Modulation of the Translation Efficiency of Heterologous mRNA and Target Protein Stability in a Plant System: The Case Study of Interferon-αA
Abstract
1. Introduction
2. Results and Discussion
2.1. Constructing Efficient Systems for Expression of Target Polypeptides in Plants
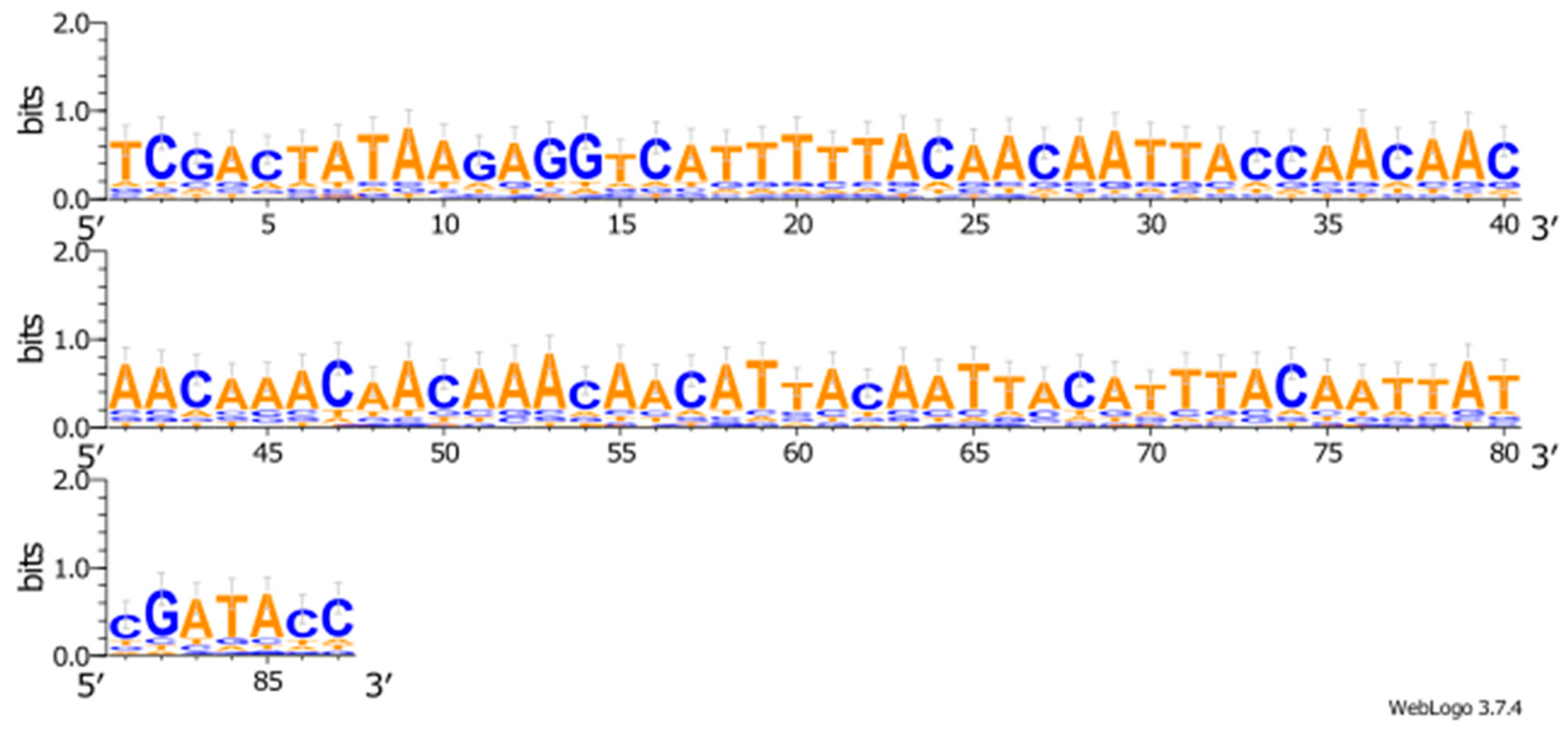
2.1.1. Modulation of Translation Efficiency: The Role of Codon Composition and 5′UTR of Heterologous Gene
In Silico Analysis
Expression Vectors for Assessing the Contribution of Optimization of Target Gene Codon Composition and 5′UTR to Heterologous Gene Translation Efficiency
Functional Estimate of the Contribution of Target Gene Optimized Codon Composition and 5′UTR to the Expression Efficiency of Heterologous Gene
2.1.2. Modulating Stability of the Protein Product: The Role of Second Amino Acid in Increasing the Target Protein Yield
Expression Vectors for Assessing the Contribution of the Second Amino Acid to the Increase in the Yield of Protein Product of Heterologous Genes
Functional Assessment of the Contribution of the Second Amino Acid to the Increase in the Yield of Protein Product of Heterologous Genes
2.1.3. Modulating Stability of Protein Product: The Role of PSP in the Increase in Target Protein Yield
Expression Vectors and Functional Assessment of the Contribution of Thermostable Lichenase as a PSP to the Increase in the Yield of the Protein Product of Heterologous Gene
Possible Purification of Recombinant Protein Using Thermostable Lichenase as a PSP
3. Materials and Methods
3.1. Methods for In Silico Analysis
3.2. Engineering Plant Expression Vectors
- Construction of the modified lichenase gene (L53) carrying two BspQI sites integrated at position 53 aa; these sites form the cloning region of the target inserts according to the GoldenGate technology. The pQE30-licBM3 plasmid was linearized using the L53_F1/L53_R1 and L53_F2/L53_R2 primer pairs; the resulting amplicons were joined and hybridized. The synthetic insert (L53dummy_S/L53dummy_AS) was integrated using a SLIC [35] approach to give the plasmid pQE30-L53.
- Integration of the modified thermostable lichenase gene into the intermediate pFGG plasmid. The pFGG vector was linearized by digestion at ApaI/SmaI sites to clone the modified thermostable lichenase gene licBM3-53 (L53) using the L_Apa_F/L_Sma_R primer pair from the pQE30 plasmid; the sticky ends were formed with ApaI and SmaI restriction endonucleases; thus, the pFGG-L53 vector was constructed.
- Construction of the variants of modified lichenase carrying destabilizing amino acid at the second position (Arg-L53) or synthetic 5′UTR (87-L-53). The pFGG-L53 plasmid was linearized by PCR with the L53_ARG_F/L53_ARG_R primer pair. Both primers carried an additional triplet coding for arginine, the destabilizing amino acid. The linearized plasmid was ligated at the blunt ends to form the circular plasmid pFGG-Arg-L53. The pFGG-L53 plasmid was linearized by PCR with the L53_87_F1/L53_87_R1 and L53_87_F2/L53_87_R2 primer pairs. The synthetic insert corresponding to the 5′UTR (UTR87S/UTR87AS) was integrated using a SLIC approach to obtain the final vector pFGG-87-L53.
- Cloning of the modified and native interferon genes to position 53 aa. The native and modified interferon genes were amplified using the INN_F1/INN_R1 and INN_F2/INN_R2 primer pairs for the native variant and INM_F1/INM_R1 and INM_F2/INM_R2 for the modified one. The resulting products were joined and hybridized to obtain sticky ends. The initial pFGG-Arg-L53 plasmid was linearized by hydrolysis with BspQI restriction endonuclease. The vector and the insert were ligated using a standard procedure (pFGG-L53-INM, pFGG-L53-87-INM, and pFGG-L53-Arg-INM). To integrate the native interferon gene, the amplicon produced by PCR with the INM_Apa_F/INM_Sma_R primer pair was hydrolyzed at the ApaI/SmaI sites and further cloned into the pFGG vector similar to different genes of thermostable lichenase.
- Transfer of the produced expression cassettes carrying hybrid genes derived from the nucleotide sequences of lichenase and interferon genes to the vector optimized for transient expression of heterologous genes in plants, pVIG-T. All expression cassettes were transferred from the vectors pFGG-L53-IN, pFGG-L53-IN-M, pFGG-87-L53-IN-M, pFGG-Arg-L53-IN-M, and pFGG-IN-M to the vectors pVIG-T (plasmids pVIG-T-L53-IN, pVIG-T-L53-IN-M, pVIG-T-87-L53-IN-M, pVIG-T-Arg-L53-IN-M, and pVIG-T-IN-M were formed) using a routine restriction–ligation cloning technique at SpeI/HpaI sites (Figure S2).
3.3. Plant Material, Agrobacterium Strains, and Agroinfiltration
3.4. Producing Plant Protein Extracts
3.5. Biochemical Methods for Determining Lichenase Activity and Interferon Biological Activity
3.6. Statistical Data Processing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shanmugaraj, B.; Bulaon, C.J.I.; Phoolcharoen, W. Plant Molecular Farming: A Viable Platform for Recombinant Biopharmaceutical Production. Plants 2020, 9, 842. [Google Scholar] [CrossRef]
- Schillberg, S.; Finnern, R. Plant Molecular Farming for the Production of Valuable Proteins—Critical Evaluation of Achievements and Future Challenges. J. Plant Physiol. 2021, 258–259, 153359. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Timko, M.P. Improving Protein Quantity and Quality—The Next Level of Plant Molecular Farming. Int. J. Mol. Sci. 2022, 23, 1326. [Google Scholar] [CrossRef]
- Chincinska, I.A. Leaf Infiltration in Plant Science: Old Method, New Possibilities. Plant Methods 2021, 17, 83. [Google Scholar] [CrossRef]
- Molina-Hidalgo, F.J.; Vazquez-Vilar, M.; D’Andrea, L.; Demurtas, O.C.; Fraser, P.; Giuliano, G.; Bock, R.; Orzáez, D.; Goossens, A. Engineering Metabolism in Nicotiana Species: A Promising Future. Trends Biotechnol. 2021, 39, 901–913. [Google Scholar] [CrossRef]
- Chung, Y.H.; Church, D.; Koellhoffer, E.C.; Osota, E.; Shukla, S.; Rybicki, E.P.; Pokorski, J.K.; Steinmetz, N.F. Integrating Plant Molecular Farming and Materials Research for Next-Generation Vaccines. Nat. Rev. Mater. 2022, 7, 372–388. [Google Scholar] [CrossRef]
- Goldenkova-Pavlova, I.V.; Pavlenko, O.S.; Mustafaev, O.N.; Deyneko, I.V.; Kabardaeva, K.V.; Tyurin, A.A. Computational and Experimental Tools to Monitor the Changes in Translation Efficiency of Plant MRNA on a Genome-Wide Scale: Advantages, Limitations, and Solutions. Int. J. Mol. Sci. 2019, 20, 33. [Google Scholar] [CrossRef]
- Kabardaeva, K.V.; Tyurin, A.A.; Pavlenko, O.S.; Gra, O.A.; Deyneko, I.V.; Kouchoro, F.; Mustafaev, O.N.; Goldenkova-Pavlova, I.V. Fine Tuning of Translation: A Complex Web of Mechanisms and Its Relevance to Plant Functional Genomics and Biotechnology. Russ. J. Plant Physiol. 2019, 66, 835–849. [Google Scholar] [CrossRef]
- Kabardaeva, K.V.; Turin, A.A.; Kouchoro, F.; Mustafaev, O.N.; Deineko, I.V.; Fadeev, V.S.; Goldenkova-Pavlova, I.V. Regulatory Contexts in the 5′-Region of MRNA from Arabidopsis Thaliana Plants and Their Role in Translation Efficiency. Russ. J. Plant Physiol. 2020, 67, 425–434. [Google Scholar] [CrossRef]
- Limkul, J.; Misaki, R.; Kato, K.; Fujiyama, K. The Combination of Plant Translational Enhancers and Terminator Increase the Expression of Human Glucocerebrosidase in Nicotiana Benthamiana Plants. Plant Sci. 2015, 240, 41–49. [Google Scholar] [CrossRef]
- Buyel, J.F.; Stöger, E.; Bortesi, L. Targeted Genome Editing of Plants and Plant Cells for Biomanufacturing. Transgenic Res. 2021, 30, 401–426. [Google Scholar] [CrossRef]
- Clemente, M.; Corigliano, M.; Pariani, S.; Sánchez-López, E.; Sander, V.; Ramos-Duarte, V. Plant Serine Protease Inhibitors: Biotechnology Application in Agriculture and Molecular Farming. Int. J. Mol. Sci. 2019, 20, 1345. [Google Scholar] [CrossRef]
- Tasaki, T.; Sriram, S.M.; Park, K.S.; Kwon, Y.T. The N-End Rule Pathway. Annu. Rev. Biochem. 2012, 81, 261–289. [Google Scholar] [CrossRef]
- Jutras, P.V.; Dodds, I.; van der Hoorn, R.A.L. Proteases of Nicotiana Benthamiana: An Emerging Battle for Molecular Farming. Curr. Opin. Biotechnol. 2020, 61, 60–65. [Google Scholar] [CrossRef]
- Pavlenko, O.S.; Gra, O.A.; Mustafaev, O.N.; Kabarbaeva, K.V.; Sadovskaya, N.S.; Tyurin, A.A.; Fadeev, V.S.; Goldenkova-Pavlova, I.V. Thermostable Lichenase from Clostridium Thermocellum as a Host Protein in the Domain Insertion Approach. Biochemistry 2019, 84, 931–940. [Google Scholar] [CrossRef]
- Tyurin, A.A.; Kabardaeva, K.V.; Mustafaev, O.N.; Pavlenko, O.S.; Sadovskaya, N.S.; Fadeev, V.S.; Zvonova, E.A.; Goldenkova-Pavlova, I.V. Expression of Soluble Active Interferon AA in Escherichia Coli Periplasm by Fusion with Thermostable Lichenase Using the Domain Insertion Approach. Biochemistry 2018, 83, 259–269. [Google Scholar] [CrossRef]
- Goldenkova-Pavlova, I.V.; Tyurin, A.; Mustafaev, O.N. The Features That Distinguish Lichenases from Other Polysaccharide-Hydrolyzing Enzymes and the Relevance of Lichenases for Biotechnological Applications. Appl. Microbiol. Biotechnol. 2018, 102, 3951–3965. [Google Scholar] [CrossRef]
- Tyurin, A.A.; Sadovskaya, N.S.; Nikiforova, K.R.; Mustafaev, O.N.; Komakhin, R.A.; Fadeev, V.S.; Goldenkova-Pavlova, I.V. Clostridium Thermocellum Thermostable Lichenase with Circular Permutations and Modifications in the N-Terminal Region Retains Its Activity and Thermostability. Biochim. Biophys. Acta Proteins Proteom. 2015, 1854, 10–19. [Google Scholar] [CrossRef]
- Kozak, M. Circumstances and Mechanisms of Inhibition of Translation by Secondary Structure in Eucaryotic MRNAs. Mol. Cell. Biol. 1989, 9, 5134–5142. [Google Scholar] [CrossRef]
- Sato, K.; Hamada, M.; Asai, K.; Mituyama, T. CENTROIDFOLD: A Web Server for RNA Secondary Structure Prediction. Nucleic Acids Res. 2009, 37, W277–W280. [Google Scholar] [CrossRef]
- Vyacheslavova, A.O.; Mustafaev, O.N.; Tyrin, A.A.; Shimshilashvili, K.R.; Berdichevets, I.N.; Shayakhmetova, D.M.; Goldenkov, M.A.; Fadeev, V.S.; Sheludko, Y.V.; Goldenkova-Pavlova, I.V. Set of Module Vectors for Stable or Transient Expression of Heterologous Genes in Plants. Russ. J. Genet. 2012, 48, 892–901. [Google Scholar] [CrossRef]
- Ortega, J.L.; Wilson, O.L.; Sengupta-Gopalan, C. The 5′ Untranslated Region of the Soybean Cytosolic Glutamine Synthetase Β1 Gene Contains Prokaryotic Translation Initiation Signals and Acts as a Translational Enhancer in Plants. Mol. Genet. Genom. 2012, 287, 881–893. [Google Scholar] [CrossRef][Green Version]
- Kanoria, S.; Burma, P.K. A 28 Nt Long Synthetic 5′UTR (SynJ) as an Enhancer of Transgene Expression in Dicotyledonous Plants. BMC Biotechnol. 2012, 12, 85. [Google Scholar] [CrossRef]
- Graciet, E.; Mesiti, F.; Wellmer, F. Structure and Evolutionary Conservation of the Plant N-End Rule Pathway. Plant J. 2010, 61, 741–751. [Google Scholar] [CrossRef]
- Tasaki, T.; Kwon, Y.T. The Mammalian N-End Rule Pathway: New Insights into Its Components and Physiological Roles. Trends Biochem. Sci. 2007, 32, 520–528. [Google Scholar] [CrossRef]
- Santos-Buelga, C.; Gonzalez-Manzano, S.; Dueñas, M.; Gonzalez-Paramas, A.M. Extraction and Isolation of Phenolic Compounds. In Natural Products Isolation. Methods in Molecular Biology, 3rd ed.; Sarker, S.D., Nahar, L., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 427–464. [Google Scholar]
- Seidel, V. Initial and Bulk Extraction of Natural Products Isolation. In Natural Products Isolation. Methods in Molecular Biology, 3rd ed.; Sarker, S.D., Nahar, L., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 27–41. [Google Scholar]
- Spiegel, H.; Stöger, E.; Twyman, R.M.; Buyel, J.F. Current Status and Perspectives of the Molecular Farming Landscape. In Molecular Pharming; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 1–23. [Google Scholar]
- Buyel, J.F. Plants as Sources of Natural and Recombinant Anti-Cancer Agents. Biotechnol. Adv. 2018, 36, 506–520. [Google Scholar] [CrossRef]
- Buyel, J.F.; Twyman, R.M.; Fischer, R. Extraction and Downstream Processing of Plant-Derived Recombinant Proteins. Biotechnol. Adv. 2015, 33, 902–913. [Google Scholar] [CrossRef]
- Keinänen, M.; Oldham, N.J.; Baldwin, I.T. Rapid HPLC Screening of Jasmonate-Induced Increases in Tobacco Alkaloids, Phenolics, and Diterpene Glycosides in Nicotiana Attenuata. J. Agric. Food Chem. 2001, 49, 3553–3558. [Google Scholar] [CrossRef]
- Buyel, J.F. Plant Molecular Farming—Integration and Exploitation of Side Streams to Achieve Sustainable Biomanufacturing. Front. Plant. Sci. 2019, 9, 1893. [Google Scholar] [CrossRef]
- Sadovskaya, N.S.; Mustafaev, O.N.; Tyurin, A.A.; Deineko, I.V.; Goldenkova-Pavlova, I.V. JetGene: Internet Resource for Analysis of Regulatory Regions or Nucleotide Contexts in Differentially Translated Plant Transcripts. Russ. J. Plant Physiol. 2021, 68, 633–640. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Li, M.Z.; Elledge, S.J. Harnessing Homologous Recombination in Vitro to Generate Recombinant DNA via SLIC. Nat. Methods 2007, 4, 251–256. [Google Scholar] [CrossRef]
- Tyurin, A.A.; Kabardaeva, K.V.; Berestovoy, M.A.; Sidorchuk, Y.V.; Fomenkov, A.A.; Nosov, A.V.; Goldenkova-Pavlova, I.V. Simple and Reliable System for Transient Gene Expression for the Characteristic Signal Sequences and the Estimation of the Localization of Target Protein in Plant Cell. Russ. J. Plant Physiol. 2017, 64, 672–679. [Google Scholar] [CrossRef]
- Berestovoy, M.; Pavlenko, O.S.; Tyurin, A.A.; Gorshkova, E.N.; Goldenkova-Pavlova, I.V. Altered Fatty Acid Composition of Nicotiana Benthamiana and Nicotiana Excelsior Leaves under Transient Overexpression of the Cyanobacterial DesC Gene. Biol. Plant 2020, 64, 167–177. [Google Scholar] [CrossRef]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Tyurin, A.A.; Suhorukova, A.V.; Deineko, I.V.; Pavlenko, O.S.; Fridman, V.A.; Goldenkova-Pavlova, I.V. A High Throughput Assay of Lichenase Activity with Congo Red Dye in Plants. Plant Methods 2021, 17, 102. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyurin, A.A.; Mustafaev, O.; Suhorukova, A.V.; Pavlenko, O.S.; Fridman, V.A.; Demyanchuk, I.S.; Goldenkova-Pavlova, I.V. Modulation of the Translation Efficiency of Heterologous mRNA and Target Protein Stability in a Plant System: The Case Study of Interferon-αA. Plants 2022, 11, 2450. https://doi.org/10.3390/plants11192450
Tyurin AA, Mustafaev O, Suhorukova AV, Pavlenko OS, Fridman VA, Demyanchuk IS, Goldenkova-Pavlova IV. Modulation of the Translation Efficiency of Heterologous mRNA and Target Protein Stability in a Plant System: The Case Study of Interferon-αA. Plants. 2022; 11(19):2450. https://doi.org/10.3390/plants11192450
Chicago/Turabian StyleTyurin, Alexander A., Orkhan Mustafaev, Aleksandra V. Suhorukova, Olga S. Pavlenko, Viktoriia A. Fridman, Ilya S. Demyanchuk, and Irina V. Goldenkova-Pavlova. 2022. "Modulation of the Translation Efficiency of Heterologous mRNA and Target Protein Stability in a Plant System: The Case Study of Interferon-αA" Plants 11, no. 19: 2450. https://doi.org/10.3390/plants11192450
APA StyleTyurin, A. A., Mustafaev, O., Suhorukova, A. V., Pavlenko, O. S., Fridman, V. A., Demyanchuk, I. S., & Goldenkova-Pavlova, I. V. (2022). Modulation of the Translation Efficiency of Heterologous mRNA and Target Protein Stability in a Plant System: The Case Study of Interferon-αA. Plants, 11(19), 2450. https://doi.org/10.3390/plants11192450







