Location of Tandem Repeats on Wheat Chromosome 5B and the Breakpoint on the 5BS Arm in Wheat Translocation T7BS.7BL-5BS Using Single-Copy FISH Analysis
Abstract
:1. Introduction
2. Results
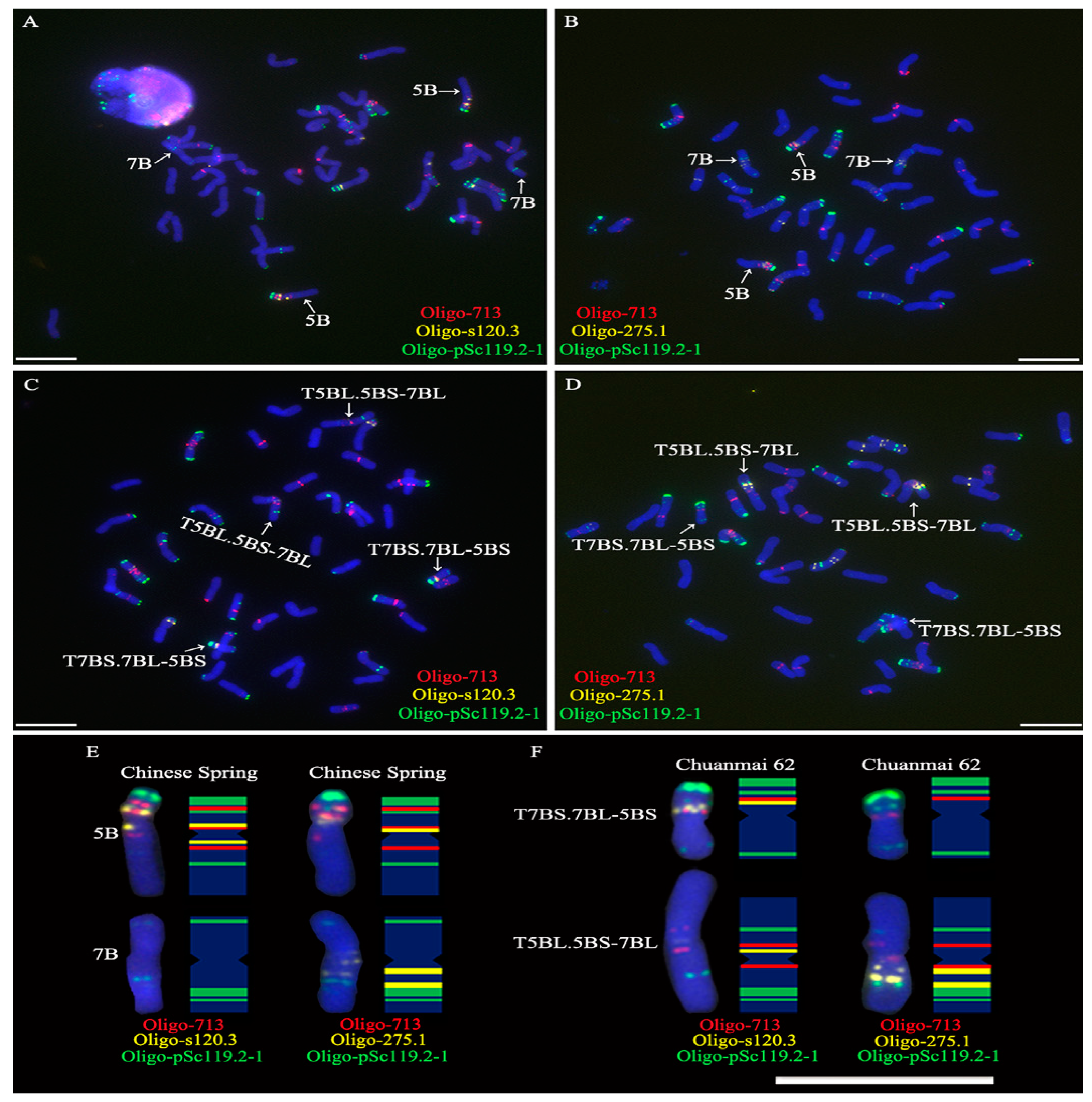
2.1. FISH Karyotype of Chromosomes 5B and 7B in Chinese Spring
2.2. ND-FISH Determining the Breakpoint Interval on 5BS Arm in T7BS.7BL-5BS
2.3. Single-Copy FISH Determining the Breakpoint Interval on the 5BS Arm in T7BS.7BL-5BS
2.4. Single-Copy FISH Determining the Position of Tandem Repeats on Chromosome 5B
3. Discussion
3.1. Methods for the Determination of Breakpoint in Wheat Translocation Chromosome
3.2. Determination of the Position of Tandem Repeats on Wheat Chromosome
4. Materials and Methods
4.1. Plant Materials
4.2. Nondenaturing Fluorescence in situ Hybridization (ND-FISH) Analysis
4.3. Single-Copy FISH Analysis
4.4. The Analysis of the Characters of the Sequence in the Breakpoint Region in 5BS Arm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garrido-Ramos, M.A. The genomics of plant satellite DNA. In Satellite DNAs in Physiology and Evolution, Progress in Molecular and Subcellular Biology; Ugarković, Ð., Ed.; Springer: Cham, Switzerland, 2021; Volume 60, pp. 103–144. [Google Scholar]
- Saxena, R.K.; Edwards, D.; Varshney, R.K. Structural variations in plant genomes. Brief. Funct. Genom. 2014, 13, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Bedbrook, J.R.; Jones, J.; O'Dell, M.; Thompson, R.D.; Flavell, R.B. A molecular description of telomeric heterochromatin in Secale species. Cell 1980, 19, 545–560. [Google Scholar] [CrossRef]
- Rayburn, A.L.; Gill, B.S. Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Rep. 1987, 4, 102–109. [Google Scholar] [CrossRef]
- Komuro, S.; Endo, R.; Shikata, K.; Kato, A. Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 2013, 56, 131–137. [Google Scholar] [CrossRef]
- Tang, S.; Tang, Z.; Qiu, L.; Yang, Z.; Li, G.; Lang, T.; Zhu, W.; Zhang, J.; Fu, S. Developing new oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front. Plant Sci. 2018, 9, 1104. [Google Scholar] [CrossRef]
- Lang, T.; Li, G.; Wang, H.; Yu, Z.; Chen, Q.; Yang, E.; Fu, S.; Tang, Z.; Yang, Z. Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 2019, 249, 663–675. [Google Scholar] [CrossRef]
- Jiang, M.; Xaio, Z.Q.; Fu, S.L.; Tang, Z.X. FISH karyotype of 85 common wheat cultivars/lines displayed by ND-FISH using oligonucleotide probes. Cereal Res. Commun. 2017, 45, 549–563. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, M.; Zhuang, L.; Zhang, S.; Wang, J.; Chen, X.; Wang, D.; Chen, J.; Bao, Y.; Guo, J.; et al. Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor. Appl. Genet. 2018, 131, 1967–1986. [Google Scholar] [CrossRef]
- Guo, J.; Gao, D.; Gong, W.; Li, H.; Li, J.; Li, G.; Song, J.; Liu, J.; Yang, Z.; Liu, C. Genetic diversity in common wheat lines revealed by fluorescence in situ hybridization. Plant Syst. Evol. 2019, 305, 247. [Google Scholar] [CrossRef]
- Hu, Z.; Luo, J.; Wan, L.; Luo, J.; Li, Y.; Fu, S.; Liu, D.; Hao, M.; Tang, Z. Chromosomes polymorphisms of Sichuan wheat cultivars displayed by ND-FISH landmarks. Cereal Res. Commun. 2022, 50, 253–262. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosomal rearrangements in wheat: Their types and distribution. Genome 2007, 50, 907–926. [Google Scholar]
- Levy, A.A.; Feldman, M. Evolution and origin of bread wheat. Plant Cell 2022, 34, 2549–2567. [Google Scholar] [CrossRef] [PubMed]
- Friebe, B.; Gill, B.S. C-band polymorphism and structural rearrangements detected in common wheat (Triticum aestivum). Euphytica 1994, 78, 1–5. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Bignell, G.R.; Santarius, T.; Pole, J.C.; Butler, A.P.; Perry, J.; Pleasance, E.; Greenman, C.; Menzies, A.; Taylor, S.; Edkins, S.; et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007, 17, 1296–1303. [Google Scholar] [CrossRef]
- Kolomietz, E.; Meyn, M.S.; Pandita, A.; Squire, J.A. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumours. Genes Chromosomes Cancer 2002, 35, 97–112. [Google Scholar] [CrossRef]
- Yang, M.; Yang, Z.; Yang, W.; Yang, E. Development and identification of new wheat varieties (lines) with multiple translocation chromosomes via cytogenetic methods. J. Triticeae Crops 2018, 38, 127–133. [Google Scholar]
- Tang, S.; Qiu, L.; Xiao, Z.; Fu, S.; Tang, Z. New Oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 2016, 7, 118. [Google Scholar] [CrossRef]
- Li, W.; Challa, G.S.; Zhu, H.; Wei, W. Recurrence of chromosome rearrangements and reuse of DNA breakpoints in the evolution of the Triticeae genomes. Genes Genom. Genet. 2016, 6, 3837–3847. [Google Scholar] [CrossRef]
- Luo, W.; Qin, N.; Mu, Y.; Tang, H.; Deng, M.; Liu, Y.; Chen, G.; Jiang, Q.; Chen, G.; Wei, Y.; et al. Variation and diversity of the breakpoint sequences on 4AL for the 4AL/5AL translocation in Triticum. Genome 2018, 61, 635–641. [Google Scholar] [CrossRef]
- Ibba, M.I.; Zhang, M.; Cai, X.; Morris, C.F. Identification of a conserved ph1b-mediated 5DS–5BS crossing over site in soft-kernel durum wheat (Triticum turgidum subsp. durum) lines. Euphytica 2019, 215, 200. [Google Scholar] [CrossRef]
- Lang, T.; La, S.; Li, B.; Yu, Z.; Chen, Q.; Li, J.; Yang, E.; Li, G.; Yang, Z. Precise identification of wheat—Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 2018, 61, 177–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokota, E.; Shibata, F.; Nagaki, K.; Murata, M. Stability of monocentric and dicentric ring minichromosomes in Arabidopsis. Chromosome Res. 2011, 19, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Cabral-de-Mello, D.C.; Marec, F. Universal fluorescence in situ hybridization (FISH) protocol for mapping repetitive DNAs in insects and other arthropods. Mol. Genet. Genom. 2021, 296, 513–526. [Google Scholar] [CrossRef]
- Danilova, T.V.; Friebe, B.; Gill, B.S. Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 2012, 121, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Danilova, T.V.; Gill, B.S. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor. Appl. Genet. 2014, 127, 715–730. [Google Scholar] [CrossRef]
- Aliyeva-Schnorr, L.; Stein, N.; Houben, A. Collinearity of homoeologous group 3 chromosomes in the genus Hordeum and Secale cereale as revealed by 3H-derived FISH analysis. Chromosome Res. 2016, 24, 231–242. [Google Scholar] [CrossRef]
- Li, Z.; Bi, Y.; Wang, X.; Wang, Y.; Yang, S.; Zhang, Z.; Chen, J.; Lou, Q. Chromosome identification in Cucumis anguria revealed by cross-species single-copy gene FISH. Genome 2018, 61, 397–404. [Google Scholar] [CrossRef]
- Said, M.; Hřibová, E.; Danilova, T.V.; Karafiátová, M.; Čížková, J.; Friebe, B.; Doležel, J.; Gill, B.S.; Vrána, J. The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor. Appl. Genet. 2018, 131, 2213–2227. [Google Scholar] [CrossRef]
- Zou, Y.; Wan, L.; Luo, J.; Tang, Z.; Fu, S. FISH landmarks reflecting meiotic recombination and structural alterations of chromosomes in wheat (Triticum aestivum L.). BMC Plant Biol. 2021, 21, 167. [Google Scholar] [CrossRef]
- Pan, Z.; Luo, J.; Tang, Z.; Fu, S. Centromere-specific single-copy sequences of Secale species. Plants 2022, 11, 2117. [Google Scholar] [CrossRef] [PubMed]
- Mclntyre, C.L.; Pereira, S.; Moran, L.B.; Appels, R. New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 1990, 33, 317–323. [Google Scholar]
- Tang, Z.; Yang, Z.; Fu, S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef]
- Fu, S.; Chen, L.; Wang, Y.; Li, M.; Yang, Z.; Qiu, L.; Yan, B.; Ren, Z.; Tang, Z. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [Green Version]


| Name of Single-Copy Probe | Name of Primers Used to Amplify Single-Copy Sequence | Nucleotide Sequence of Primer (5′-3′) | The Position on the Chromosome 5B of Chinese Spring (IWGSC RefSeq V2.1) (bp) |
|---|---|---|---|
| SC5B-157 | 157Mb-5BF | ACCCGCCAGAATCATCACTG | 157,749,421–157,752,276 |
| 157Mb-5BR | GGGAATCCAAGCCACGATCT | ||
| SC5B-158 | 158Mb-5BF | TTCCAATTCCAGACCCTGCC | 158,552,855–158,555,080 |
| 158Mb-5BR | CTGTTCTTGCTATTCCGCGC | ||
| SC5B-160 | 160Mb-5BF | ACTCGGTGGTGCATGAAACT | 160,001,176–160,003,706 |
| 160Mb-5BR | TGGACTAGTCGAGGGGTCTG | ||
| SC5B-201 | 201Mb-5BF | TCCCTCACTGGTACCATCCC | 201,713,962–201,711,028 |
| 201Mb-5BR | CCTGGTATGCAGGAATCCCC | ||
| SC5B-234 | 234Mb-5BF | AAAACCATTGCCACAGGTGC | 234,929,992–234,932,621 |
| 234Mb-5BR | ATTCGGTCGATCCTACACGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Tang, Z.; Luo, J.; Li, G.; Yang, Z.; Yang, M.; Yang, E.; Fu, S. Location of Tandem Repeats on Wheat Chromosome 5B and the Breakpoint on the 5BS Arm in Wheat Translocation T7BS.7BL-5BS Using Single-Copy FISH Analysis. Plants 2022, 11, 2394. https://doi.org/10.3390/plants11182394
Zhang W, Tang Z, Luo J, Li G, Yang Z, Yang M, Yang E, Fu S. Location of Tandem Repeats on Wheat Chromosome 5B and the Breakpoint on the 5BS Arm in Wheat Translocation T7BS.7BL-5BS Using Single-Copy FISH Analysis. Plants. 2022; 11(18):2394. https://doi.org/10.3390/plants11182394
Chicago/Turabian StyleZhang, Wei, Zongxiang Tang, Jie Luo, Guangrong Li, Zujun Yang, Manyu Yang, Ennian Yang, and Shulan Fu. 2022. "Location of Tandem Repeats on Wheat Chromosome 5B and the Breakpoint on the 5BS Arm in Wheat Translocation T7BS.7BL-5BS Using Single-Copy FISH Analysis" Plants 11, no. 18: 2394. https://doi.org/10.3390/plants11182394
APA StyleZhang, W., Tang, Z., Luo, J., Li, G., Yang, Z., Yang, M., Yang, E., & Fu, S. (2022). Location of Tandem Repeats on Wheat Chromosome 5B and the Breakpoint on the 5BS Arm in Wheat Translocation T7BS.7BL-5BS Using Single-Copy FISH Analysis. Plants, 11(18), 2394. https://doi.org/10.3390/plants11182394








