Combined Computational and Experimental Studies of Anabasine Encapsulation by Beta-Cyclodextrin
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Silico Studies
2.1.1. Molecular Docking of Anabasine against α-, β-, γ-cyclodextrins
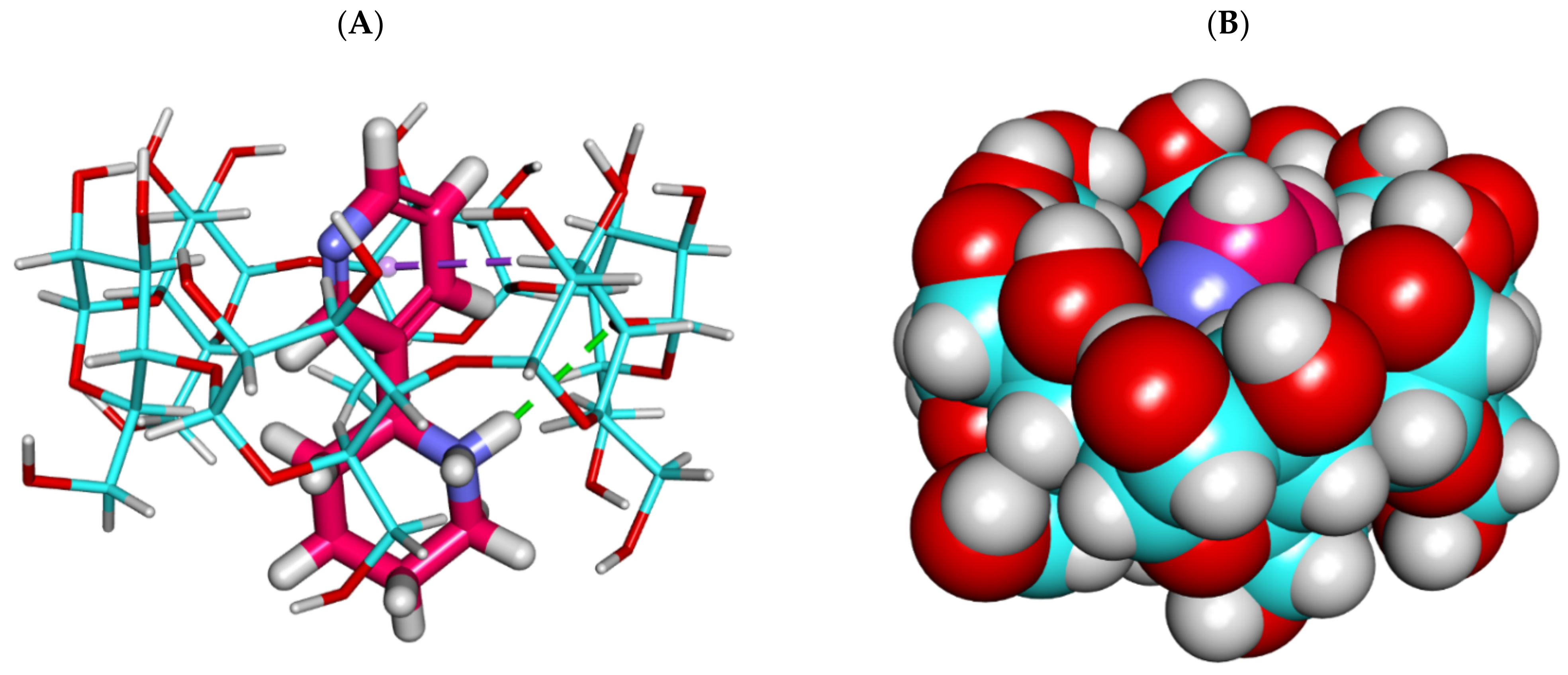
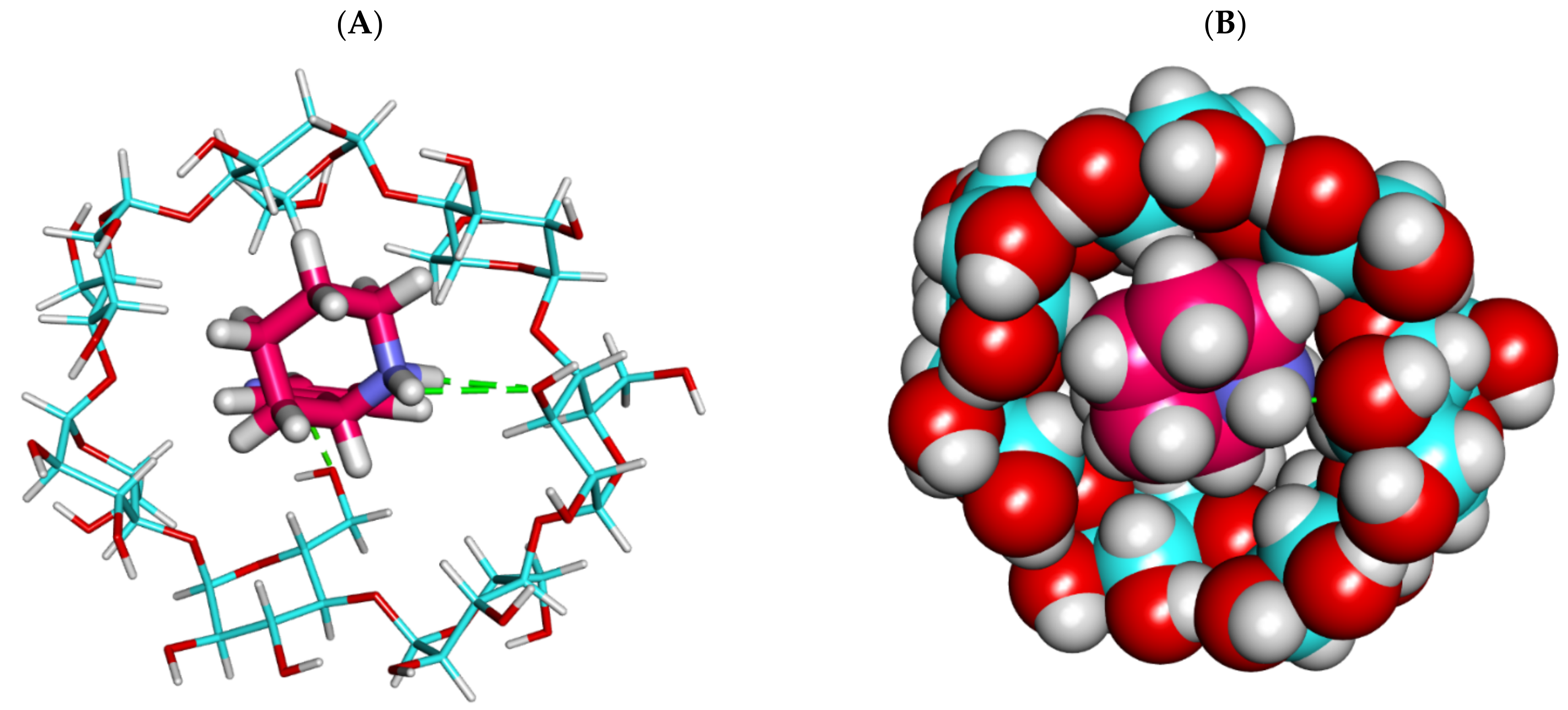
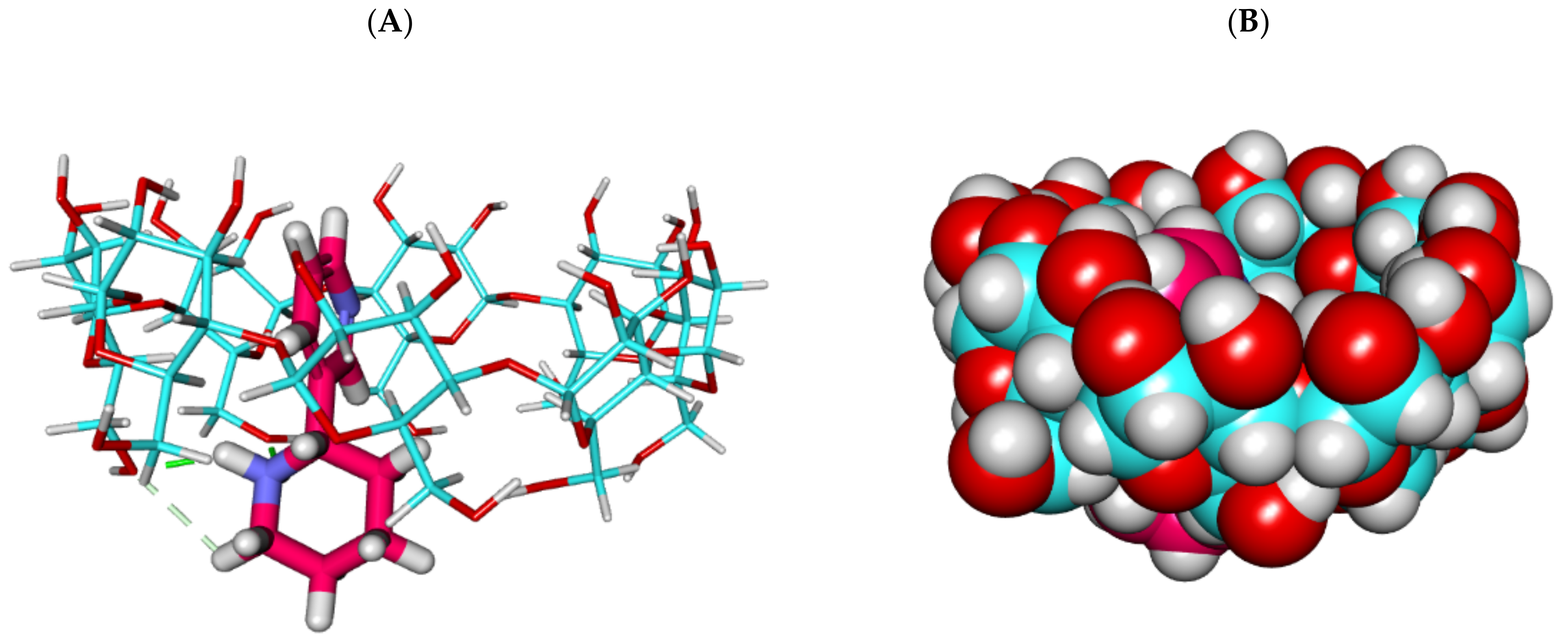
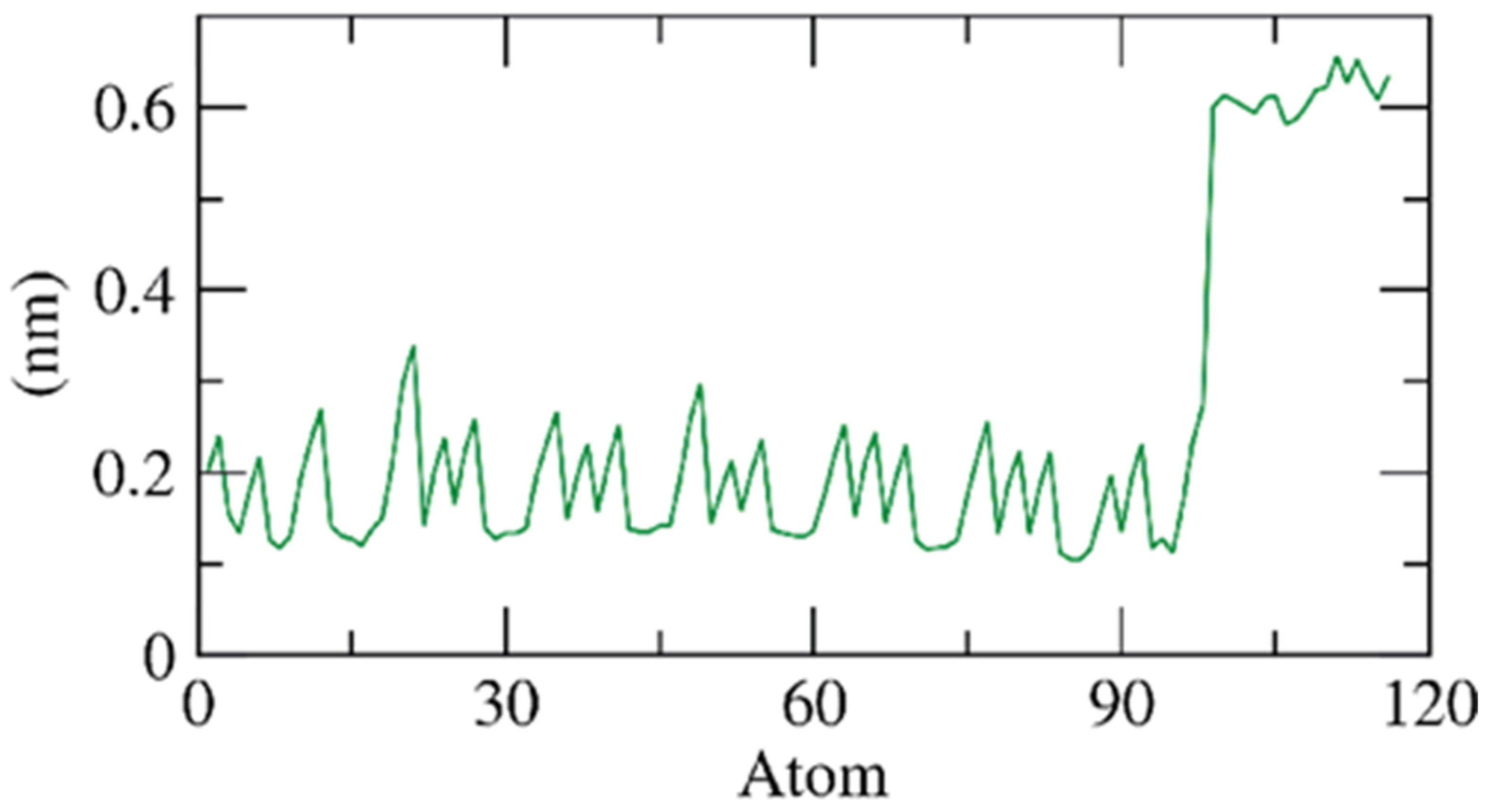
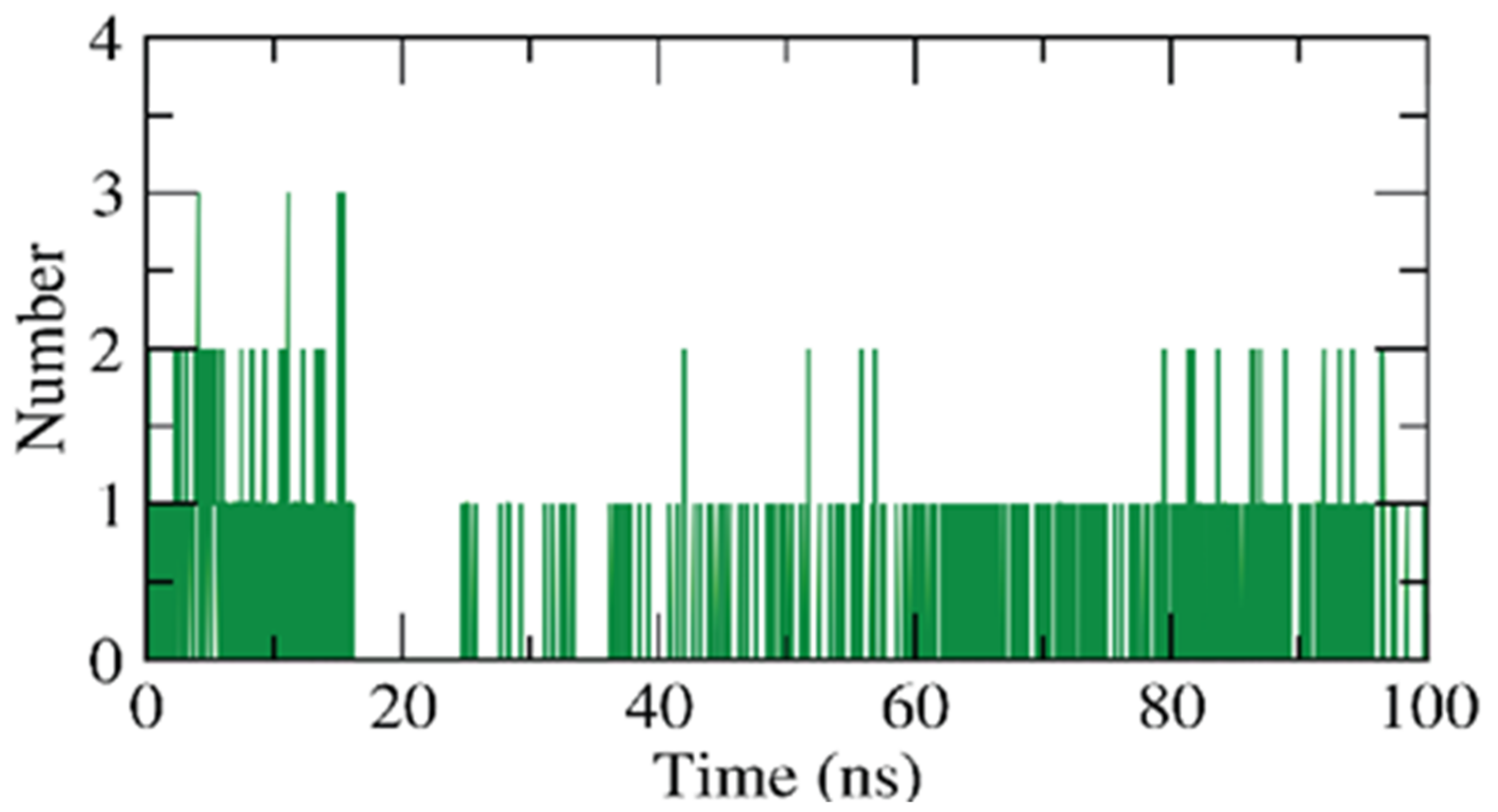
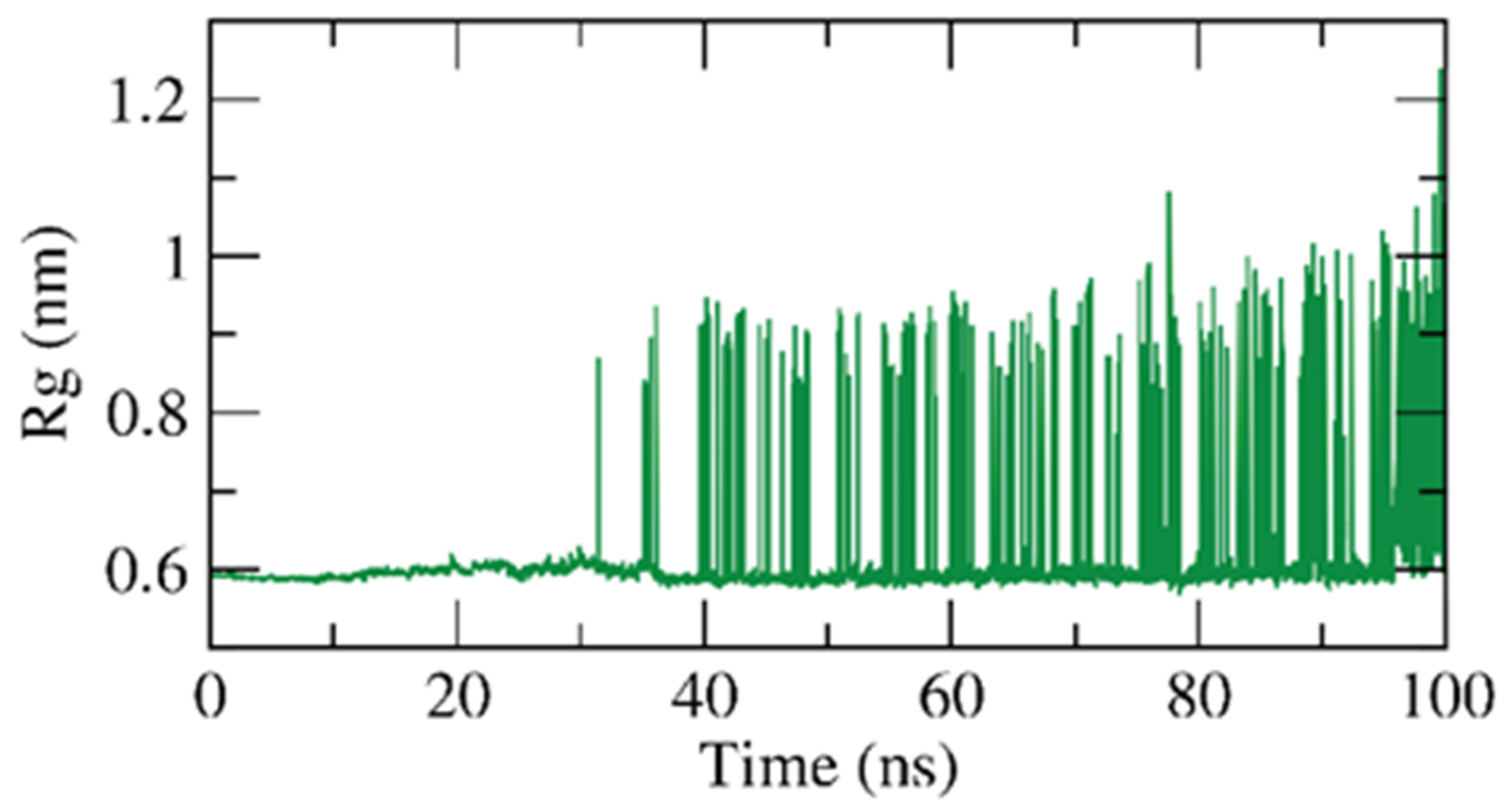
2.1.2. Molecular Dynamics (MD) Simulations
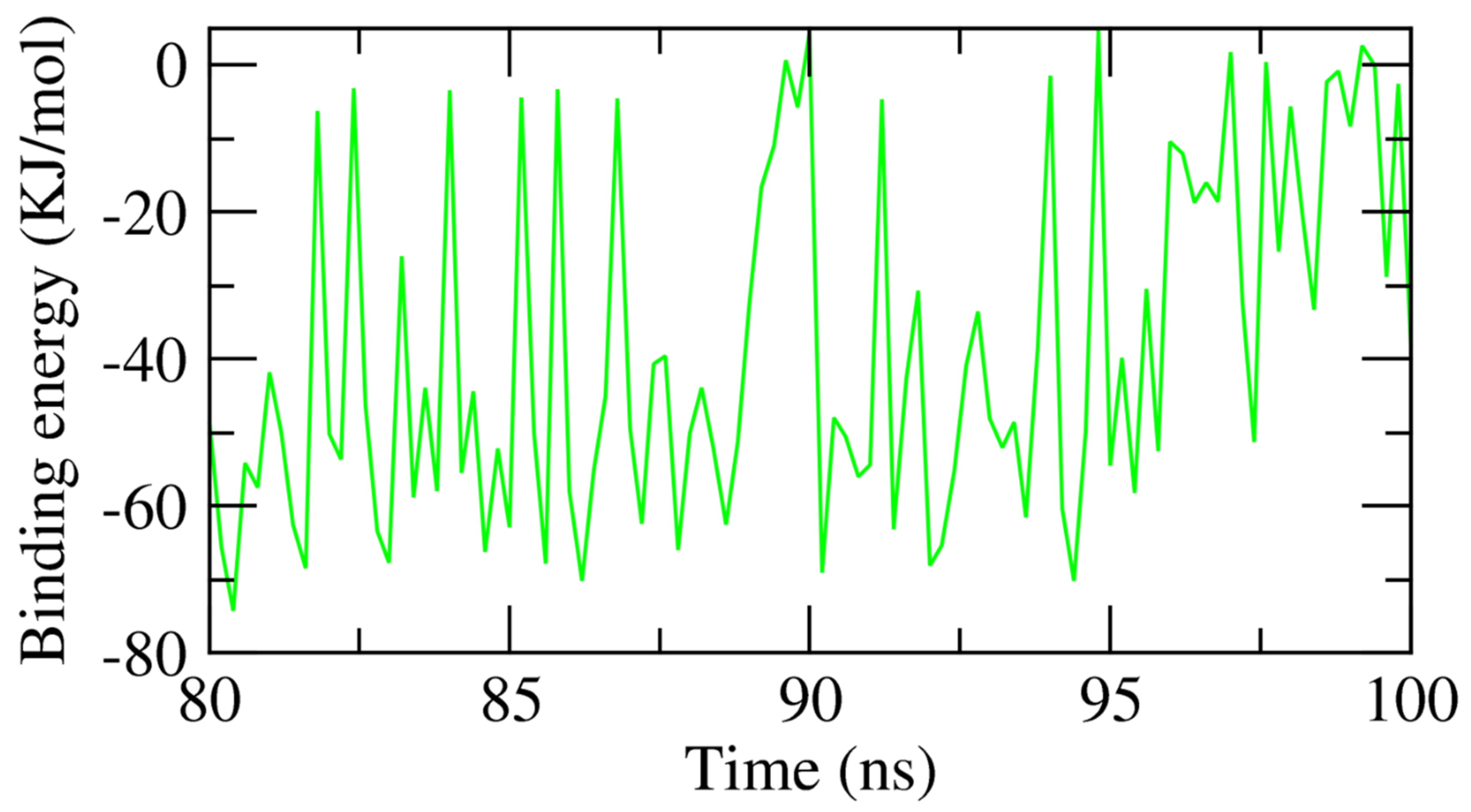
2.1.3. MMPBSA of the β CD-Anabasine Complex
2.2. Experimental Studies
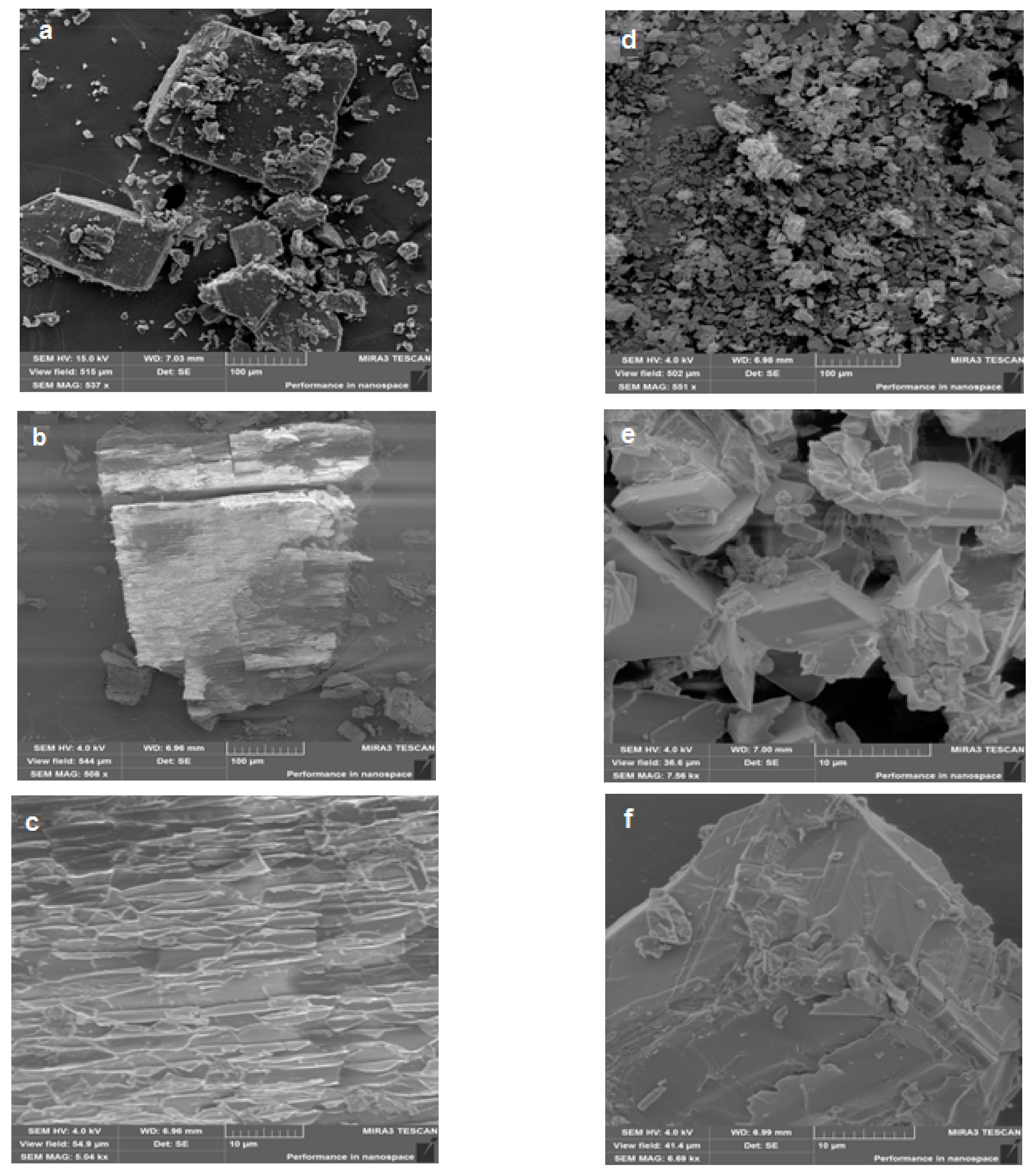
2.2.1. Surface Morphology Study of the Anabasine-β-CD Complexes Samples
2.2.2. Study of the Structure of the Obtained Anabasine-β-CD Supramolecular Inclusion Complexes
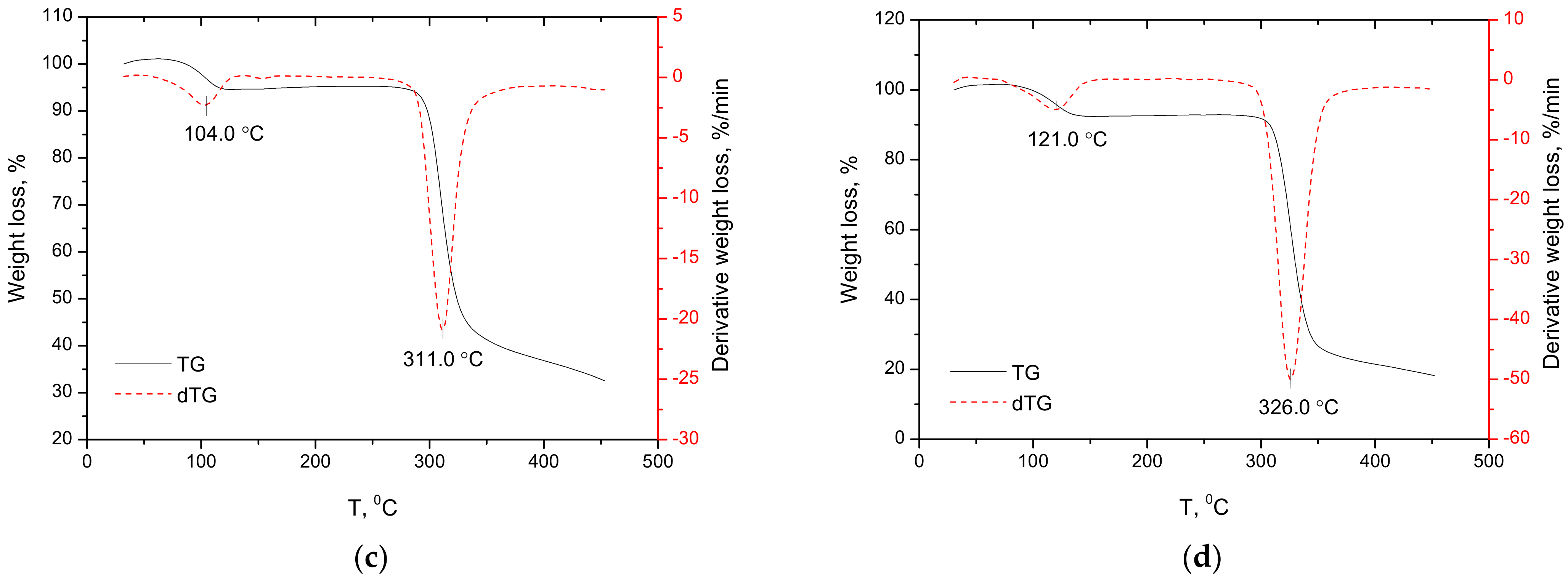
2.2.3. Thermal Analysis of Anabasine-β CD Inclusion Complexes Samples by Differential Thermogravimetry and Differential Scanning Calorimetry
3. Experimental
3.1. Geometric Parameters of α-, β-, γ-cyclodextrins and Anabasine
3.2. Molecular Docking
3.3. MD Simulations
3.4. Complex Preparation
3.5. Spectroscopic Measurements
3.6. Surface Morphology
3.7. Thermal Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Metwaly, A.M.; Ghoneim, M.M.; Eissa, I.H.; Elsehemy, I.A.; Mostafa, A.E.; Hegazy, M.M.; Afifi, W.M.; Dou, D. Traditional ancient Egyptian medicine: A review. Saudi J. Biol. Sci. 2021, 28, 5823–5832. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, Y.; Metwaly, A.M.; Xue, Y.; Shi, Y.; Dou, D. The Chinese herbal formulae (Yitangkang) exerts an antidiabetic effect through the regulation of substance metabolism and energy metabolism in type 2 diabetic rats. J. Ethnopharmacol. 2019, 239, 111942. [Google Scholar] [CrossRef] [PubMed]
- Conroy, M.S. Russian-American pharmaceutical relations, 1900–1945. Pharm. Hist. 2004, 46, 143–166. [Google Scholar] [PubMed]
- Sadykov, A.; Tumur, B. The alkaloids of Anabasis aphylla var. iljinii. Dokl. Akad. Nauk Uzb. SSR 1960, 1, 27–29. [Google Scholar]
- Mastropaolo, J.; Rosse, R.B.; Deutsch, S.I. Anabasine, a selective nicotinic acetylcholine receptor agonist, antagonizes MK-801-elicited mouse popping behavior, an animal model of schizophrenia. Behav. Brain Res. 2004, 153, 419–422. [Google Scholar] [CrossRef]
- Gong, N.-B.; Du, L.-D.; Lu, Y. Anabasine. In Natural Small Molecule Drugs from Plants; Springer: Berlin/Heidelberg, Germany, 2018; pp. 353–356. [Google Scholar]
- Trofimov, B.A.; Andriyankova, L.V.; Tlegenov, R.T.; Mal’kina, A.G.; Afonin, A.V.; Il’icheva, L.N.; Nikitina, L.P. Reaction of anabasine with 3-(1-hydroxycyclohexyl)-2-propynenitrile: A new route to functionalised anabasine alkaloids. Mendeleev Commun. 2005, 15, 33–35. [Google Scholar] [CrossRef]
- Babaev, B.N.; Dalimov, D.N.; Tiliabaev, Z.; Tylegenov, R.T. Synthesis, structure and biological properties of phosphorylated anabazine derivatives. Chem. Plant Raw Mater. 2010, 2, 57–62. [Google Scholar]
- Gessner, G.; Larrañaga, M.D.; Lewis Sr, R.J.; Lewis, R.A. Hawley’s Condensed Chemical Dictionary; John Wiley & Sons: Hoboken, NJ, USA, 1997. [Google Scholar]
- Haag, H. A contribution to the pharmacology of anabasine. J. Pharmacol. Exp. Ther. 1933, 48, 95–104. [Google Scholar]
- Kurkov, S.V.; Loftsson, T. Cyclodextrins. Int. J. Pharm. 2013, 453, 167–180. [Google Scholar] [CrossRef]
- Astray, G.; Gonzalez-Barreiro, C.; Mejuto, J.C.; Rial-Otero, R.; Simal-Gandara, J. A review on the use of cyclodextrins in foods. Food Hydrocoll. 2009, 23, 1631–1640. [Google Scholar] [CrossRef]
- Larsen, K.L. Large cyclodextrins. J. Incl. Phenom. Macrocycl. Chem. 2002, 43, 1–13. [Google Scholar] [CrossRef]
- Loftsson, T.; Másson, M.; Brewster, M.E. Self-association of cyclodextrins and cyclodextrin complexes. J. Pharm. Sci. 2004, 93, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Crini, G. A history of cyclodextrins. Chem. Rev. 2014, 114, 10940–10975. [Google Scholar] [CrossRef]
- Del Valle, E.M. Cyclodextrins and their uses: A review. Process Biochem. 2004, 39, 1033–1046. [Google Scholar] [CrossRef]
- Marques, H.M.C. A review on cyclodextrin encapsulation of essential oils and volatiles. Flavour Fragr. J. 2010, 25, 313–326. [Google Scholar] [CrossRef]
- Iskineyeva, A.; Fazylov, S.; Bakirova, R.; Sarsenbekova, A.; Pustolaikina, I.; Seilkhanov, O.; Alsfouk, A.A.; Elkaeed, E.B.; Eissa, I.H.; Metwaly, A.M. Combined In Silico and Experimental Investigations of Resveratrol Encapsulation by Beta-Cyclodextrin. Plants 2022, 11, 1678. [Google Scholar] [CrossRef]
- Palli, V.; Leonis, G.; Zoupanou, N.; Georgiou, N.; Chountoulesi, M.; Naziris, N.; Tzeli, D.; Demetzos, C.; Valsami, G.; Marousis, K.D.J.M. Losartan Interactions with 2-Hydroxypropyl-β-CD. Molecules 2022, 27, 2421. [Google Scholar] [CrossRef]
- Hansson, T.; Oostenbrink, C.; van Gunsteren, W. Molecular dynamics simulations. Curr. Opin. Struct. Biol. 2002, 12, 190–196. [Google Scholar] [CrossRef]
- Serra, R.; Sempere, J.; Nomen, R. A new method for the kinetic study of thermoanalytical data:: The non-parametric kinetics method. Thermochim. Acta 1998, 316, 37–45. [Google Scholar] [CrossRef]
- Burkeev, M.; Fazylov, S.; Bakirova, R.; Iskineyeva, A.; Sarsenbekova, A.; Tazhbaev, E.; Davrenbekov, S. Thermal decomposition of β-cyclodextrin and its inclusion complex with vitamin E. Mendeleev Commun. 2021, 31, 76–78. [Google Scholar] [CrossRef]
- Coats, A.W.; Redfern, J. Kinetic parameters from thermogravimetric data. Nature 1964, 201, 68–69. [Google Scholar] [CrossRef]
- Freeman, E.S.; Carroll, B. The application of thermoanalytical techniques to reaction kinetics: The thermogravimetric evaluation of the kinetics of the decomposition of calcium oxalate monohydrate. J. Phys. Chem. 1958, 62, 394–397. [Google Scholar] [CrossRef]
- Sharp, J.H.; Wentworth, S.A. Kinetic analysis of thermogravimetric data. Anal. Chem. 1969, 41, 2060–2062. [Google Scholar] [CrossRef]
- Achar, B.N.; Brindley, G.; Sharp, J. Kinetics and mechanism of dehydroxylation processes. III. Applications and limitations of dynamic methods. In Proceedings of the International Clay Conference 1966, Jerusalem, Israel, 20–24 June 1966; pp. 67–73. [Google Scholar]
- Alanazi, M.M.; Elkady, H.; Alsaif, N.A.; Obaidullah, A.J.; Alkahtani, H.M.; Alanazi, M.M.; Alharbi, M.A.; Eissa, I.H.; Dahab, M.A. New quinoxaline-based VEGFR-2 inhibitors: Design, synthesis, and antiproliferative evaluation with in silico docking, ADMET, toxicity, and DFT studies. RSC Adv. 2021, 11, 30315–30328. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks III, C.L.; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]






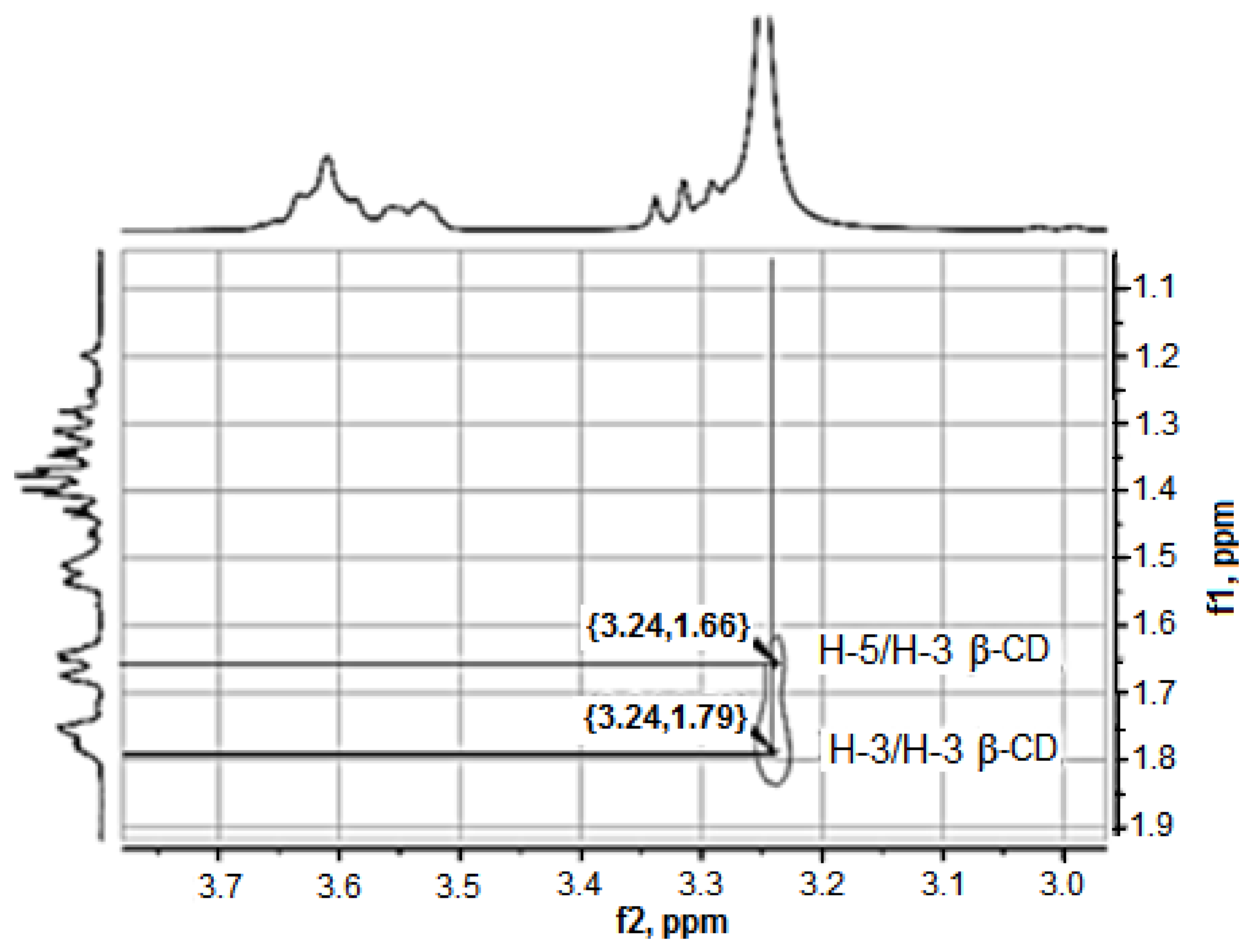
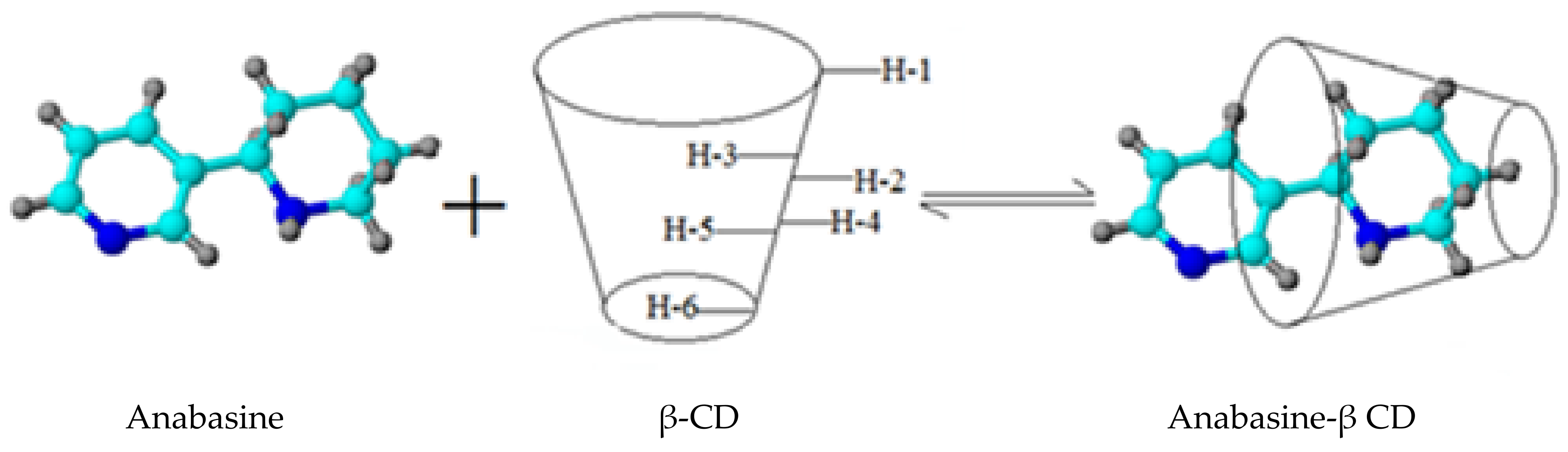

| Carbon Atom Number | Group | The Value of δ0 in the Free State, ppm | The Value of δ in the Composition of the Complex, ppm | The Value of δ in the Composition of the Complex, ∆δ = δ − δ0, ppm | |||
|---|---|---|---|---|---|---|---|
| CHx | δ(1H) | δ(13C) | δ(1H) | δ(13C) | ∆δ(1H) | ∆δ(13C) | |
| Anabasine | |||||||
| 2 | -CHax | 2.610 | 47.203 | 2.610 | 47.163 | 0 | −0.04 |
| -CHeq | 2.991 | 2.991 | 0 | ||||
| 3 | -CH2- | 1.882 | 25.973 | 1.750 | 25.952 | −0.132 | −0.021 |
| 4 | -CH2- | 1.452 | 25.510 | 1.442 | 25.480 | −0.010 | −0.030 |
| 5 | -CH2- | 1.471 | 35.245 | 1.374 | 35.212 | −0.097 | −0.033 |
| 6 | >CH- | 3.542 | 59.272 | 3.556 | 59.335 | 0.014 | −0.06 |
| 7 | >C= | – | 141.602 | – | 141.571 | – | −0.031 |
| 8 | -CH=N | 8.500 | 148.263 | 8.490 | 148.285 | −0.010 | 0.022 |
| 10 | -CH=N | 8.380 | 148.620 | 8.380 | 148.62 | 0 | 0 |
| 11 | -CH= | 7.263 | 123.660 | 7.274 | 123.702 | 0.011 | 0.042 |
| 12 | -CH= | 7.680 | 134.522 | 7.680 | 134.665 | 0 | 0.143 |
| β-CD | |||||||
| 1 | >CH- | 4.773 | 102.432 | 4.796 | 102.667 | 0.023 | 0.235 |
| 2 | >CH- | 3.270 | 72.875 | 3.303 | 72.965 | 0.033 | 0.091 |
| 3 | >CH- | 3.49 | 73.545 | 3.615 | 73.685 | 0.123 | 0.140 |
| 4 | >CH- | 3.302 | 82.005 | 3.345 | 82.168 | 0.042 | 0.163 |
| 5 | >CH- | 3.450 | 72.525 | 3.560 | 72.658 | 0.110 | 0.133 |
| 6 | -CH2- | 3.572 | 60.402 | 3.635 | 60.576 | 0.063 | 0.174 |
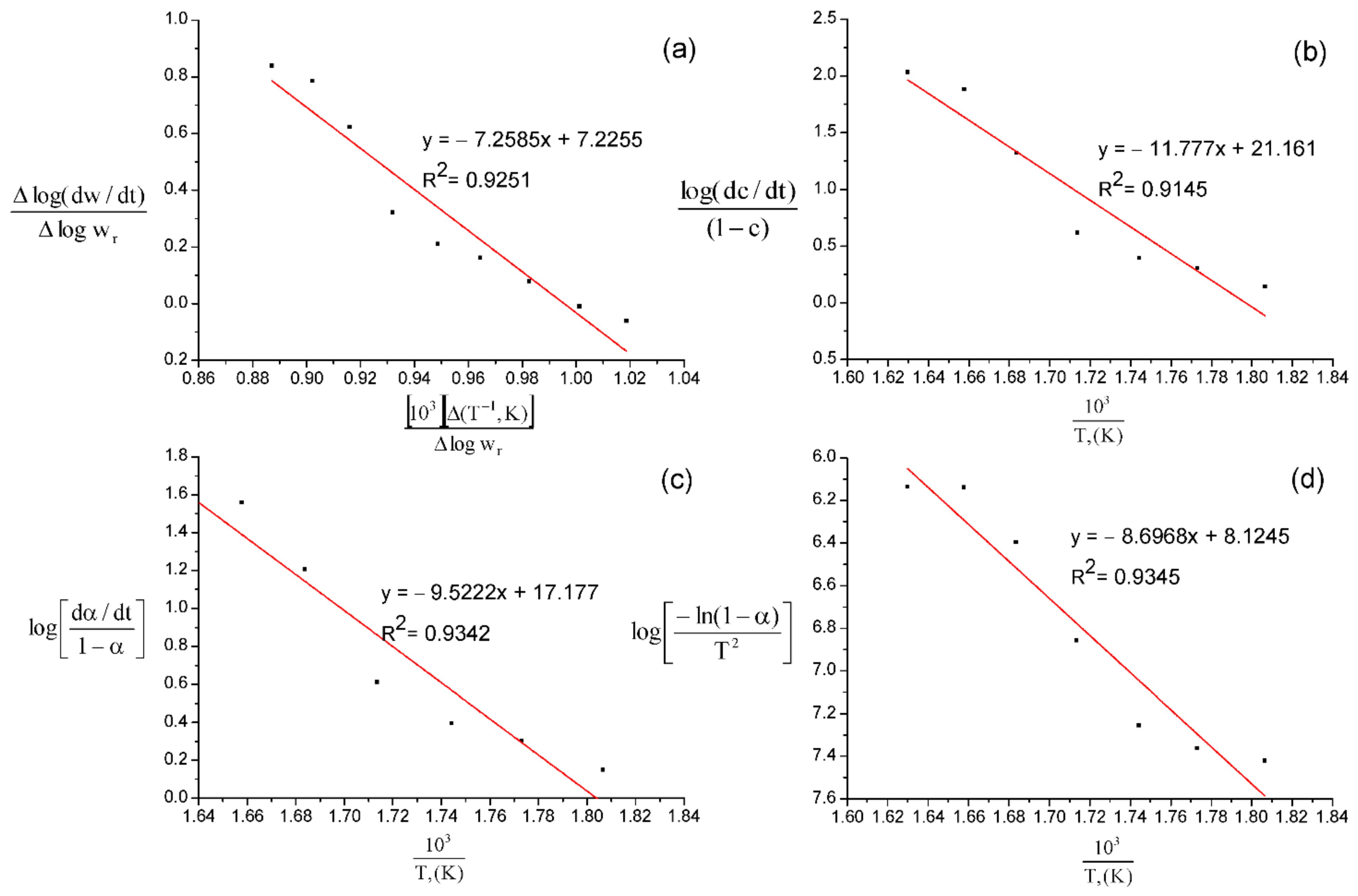
| Method | Freeman–Carroll | Sharp–Wentworth | Ahar | Coates–Redfern | |||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | E, kJ/mol | n | E, kJ/mol | 109, min−1 | E, kJ/mol | 107, min−1 | E, kJ/mol | 103, min−1 | |
| β-CD * | 548.71 | 1.40 | 712.24 | 1.27 | 600.84 | 1.10 | 458.70 | 1.32 | |
| Anabasine-β CD 1:1 | 138.80 | 1.90 | 225.20 | 1.55 | 182.09 | 2.88 | 166.30 | 3.38 | |
| Anabasine-β CD 1:2 | 328.83 | 2.81 | 356.06 | 4.13 | 336.70 | 6.13 | 281.36 | 2.40 | |
| Anabasine-β CD 1:3 | 217.25 | 2.19 | 312.34 | 8.06 | 249.60 | 4.73 | 234.92 | 4.19 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muldakhmetov, Z.; Fazylov, S.; Nurkenov, O.; Gazaliev, A.; Sarsenbekova, A.; Pustolaikina, I.; Nurmaganbetov, Z.; Seilkhanov, O.; Alsfouk, A.A.; Elkaeed, E.B.; et al. Combined Computational and Experimental Studies of Anabasine Encapsulation by Beta-Cyclodextrin. Plants 2022, 11, 2283. https://doi.org/10.3390/plants11172283
Muldakhmetov Z, Fazylov S, Nurkenov O, Gazaliev A, Sarsenbekova A, Pustolaikina I, Nurmaganbetov Z, Seilkhanov O, Alsfouk AA, Elkaeed EB, et al. Combined Computational and Experimental Studies of Anabasine Encapsulation by Beta-Cyclodextrin. Plants. 2022; 11(17):2283. https://doi.org/10.3390/plants11172283
Chicago/Turabian StyleMuldakhmetov, Zeinolla, Serik Fazylov, Oral Nurkenov, Arstan Gazaliev, Akmaral Sarsenbekova, Irina Pustolaikina, Zhangeldy Nurmaganbetov, Olzhas Seilkhanov, Aisha A. Alsfouk, Eslam B. Elkaeed, and et al. 2022. "Combined Computational and Experimental Studies of Anabasine Encapsulation by Beta-Cyclodextrin" Plants 11, no. 17: 2283. https://doi.org/10.3390/plants11172283
APA StyleMuldakhmetov, Z., Fazylov, S., Nurkenov, O., Gazaliev, A., Sarsenbekova, A., Pustolaikina, I., Nurmaganbetov, Z., Seilkhanov, O., Alsfouk, A. A., Elkaeed, E. B., Eissa, I. H., & Metwaly, A. M. (2022). Combined Computational and Experimental Studies of Anabasine Encapsulation by Beta-Cyclodextrin. Plants, 11(17), 2283. https://doi.org/10.3390/plants11172283











