thaliana. Plant J. 2017, 89, 1079–1092. [Google Scholar] [CrossRef]
- Sugita, C.; Komura, Y.; Tanaka, K.; Kometani, K.; Satoh, H.; Sugita, M. Molecular characterization of three PRORP proteins in the moss Physcomitrella patens: Nuclear PRORP protein is not essential for moss viability. PLoS ONE 2014, 9, e108962-10. [Google Scholar] [CrossRef]
- Gobert, A.; Gutmann, B.; Taschner, A.; Gößringer, M.; Holzmann, J.; Hartmann, R.K.; Rossmanith, W.; Giegé, P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010, 17, 740–744. [Google Scholar] [CrossRef]
- Pfalz, J.; Liere, K.; Kandlbinder, A.; Dietz, K.J.; Oelmüller, R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 2006, 18, 176–197. [Google Scholar] [CrossRef]
- Takahashi, A.; Sugita, C.; Ichinose, M.; Sugita, M. Moss PPR-SMR protein PpPPR_64 influences the expression of a psaA-psaB-rps14 gene cluster and processing of the 23S–4.5S rRNA precursor in chloroplasts. Plant Mol. Biol. 2021, 107, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Sugita, C.; Yagi, Y.; Nakamura, T.; Sugita, M. Two DYW subclass PPR proteins are involved in RNA editing of ccmFc and atp9 transcripts in the moss Physcomitrella patens: First complete set of PPR editing factors in plant mitochondria. Plant Cell Physiol. 2013, 54, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Schallenberg-Rüdinger, M.; Kindgren, P.; Zehrmann, A.; Small, I.; Knoop, V. A DYW-protein knockout in Physcomitrella affects two closely spaced mitochondrial editing sites and causes a severe developmental phenotype. Plant J. 2013, 76, 420–432. [Google Scholar] [CrossRef]
- Ito, A.; Sugita, C.; Ichinose, M.; Kato, Y.; Yamamoto, H.; Shikanai, T.; Sugita, M. An evolutionarily conserved P-subfamily pentatricopeptide repeat protein is required to splice the plastid ndhA transcript in the moss Physcomitrella patens and Arabidopsis thaliana. Plant J. 2018, 94, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.D.; Cui, Y.L.; Huang, C.; Yin, Q.-Q.; Qin, X.-M.; Xu, T.; He, X.-F.; Zhang, Y.; Li, Z.-R.; Yang, Z.-N. PPR protein PDM1/SEL1 is involved in RNA editing and splicing of plastid genes in Arabidopsis thaliana. Photosynth. Res. 2015, 126, 311–321. [Google Scholar] [CrossRef]
- Kushwaha, H.R.; Singh, A.K.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genom. 2009, 10, 200. [Google Scholar] [CrossRef]
- Tasaki, E.; Hattori, M.; Sugita, M. The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J. 2010, 62, 560–570. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Uchida, M.; Ohtani, S.; Ichinose, M.; Sugita, C.; Sugita, M. The PPR-DYW proteins are required for RNA editing of rps14, cox1 and nad5 transcripts in Physcomitrella patens mitochondria. FEBS Lett. 2011, 585, 2367–2371. [Google Scholar] [CrossRef] [PubMed]
- Rüdinger, M.; Szövényi, P.; Rensing, S.A.; Knoop, V. Assigning DYW-type PPR proteins to RNA editing sites in the funariid mosses Physcomitrella patens and Funaria hygrometrica. Plant J. 2011, 67, 370–380. [Google Scholar] [CrossRef]
- Aryamanesh, N.; Ruwe, H.; Sanglard, L.V.P.; Eshraghi, L.; Bussell, J.D.; Howell, K.A.; Small, I. Colas des Francs-Small, C. The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 2017, 173, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, S.J.; Colas des Francs-Small, C.; Whitby, M.; Small, I.; Kang, H. The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 2019, 100, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Linneweber, C.; Williams-Carrier, R.; Barkan, A. RNA Immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation It activates. Plant Cell 2005, 17, 2791–2804. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; Tadini, L.; Moratti, F.; Lehniger, M.-K.; Costa, A.; Rossi, F.; Colombo, M.; Masiero, S.; Schmitz-Linneweber, C.; Pesaresi, P. CRP1 Protein: (dis)similarities between Arabidopsis thaliana and Zea mays. Front. Plant Sci. 2017, 8, 163. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, W.; Che, L.; Rochaix, J.D.; Lu, C.; Li, W.; Peng, L. PPR protein BFA2 is essential for the accumulation of the atpH/F transcript in chloroplasts. Front. Plant Sci. 2019, 10, 446. [Google Scholar] [CrossRef]
- Zhelyazkova, P.; Hammani, K.; Rojas, M.; Voelker, R.; Vargas-Suárez, M.; Börner, T.; Barkan, A. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012, 40, 3092–3105. [Google Scholar] [CrossRef]
- Yagi, Y.; Ishizaki, Y.; Nakahira, Y.; Tozawa, Y.; Shiina, T. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl. Acad. Sci. USA 2012, 109, 7541–7546. [Google Scholar] [CrossRef]
- Pfalz, J.; Bayramtar, O.A.; Prikryl, J.; Barkan, A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009, 28, 2042–2052. [Google Scholar] [CrossRef]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.S.; Bond, C.S.; Small, I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012, 8, e1002910. [Google Scholar] [CrossRef]
- Yagi, Y.; Hayashi, S.; Kobayashi, K.; Hirayama, T.; Nakamura, T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 2013, 8, e57286. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Brennicke, A.; Graichen, K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS ONE 2013, 8, e65343. [Google Scholar] [CrossRef]
- Yin, P.; Li, Q.; Yan, C.; Liu, Y.; Liu, J.; Yu, F.; Wang, Z.; Long, J.; He, J.; Wang, H.-W.; et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 2013, 504, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Chen, R.-Z.; Ban, T.; Zhou, X.E.; Gu, X.; Tan, M.H.E.; Chen, C.; Kang, Y.; Brunzelle, J.B.; Zhu, J.-K.; et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 2013, 20, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Colas des Francs-Small, C.; Vincis Pereira Sanglard, L.; Small, I. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun. Biol. 2018, 1, 166. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Yu, Q.; Williams-Carrier, R.; Maliga, P.; Barkan, A. Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat. Plants 2019, 5, 505–511. [Google Scholar] [CrossRef]
- Yu, Q.; Barkan, A.; Maliga, P. Engineered RNA-binding protein for transgene activation in non-green plastids. Nat. Plants 2019, 5, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Künstner, P.; Guardiola, A.; Takahashi, Y.; Rochaix, J.D. A mutant strain of Chlamydomonas reinhardtii lacking the chloroplast photosystem II psbI gene grows photoautotrophically. J. Biol. Chem. 1995, 270, 9651–9654. [Google Scholar] [CrossRef]
- Schwenkert, S.; Umate, P.; Dal Bosco, C.; Volz, S.; Mlçochová, L.; Zoryan, M.; Eichacker, L.A.; Ohad, I.; Herrmann, R.G.; Meuer, J. PsbI affects the stability, function, and phosphorylation patterns of photosystem II assemblies in tobacco. J. Biol. Chem. 2006, 281, 34227–34238. [Google Scholar] [CrossRef] [Green Version]
- del Campo, E.M.; Sabater, B.; Martin, M. Post-transcriptional control of chloroplast gene expression: Accumulation of stable psaC mRNA is due to downstream RNA cleavages in the ndhD gene. J. Biol. Chem. 2002, 277, 36457–36464. [Google Scholar] [CrossRef]
- Li, X.; Luo, W.; Zhou, W.; Yin, X.; Wang, X.; Li, X.; Jiang, C.; Zhang, Q.; Kang, X.; Zhang, A.; et al. CAF proteins help SOT1 regulate the stability of chloroplast ndhA transcripts. Int. J. Mol. Sci. 2021, 22, 12639. [Google Scholar] [CrossRef]
- Hirose, T.; Sugiura, M. Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: A possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997, 16, 6804–6811. [Google Scholar] [CrossRef] [PubMed]
- Burrows, P.A.; Sazanov, L.A.; Svab, Z.; Maliga, P.; Nixon, P.J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998, 17, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Goldschmidt-Clermont, M.; Soen, S.Y.; Franzén, L.G.; Rochaix, J.D. Directed chloroplast transformation in Chlamydomonas reinhardtii: Insertional inactivation of the psaC gene encoding the iron sulfur protein destabilizes photosystem I. EMBO J. 1991, 10, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Schöttler, M.A.; Albus, C.A.; Bock, R. Photosystem I: Its biogenesis and function in higher plants. J. Plant Physiol. 2011, 168, 1452–1461. [Google Scholar] [CrossRef]
- Watkins, K.P.; Kroeger, T.S.; Cooke, A.M.; Williams-Carrier, R.E.; Friso, G.; Belcher, S.E.; van Wijk, K.J.; Barkan, A. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 2007, 19, 2606–2623. [Google Scholar] [CrossRef]
- Kroeger, T.S.; Watkins, K.P.; Friso, G.; van Wijk, K.J.; Barkan, A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 2006, 106, 4537–4542. [Google Scholar] [CrossRef]
- Asakura, Y.; Galarneau, E.; Watkins, K.P.; Barkan, A.; van Wijk, K.J. Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 2012, 159, 961–974. [Google Scholar] [CrossRef]
- Hammani, K.; Barkan, A. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 2014, 42, 5033–5042. [Google Scholar] [CrossRef] [Green Version]
- Zoschke, R.; Nakamura, M.; Liere, K.; Sugiura, M.; Börner, T.; Schmitz-Linneweber, C. An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA 2010, 107, 3245–3250. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Lampe, M.-K.; Sultan, L.D.; Ostersetzer-Biran, O. Organellar maturases: A window into the evolution of the spliceosome. Biochim. Biophys. Acta 2015, 1847, 798–808. [Google Scholar] [CrossRef]
- Jenkins, B.; Kulhanek, D.; Barkan, A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 1997, 9, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Ostheimer, G.J.; Williams-Carrier, R.; Belcher, S.; Osborne, E.; Gierke, J.; Barkan, A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003, 22, 3919–3929. [Google Scholar] [CrossRef] [PubMed]
- Asakura, Y.; Barkan, A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell 2007, 19, 3864–3875. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; Pace, N.R. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 1998, 67, 153–180. [Google Scholar] [CrossRef]
- Gutmann, B.; Gobert, A.; Giegé, P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012, 26, 1022–1027. [Google Scholar] [CrossRef]
- Anantharaman, V.; Aravind, L. The NYN domains. Novel predicted RNAses with a PIN domain-like fold. RNA Biol. 2006, 3, 18–27. [Google Scholar] [CrossRef]
- Moreira, D.; Philippe, H. Smr: A bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem. Sci. 1999, 24, 298–300. [Google Scholar] [CrossRef]
- Liu, S.; Melonek, J.; Boykin, L.M.; Small, I.; Howell, K.A. PPR-SMRs. Ancient proteins with enigmatic functions. RNA Biol. 2013, 10, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Yu, F.; Rodermel, S. An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 2010, 154, 1588–1601. [Google Scholar] [CrossRef]
- Wu, W.; Liu, S.; Ruwe, H.; Zhang, D.; Melonek, J.; Zhu, Y.; Hu, X.; Gusewski, S.; Yin, P.; Small, I.; et al. SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S–4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 2016, 85, 607–621. [Google Scholar] [CrossRef]
- Zoschke, R.; Watkins, K.P.; Miranda, R.G.; Barkan, A. The PPR-SMR protein PPR 53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 2016, 85, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lu, Q.; Li, Q.; Wang, L.; Ding, S.; Zhang, A.; Wen, X.; Zhang, L.; Lu, C. PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl. Acad. Sci. USA 2017, 114, E1554–E1563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, C. The enigmatic roles of PPR-SMR proteins in plants. Adv. Sci. 2019, 6, 1900361. [Google Scholar] [CrossRef] [PubMed]
- Sugita, M.; Ichinose, M.; Ide, M.; Sugita, C. Architecture of the PPR gene family in the moss Physcomitrella patens. RNA Biol. 2013, 10, 1439–1445. [Google Scholar] [CrossRef]
- Wang, D.; Liu, H.; Zhai, G.; Wang, L.; Shao, J.; Tao, Y. OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. J. Genet. Genom. 2016, 43, 601–608. [Google Scholar] [CrossRef]
- Pyo, Y.J.; Kwon, K.C.; Kim, A.; Cho, M.H. Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 2013, 163, 1844–1858. [Google Scholar] [CrossRef]
- Chen, L.; Huang, L.; Dai, L.; Gao, Y.; Zou, W.; Lu, X.; Wang, C.; Zhang, G.; Ren, D.; Hu, J.; et al. PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in rice. Plant Cell Physiol. 2019, 60, 587–598. [Google Scholar] [CrossRef]
- Knoop, V.; Rüdinger, M. DYW-type PPR proteins in a heterolobosean protist: Plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 2010, 584, 4287–4291. [Google Scholar] [CrossRef] [Green Version]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Härtel, B.; Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. RNA editing and its molecular mechanism in plant organelles. Genes 2017, 8, 5. [Google Scholar] [CrossRef]
- Okuda, K.; Nakamura, T.; Sugita, M.; Shimizu, T.; Shikanai, T. A pentatricopeptide repeat protein is a site-recognition factor in the plastid RNA editing. J. Biol. Chem. 2006, 281, 37661–37667. [Google Scholar] [CrossRef] [PubMed]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Osteretzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef] [PubMed]
- Plant RNA Editing-Prediction & Analysis Computer Tool (PREPACT). Available online: http://www.prepact.de/prepact-main.php (accessed on 1 July 2022).
- Miyata, Y.; Sugiura, C.; Kobayashi, Y.; Hagiwara, M.; Sugita, M. Chloroplast ribosomal S14 protein transcript is edited to create a translation initiation codon in the moss Physcomitrella patens. Biochim. Biophys. Acta 2002, 1576, 346–349. [Google Scholar] [CrossRef]
- Rüdinger, M.; Funk, H.T.; Rensing, S.A.; Maier, U.G.; Knoop, V. RNA editing: Only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol. Genet. Genom. 2009, 281, 473–481. [Google Scholar] [CrossRef]
- Hayes, M.L.; Giang, K.; Mulligan, R.M. Molecular evolution of pentatricopeptide repeat genes reveals truncation in species lacking an editing target and structural domains under distinct selective pressures. BMC Evol. Biol. 2012, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Miyata, Y.; Sugita, M. Tissue- and stage-specific RNA editing of rps14 transcripts in moss (Physcomitrella patens) chloroplasts. J. Plant Physiol. 2004, 161, 113–115. [Google Scholar] [CrossRef]
- Salone, V.; Rüdinger, M.; Polsakiewicz, M.; Hoffmann, B.; Groth-Malonek, M.; Szurek, B.; Small, I.; Knoop, V.; Lurin, C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007, 581, 4132–4138. [Google Scholar] [CrossRef]
- Rüdinger, M.; Polsakiewicz, M.; Knoop, V. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol. Biol. Evol. 2008, 25, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Oldenkott, B.; Yang, Y.; Lesch, E.; Knoop, V.; Schallenberg-Rüdinger, M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019, 2, 85. [Google Scholar] [CrossRef]
- Hayes, M.L.; Santibanez, P.I. A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J. Biol. Chem. 2020, 295, 3497–3505. [Google Scholar] [CrossRef]
- Takenaka, M.; Takenaka, S.; Barthel, T.; Frink, B.; Haag, S.; Verbitskiy, D.; Oldenkott, B.; Schallenberg-Rüdinger, M.; Feiler, C.G.; Weiss, M.S.; et al. DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis. Nat. Catal. 2021, 4, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Schuster, G.; Gruissem, W. Chloroplast mRNA 3’ end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991, 10, 493–1502. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Ohta, M.; Sugiura, M.; Sugita, M. Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome free mRNAs in the stroma. J. Biol. Chem. 2001, 276, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Kupsch, C.; Ruwe, H.; Gusewski, S.; Tillich, M.; Small, I.; Schmitz-Linneweber, C. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 2012, 24, 4266–4280. [Google Scholar] [CrossRef] [PubMed]
- Teubner, M.; Fuß, J.; Kühn, K.; Krause, K.; Schmitz-Linneweber, C. The RNA recognition motif protein CP33A is a global ligand of chloroplast mRNAs and is essential for plastid biogenesis and plant development. Plant J. 2017, 89, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Sugiura, M. Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: Development of a chloroplast in vitro RNA editing system. EMBO J. 2001, 20, 1144–1152. [Google Scholar] [CrossRef]
- Tillich, M.; Hardel, S.L.; Kupsch, C.; Armbruster, U.; Delannoy, E.; Gualberto, M.; Lehwark, P.; Leister, D.; Small, I.D.; Schmitz-Linneweber, C. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 6002–6007. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.J.; Lang, D.; Hoernstein, S.N.W.; Lang, E.G.E.; Schuessele, C.; Schmidt, A.; Fluck, M.; Leisibach, D.; Niegl, C.; Zimmer, A.D.; et al. Quantitative analysis of the mitochondrial and plastid proteomes of the moss Physcomitrella patens reveals protein macrocompartmentation and microcompartmentation. Plant Physiol. 2014, 164, 2081–2095. [Google Scholar] [CrossRef] [Green Version]
- Uchiyama, H.; Ichinose, M.; Sugita, M. Chloroplast ribonucleoprotein-like proteins of the moss Physcomitrella patens are not involved in RNA stability and RNA editing. Photosynthetica 2018, 56, 62–66. [Google Scholar] [CrossRef]
- Sun, T.; Germain, A.; Giloteaux, L.; Hammani, K.; Barkan, A.; Hanson, M.R.; Bentolila, S. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. USA 2013, 110, E1169–E1178. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Kugelmann, M.; Härtel, B.; Brennicke, A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Bentolila, S.; Heller, W.P.; Sun, T.; Babina, A.M.; Friso, G.; van Wijk, K.J.; Hanson, M. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. USA 2012, 109, E1453–E1461. [Google Scholar] [CrossRef] [PubMed]
- Zehrmann, A.; Härtel, B.; Glass, F.; Bayer-Császár, E.; Obata, T.; Meyer, E.; Brennicke, A.; Takenaka, M. Selective homo- and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 6445–6456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Tang, W.; Hedtke, B.; Zhong, L.; Liu, L.; Peng, L.; Lu, C.; Grimm, B.; Lin, R. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. USA 2014, 111, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Shi, X.; Friso, G.; van Wijk, K.; Bentolila, S.; Hanson, M.R. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015, 11, e1005028. [Google Scholar] [CrossRef]
- Guillaumot, D.; Lopez-Obando, M.; Baudry, K.; Avon, A.; Rigaill, G.; Falcon De Longevialle, A.; Broche, B.; Takenaka, M.; Berthomeé, R.; De Jaeger, G.; et al. Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc. Natl. Acad. Sci. USA 2017, 114, 8877–8882. [Google Scholar] [CrossRef]
- Andreés-Colaás, N.; Zhu, Q.; Takenaka, M.; De Rybel, B.; Weijers, D.; Van Der Straeten, D. Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids. Proc. Natl. Acad. Sci. USA 2017, 114, 8883–8888. [Google Scholar] [CrossRef]
- García-Andrade, J.; Ramiírez, V.; Loópez, A.; Vera, P. Mediated plastid RNA editing in plant immunity. PLoS Pathog. 2013, 9, e1003713. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Yu, Q.-B.; Li, Z.-R.; Ye, L.-S.; Xu, L.; Yang, Z.-N. Porphobilinogen deaminase HEMC interacts with the PPR-protein AtECB2 for chloroplast RNA editing. Plant J. 2017, 92, 546–556. [Google Scholar] [CrossRef]
- Gutmann, B.; Royan, S.; Small, I. Protein complexes implicated in RNA editing in plant organelles. Mol. Plant 2017, 10, 1255–1257. [Google Scholar] [CrossRef]
- Okuda, K.; Chateigner-Boutin, A.L.; Nakamura, T.; Delannoy, E.; Sugita, M.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Small, I.; Shikanai, T. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 2009, 21, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Sugita, M. A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett. 2008, 582, 4163–4168. [Google Scholar] [CrossRef] [PubMed]
- Waltz, F.; Nguyen, T.T.; Arrivé, M.; Bochler, A.; Chicher, J.; Hammann, P.; Kuhn, L.; Quadrado, M.; Mireau, H.; Hashem, Y.; et al. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 2019, 5, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Honkanen, S.; Small, I. The GENOME UNCOUPLED1 protein has an ancient, highly conserved role but not in retrograde signaling. New Phytol. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Duckett, J.G.; Renzaglia, K.S. Ultrastructure and development of plastids in bryophytes. Adv. Bryol. 1988, 3, 33–93. [Google Scholar]
- Sugita, M.; Aoki, S. Chloroplasts. In The Moss Physcomitrella patens. Annual Plant Reviews; Knight, C.D., Perroud, P.-F., Cove, D.J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2009; Chapter 8; Volume 36, pp. 182–210. ISBN 978-1-4051-8189-1. [Google Scholar]
| Protein Name | Type | Additional Domain | Amino Acid Length (aa) | Amino Acid Identity (%) | Arabidopsis Homolog |
|---|---|---|---|---|---|
| PpPPR_3 | P | RRM | 939 | 62 | At5g04810 (AtPPR4) |
| PpPPR_76 | 947 | ||||
| PpPPR_5 | P | 611 | 79.2 | ||
| PpPPR_88 | 611 | ||||
| PpPPR_6 | P | 451 | 62.5 | ||
| PpPPR_102 | 395 | ||||
| PpPPR_7 | P | LAGLIDADG | 1195 | 71.5 | At2g15820 (OTP51) |
| PpPPR_22 | 883 | ||||
| PpPPR_13 | P | 657 | 75.4 | ||
| PpPPR_101 | 670 | ||||
| PpPPR_16 | P | 637 | 70.8 | ||
| PpPPR_89 | 630 | ||||
| PpPPR_17 | P | 489 | 58.5 | At4g39620 (AtPPR5) | |
| PpPPR_80 | 482 | ||||
| PpPPR_19 | P | 958 | 65.8 | At4g34830 (MRL1) | |
| PpPPR_51 | 982 | ||||
| PpPPR_26 | P | 1110 | 71.2 | ||
| PpPPR_40 | 961 | ||||
| PpPPR_27 | P | 487 | 68.2 | At3g53170 | |
| PpPPR_35 | 532 | ||||
| PpPPR_39 | P | 582 | 58.3 | At3g42630 (PBF2) | |
| PpPPR_73 | 603 | ||||
| PpPPR_42 | P | SMR | 936 | 56.1 | At5g02830 |
| PpPPR_59 | 935 | ||||
| PpPPR_58 | P | 530 | 74.2 | At4g35850 | |
| PpPPR_61 | 522 | ||||
| PpPPR_66 | P | 578 | 77.3 | At2g35130 (AtPPR66L) | |
| PpPPR_72 | 578 | ||||
| PpPPR_63 | P | NYN | 655 | 60.7–79.1 | At2g32230 (PRORP1), At2g16650 (PRORP2), At4g21900 (PRORP3) |
| PpPPR_67 | 791 | ||||
| PpPPR_104 | 993 | ||||
| PpPPR_75 | P | SMR | 871 | 91.5 | At2g31400 (GUN1) |
| PpPPR_85 | 871 | ||||
| PpPPR_82 | P | 717 | 76.8 | ||
| PpPPR_84 | 717 | ||||
| PpPPR_92 | P | 1010 | 73.7 | At4g30825 (BFA2) | |
| PpPPR_94 | 1000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugita, M. An Overview of Pentatricopeptide Repeat (PPR) Proteins in the Moss Physcomitrium patens and Their Role in Organellar Gene Expression. Plants 2022, 11, 2279. https://doi.org/10.3390/plants11172279
Sugita M. An Overview of Pentatricopeptide Repeat (PPR) Proteins in the Moss Physcomitrium patens and Their Role in Organellar Gene Expression. Plants. 2022; 11(17):2279. https://doi.org/10.3390/plants11172279
Chicago/Turabian StyleSugita, Mamoru. 2022. "An Overview of Pentatricopeptide Repeat (PPR) Proteins in the Moss Physcomitrium patens and Their Role in Organellar Gene Expression" Plants 11, no. 17: 2279. https://doi.org/10.3390/plants11172279
APA StyleSugita, M. (2022). An Overview of Pentatricopeptide Repeat (PPR) Proteins in the Moss Physcomitrium patens and Their Role in Organellar Gene Expression. Plants, 11(17), 2279. https://doi.org/10.3390/plants11172279






