Proteomic Changes in Paspalum fasciculatum Leaves Exposed to Cd Stress
Abstract
1. Introduction
2. Results
2.1. Quantitative Analysis of Proteins in P. fasciculatum Leaves Exposed to Cd Stress
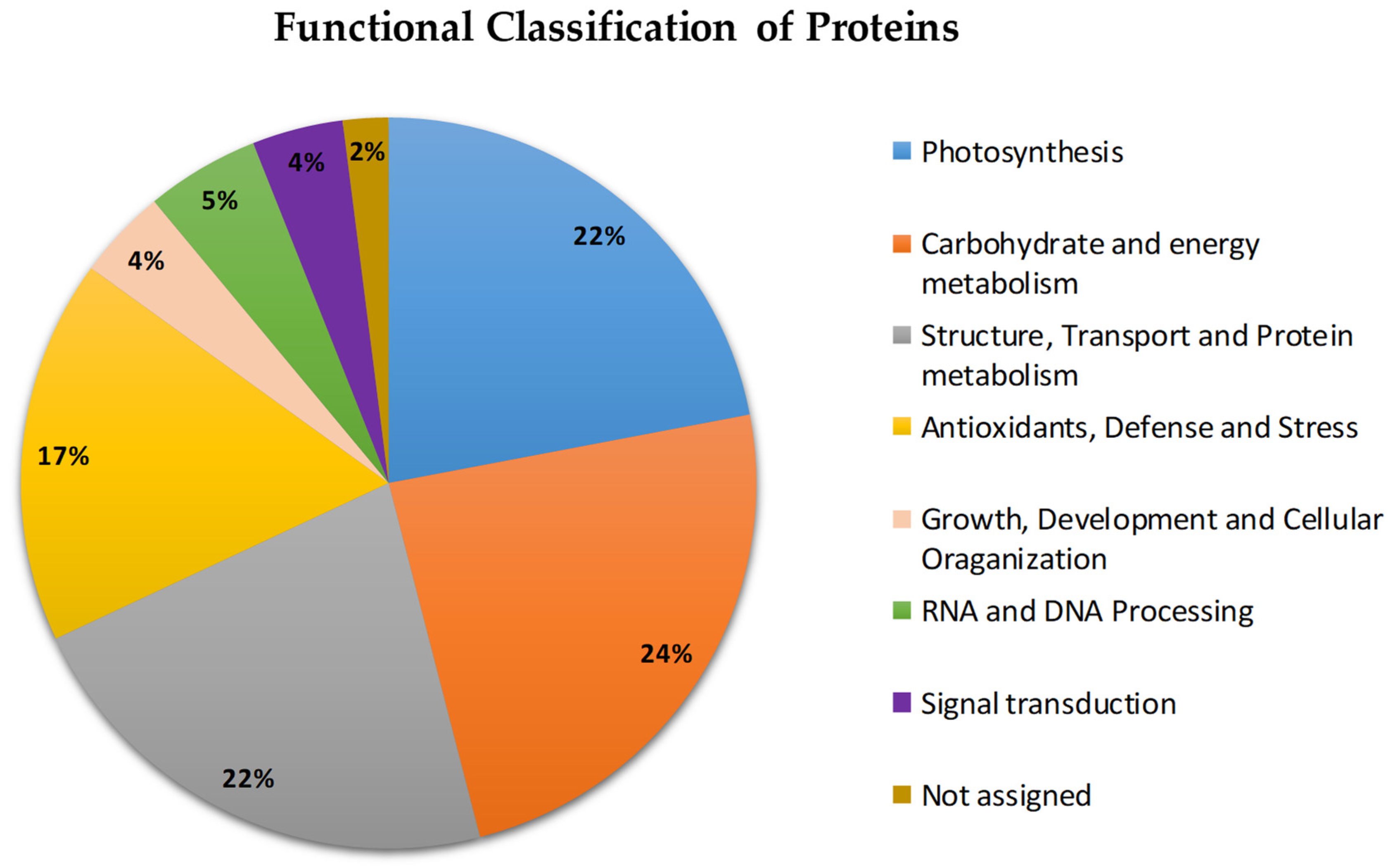
2.2. Functional Classification of Proteins
3. Discussion
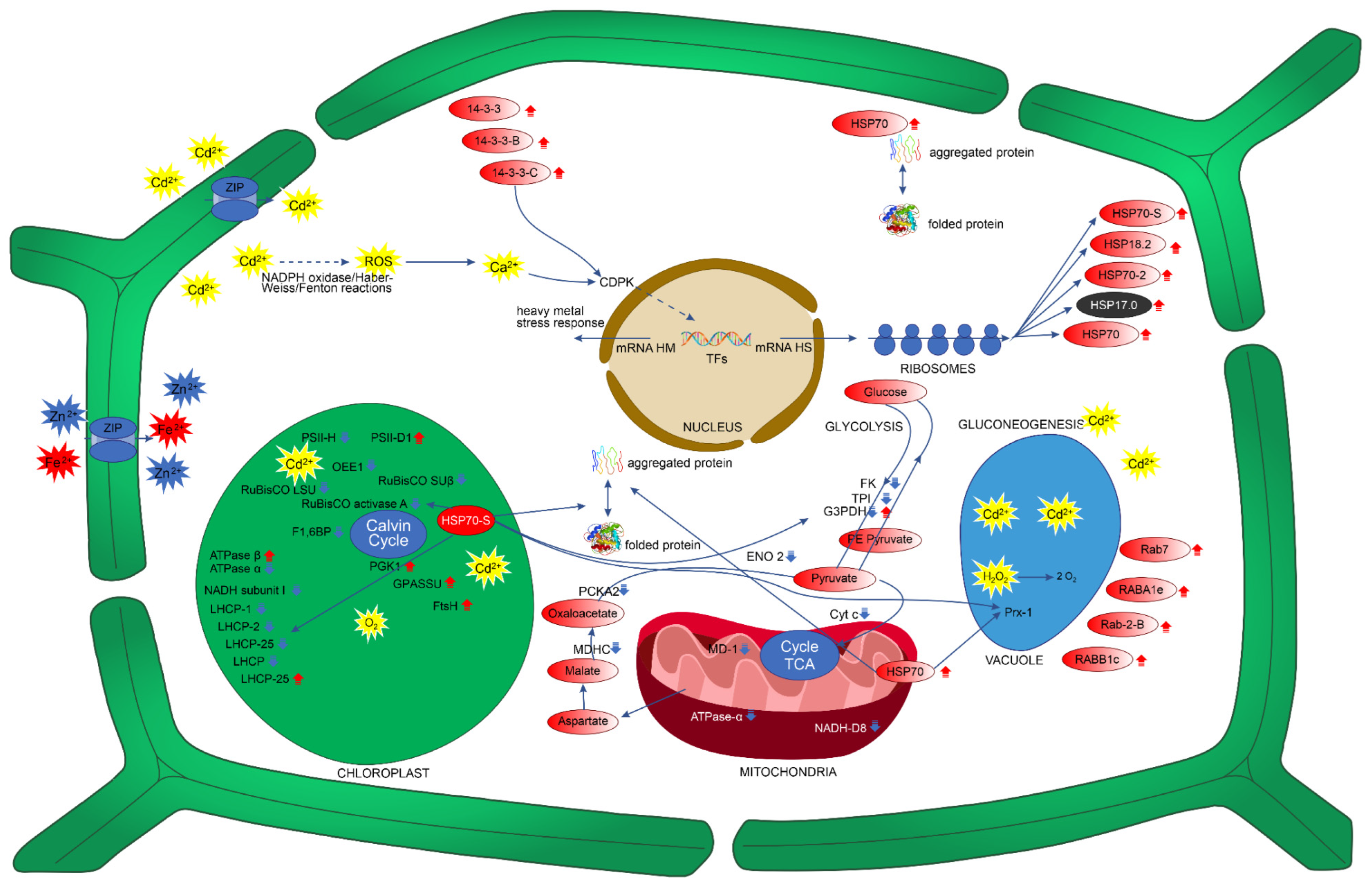
3.1. P. fasciculatum Overcomes the Deleterious Effects of Cadmium on Photosynthesis Proteins and Carbohydrate Energy Metabolism
3.2. Signal-Transduction-Associated Protein Changes during Cd Stress
3.3. Protein Changes Associated with Growth and Development during Cd Stress
3.4. Antioxidant-Defense- and Stress-Response-Involved Protein Changes during Cd Stress
4. Materials and Methods
4.1. Plant Growth Conditions and Tissue Sampling
4.2. Protein Extraction and Digestion
4.3. Shotgun LC–MS/MS Analysis
4.4. Protein Identification and Functional Classification
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- ATSDR. ATSDR Substance Priority List. Available online: https://www.atsdr.cdc.gov/spl/ (accessed on 26 September 2018).
- Talebzadeh, F.; Valeo, C.; Gupta, R. Cadmium Water Pollution Associated with Motor Vehicle Brake Parts. IOP Conf. Ser. Earth Environ. Sci. 2021, 691, 012001. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, D.; Wu, J.; Cheng, Z.; Yan, X.; Deng, X.; Yan, Y. Identification of differentially accumulated proteins involved in regulating independent and combined osmosis and cadmium stress response in Brachypodium seedling roots. Sci. Rep. 2018, 8, 7790. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Sun, L.; Yang, X.; Liu, J.X. Transcriptomic analysis of cadmium stress response in the heavy metal hyperaccumulator Sedum alfredii Hance. PLoS ONE 2013, 8, e64643. [Google Scholar] [CrossRef]
- Tinkov, A.A.; Filippini, T.; Ajsuvakovae, O.P.; Skalnaya, M.G.; Aasethf, J.; Bjørklundh, G.; Gatiatulinai, E.R.; Popova, E.V.; Nemereshinai, O.N.; Huangk, P.T.; et al. Cadmium and atherosclerosis: A review of toxicological mechanisms and a meta-analysis of epidemiologic studies. Environ. Res. 2018, 162, 240–260. [Google Scholar] [CrossRef] [PubMed]
- Ismael, M.A.; Elyamine, A.M.; Moussa, M.G.; Cai, M.; Zhao, X.; Hu, C. Cadmium in plants: Uptake, toxicity, and its interactions with selenium fertilizers. Metallomics 2019, 11, 255–277. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, Y.-L.; Cui, S.-X.; Chen, M.; Yang, H.-M.; Liu, H.-M.; Chai, T.-Y.; Huang, F. Cd-induced changes in leaf proteome of the hyperaccumulator plant Phytolacca americana. Chemosphere 2011, 85, 56–66. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Qayyum, M.F.; Ok, Y.S.; Zia-ur-Rehman, M.; Abbas, Z.; Hannan, F. Use of maize (Zea mays L.) for phytomanagement of Cd-contaminated soils: A critical review. Environ. Geochem. Health 2017, 39, 259–277. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Zia Ur Rehman, M.; Rinklebe, J.; Tsang, D.C.W.; Bashir, A.; Maqbool, A.; Tack, F.M.G.; Ok, Y.S. Cadmium phytoremediation potential of Brassica crop species: A review. Sci. Total Environ. 2018, 631–632, 1175–1191. [Google Scholar] [CrossRef]
- Bechtold, U.; Benjamin, F. Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 2018, 69, 2753–2758. [Google Scholar] [CrossRef]
- Oliveira, B.R.M.; de Almeida, A.-A.F.; Pirovani, C.P.; Barroso, J.P.; de C.Neto, C.H.; Santos, N.A.; Ahnert, D.; Balegar, V.C.; Mangabeira, P.A.O. Mitigation of Cd toxicity by Mn in young plants of cacao, evaluated by the proteomic profiles of leaves and roots. Ecotoxicology 2020, 29, 340–358. [Google Scholar] [CrossRef]
- Sun, X.; Huang, N.; Li, X.; Zhu, J.; Bian, X.; Li, H.; Wang, L.; Hu, Q.; Luo, H. A chloroplast heat shock protein modulates growth and abiotic stress response in creeping bentgrass. Plant Cell Environ. 2021, 44, 1769–1787. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Yu, G.; Li, H.; Li, X.; Mu, C. Overexpression of small heat shock protein LimHSP16. 45 in Arabidopsis hsp17. 6II mutant enhances tolerance to abiotic stresses. Russ. J. Plant Physiol. 2020, 67, 231–241. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, T.; Wang, Y.; Lei, X.; Zou, Y.; Yao, L.; Guo, W. Identification and Expression of Heat Shock Protein 60 (HSP60) from the Ark Shell Scapharca broughtonii. J. Shellfish Res. 2019, 38, 611–618. [Google Scholar] [CrossRef]
- ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-Z.; Zhang, H.-Z.; Wei, A.-M.; Gong, Z.-H. Heat Shock Proteins: Dynamic Biomolecules to Counter Plant Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Cruz, E.Y.; Arancibia-Hernández, Y.L.; Loyola-Mondragón, D.Y.; Pedraza-Chaverri, J. Oxidative Stress and Its Role in Cd-Induced Epigenetic Modifications: Use of Antioxidants as a Possible Preventive Strategy. Oxygen 2022, 2, 177–210. [Google Scholar] [CrossRef]
- Lan, X.Y.; Yan, Y.Y.; Yang, B.; Li, X.Y.; Xu, F.L. Differential expression of proteins in the leaves and roots of cadmium-stressed Microsorum pteropus, a novel potential aquatic cadmium hyperaccumulator. Sci. Total Environ. 2018, 642, 1369–1377. [Google Scholar] [CrossRef]
- Gallo, V.; Zappettini, A.; Villani, M.; Marmiroli, N.; Marmiroli, M. Comparative analysis of proteins regulated during cadmium sulfide quantum dots response in Arabidopsis thaliana wild type and tolerant mutants. Nanomaterials 2021, 11, 615. [Google Scholar] [CrossRef]
- Zhong, M.; Li, S.; Huang, F.; Qiu, J.; Zhang, J.; Sheng, Z.; Tang, X.; Wei, X.; Hu, P. The Phosphoproteomic Response of Rice Seedlings to Cadmium Stress. Int. J. Mol. Sci. 2017, 18, 2055. [Google Scholar] [CrossRef]
- Cao, F.; Dai, H.; Hao, P.-F.; Wu, F. Silicon regulates the expression of vacuolar H+-pyrophosphatase 1 and decreases cadmium accumulation in rice (Oryza sativa L.). Chemosphere 2019, 240, 124907. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Yang, S.; Zhou, Y.; Dong, C.; Ren, J.; Sun, X.; Yang, Y. Comparative physiological and proteomic analysis Reveals the leaf response to cadmium-induced stress in poplar (Populus yunnanensis). PLoS ONE 2015, 10, e0137396. [Google Scholar] [CrossRef]
- Chowardhara, B.; Borgohain, P.; Saha, B.; Awasthi, J.P.; Panda, S.K. Differential oxidative stress responses in Brassica juncea (L.) Czern and Coss cultivars induced by cadmium at germination and early seedling stage. Acta Physiol. Plant 2020, 42, 105. [Google Scholar] [CrossRef]
- Luo, J.-S.; Zhang, Z. Proteomic changes in the xylem sap of Brassica napus under cadmium stress and functional validation. BMC Plant Biol. 2019, 19, 280. [Google Scholar] [CrossRef] [PubMed]
- Jian, M.; Zhang, D.; Wang, X.; Wei, S.; Zhao, Y.; Ding, Q.; Han, Y.; Ma, L. Differential expression pattern of the proteome in response to cadmium stress based on proteomics analysis of wheat roots. BMC Genom. 2020, 21, 343. [Google Scholar] [CrossRef]
- Salas-Moreno, M.; Contreras-Puentes, N.; Rodríguez-Cavallo, E.; Jorrín-Novo, J.; Marrugo-Negrete, J.; Méndez-Cuadro, D. Protein Carbonylation As a Biomarker of Heavy Metal, Cd and Pb, Damage in Paspalum fasciculatum Willd. ex Flüggé. Plant 2019, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Salas-Moreno, M.; Marrugo-Negrete, J. Phytoremediation potential of Cd and Pb-contaminated soils by Paspalum fasciculatum Willd. ex Flüggé. Int. J. Phytoremediat. 2019, 22, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Asplund-Samuelsson, J.; Hudson, E.P. Wide range of metabolic adaptations to the acquisition of the Calvin cycle revealed by comparison of microbial genomes. PLoS Comput. Biol. 2021, 17, e1008742. [Google Scholar] [CrossRef]
- Bagheri, R.; Ahmad, J.; Bashir, H.; Iqbal, M.; Qureshi, M.I. Changes in rubisco, cysteine-rich proteins and antioxidant system of spinach (Spinacia oleracea L.) due to sulphur deficiency, cadmium stress and their combination. Protoplasma 2016, 254, 1031–1043. [Google Scholar] [CrossRef]
- Wientjes, E.; Philippi, J.; Borst, J.W.; van Amerongen, H. Imaging the Photosystem I/Photosystem II chlorophyll ratio inside the leaf. BBA Bioenerg. 2017, 1858, 259–265. [Google Scholar] [CrossRef]
- Morina, F.; Küpper, H. Direct inhibition of photosynthesis by Cd dominates over inhibition caused by micronutrient deficiency in the Cd/Zn hyperaccumulator Arabidopsis halleri. Plant Physiol. Biochem. 2020, 155, 252–261. [Google Scholar] [CrossRef]
- Lefèvre, I.; Vogel-Mikuš, K.; Jeromel, L.; Vavpetič, P.; Planchon, S.; Arčon, I.; van Elteren, J.T.; Lepoint, G.; Gobert, S.; Renaut, J.; et al. Differential cadmium and zinc distribution in relation to their physiological impact in the leaves of the accumulating Zygophyllum fabago L. Plant Cell. Environ. 2013, 37, 1299–1320. [Google Scholar] [CrossRef]
- Wang, L.; Yang, H.; Liu, R.; Fan, G. Detoxification strategies and accumulation of oxygen production and flowering of Platanus acerifolia under lead (Pb) stress by transcriptome analysis. Environ. Sci. Pollut. Res. 2015, 22, 12747–12758. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, H.; Bukhat, S.; Rasul, S.; Rehmani, M.I.A.; Noreen, S.; Zafar, Z.U.; Skalicky, M.; Soufan, W.; Brestic, M.; Habib-ur-Rahman, M.; et al. Methyl Jasmonate Alleviated the Adverse Effects of Cadmium Stress in Pea (Pisum sativum L.): A Nexus of Photosystem II Activity and Dynamics of Redox Balance. Front. Plant Sci. 2022, 13, 860664. [Google Scholar] [CrossRef] [PubMed]
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 2021, 11, 111887. [Google Scholar] [CrossRef] [PubMed]
- Bashir, H.; Qureshi, M.I.; Ibrahim, M.M.; Iqbal, M. Chloroplast and photosystems: Impact of cadmium and iron deficiency. Photosynthetica 2015, 53, 321–335. [Google Scholar] [CrossRef]
- Hahn, A.; Vonck, J.; Mills, D.J.; Meier, T.; Kühlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 2018, 360, eaat4318. [Google Scholar] [CrossRef]
- Kato, Y.; Sakamoto, W. FtsH protease in the thylakoid membrane: Physiological functions and the accumulation of protease activity. Front. Plant Sci. 2018, 9, 855. [Google Scholar] [CrossRef]
- Bohler, S.; Sergeant, K.; Jolivet, Y.; Hoffmann, L.; Hausman, J.-F.; Dizengremel, P.; Renaut, J. A physiological and proteomic study of poplar leaves during ozone exposure combined with mild drought. Proteomics 2013, 13, 1737–1754. [Google Scholar] [CrossRef]
- Luo, Y.; Ge, C.; Yang, M.; Long, Y.; Li, M.; Zhang, Y.; Chen, Q.; Sun, B.; Wang, Y.; Wang, X.; et al. Cytosolic/Plastid Glyceraldehyde-3-Phosphate Dehydrogenase Is a Negative Regulator of Strawberry Fruit Ripening. Genes 2020, 11, 580. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Hong, L.; Yang, Y.; Yanping, X.; Xing, H.; Gang, D. Protein Changes in Response to Lead Stress of Lead-Tolerant and Lead-Sensitive Industrial Hemp Using SWATH Technology. Genes 2019, 10, 396. [Google Scholar] [CrossRef]
- Eprintsev, A.T.; Fedorin, D.N.; Cherkasskikh, M.V.; Igamberdiev, A.U. Accumulation of expression of the mitochondrial and cytosolic forms of aconitase in maize leaves via phytochrome. Plant Physiol. Biochem. 2019, 146, 157–162. [Google Scholar] [CrossRef]
- Moeder, W.; del Pozo, O.; Navarre, D.A.; Martin, G.B.; Klessig, D.F. Aconitase plays a role in regulating resistance to oxidative stress and cell death in Arabidopsis and Nicotiana benthamiana. Plant Mol. Biol. 2006, 63, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Jia, X.; Wu, Y.; Cheng, L.; Zhao, T.; Huang, Z.; Wang, Y. Quantitative proteomic analysis of Malus halliana exposed to salt-alkali mixed stress reveals alterations in energy metabolism and stress accumulation. Plant Growth Regul. 2020, 90, 205–222. [Google Scholar] [CrossRef]
- Zhao, X.; Li, F.; Li, K. The 14-3-3 proteins: Regulators of plant metabolism and stress responses. Plant Biol. 2021, 23, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 proteins in plant hormone signaling: Doing several things at once. Front. Plant Sci. 2018, 9, 297. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.; Yu, H.; Peng, J.; Hu, Z.; Chen, L. The role of 14-3-3 proteins in plant growth and response to abiotic stress. Plant Cell Rep. 2021, 41, 833–852. [Google Scholar] [CrossRef]
- Elena-Real, C.A.; González-Arzola, K.; Pérez-Mejías, G.; Díaz-Quintana, A.; Velázquez-Campoy, A.; Desvoyes, B.; Díaz-Moreno, I. Proposed mechanism for regulation of H2O2-induced programmed cell death in plants by binding of cytochrome c to 14-3-3 proteins. Plant J. 2021, 106, 74–85. [Google Scholar] [CrossRef]
- Weckwerth, P.; Ehlert, B.; Romeis, T. ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2014, 38, 544–558. [Google Scholar] [CrossRef]
- Cheng, L.; Gao, X.; Li, S.; Shi, M.; Javeed, H.; Jing, X.; Yang, G.; He, G. Proteomic analysis of soybean [Glycine max (L.) Meer.] seeds during imbibition at chilling temperature. Mol. Breed. 2010, 26, 1–17. [Google Scholar] [CrossRef]
- Futai, M.; Sun-Wada, G.H.; Wada, Y.; Matsumoto, N.; Nakanishi-Matsui, M. Vacuolar-type ATPase: A proton pump to lysosomal trafficking. Proc. Jpn. Acad. Ser. B 2019, 95, 261–277. [Google Scholar] [CrossRef]
- Li, P.; Guo, W. Genome-wide characterization of the Rab gene family in Gossypium by comparative analysis. Bot. Stud. 2017, 58, 26. [Google Scholar] [CrossRef][Green Version]
- Wang, C.Y.; Shen, R.F.; Wang, C.; Wang, W. Root protein profile changes induced by Al exposure in two rice cultivars differing in Al tolerance. J. Proteom. 2013, 78, 281–293. [Google Scholar] [CrossRef]
- Chebli, Y.; Bidhendi, A.J.; Kapoor, K.; Geitmann, A. Cytoskeletal regulation of primary plant cell wall assembly. Curr. Biol. 2021, 31, R681–R695. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.N.; Martin, A.C. Actin-based force generation and cell adhesion in tissue morphogenesis. Curr. Biol. 2021, 31, R667–R680. [Google Scholar] [CrossRef]
- Liu, J.; Geisler, M. Cooperation Between Auxin and Actin During the Process of Plant Polar Growth. In The Cytoskeleton. Plant Cell Monographs; Sahi, V., Baluška, F., Eds.; Springer: Cham, Switzerland, 2019; Volume 24, pp. 101–123. [Google Scholar]
- Hasan, M.K.; Cheng, Y.; Kanwar, M.K.; Chu, X.-Y.; Ahammed, G.J.; Qi, Z.-Y. Responses of plant proteins to heavy metal stress—A review. Front. Plant Sci. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Wu, X.; Chen, J.; Yue, X.; Wei, X.; Zou, J.; Chen, Y.; Su, N.; Cui, J. The zinc-regulated protein (ZIP) family genes and glutathione s-transferase (GST) family genes play roles in Cd resistance and accumulation of pak choi (Brassica campestris ssp. chinensis). Ecotoxicol. Environ. Saf. 2019, 183, 109571. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Fu, Y.; Li, C.; Chen, M.; Gu, Z.; Shan, Y.; Tan, X. Cadmium decreased superoxide anion derived from nadph oxidase through overload of calcium in wheat seedling. Pak. J. Bot. 2020, 52, 1589–1594. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hamed, B.A.; Selim, S.; Beemster, G.; Hozzein, W.N.; Wadaan, M.; Asard, D.; Abuelsoud, W. Maize roots and shoots show distinct profiles of oxidative stress and antioxidant defense under heavy metal toxicity. Environ. Pollut. 2020, 258, 113705. [Google Scholar] [CrossRef]
- Veljović Jovanović, S.; Kukavica, B.; Vidović, M.; Morina, F.; Menckhoff, L. Class III Peroxidases: Functions, Localization and Redox Regulation of Isoenzymes. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D., Palma, J., Corpas, F., Eds.; Springer: Cham, Switzerland, 2018; Volume 1, pp. 269–300. [Google Scholar]
- Zipor, G.; Oren-Shamir, M. Do vacuolar peroxidases act as plant caretakers? Plant Sci. 2013, 199–200, 41–47. [Google Scholar] [CrossRef]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pascual, J.; Canal, M.J.; Escandon, M.; Meijon, M.; Weckwerth, W.; Valledor, L. Integrated physiological, proteomic, and metabolomic analysis of ultra violet (UV) stress responses and adaptation mechanisms in pinus radiata. Mol. Cell. Proteom. 2017, 16, 485–501. [Google Scholar] [CrossRef] [PubMed]
- Romero-Rodriguez, M.C.; Jorrin-Novo, J.V.; Castillejo, M.A. Toward characterizing germination and early growth in the non-orthodox forest tree species Quercus ilex through complementary gel and gel-free proteomic analysis of embryo and seedlings. J. Proteom. 2019, 197, 60–70. [Google Scholar] [CrossRef] [PubMed]
| Treatments | Fresh Weight of Protein (mg g−1) ± SD |
|---|---|
| TC 1 | 2.23 ± 0.2 |
| TC30 2 | 2.43 ± 0.60 |
| TC50 3 | 2.54 ± 0.35 |
| No. | Accession 1 | Protein Name | MW (kDa) 2 | Coverage 3 | Source | Fold Change 4 TC30/TC TC50/TC | Subcellular 5 Localization | |
|---|---|---|---|---|---|---|---|---|
| Photosynthesis | ||||||||
| 1 | B1NWD5 | ATP synthase subunit alpha | 55.564 | 29,7830374 | Manihot esculenta | 0.4 | −7.9 | Chloroplast |
| 2 | P04782 | Chlorophyll a–b binding protein 25 | 28.149 | 28.1954887 | Petunia sp. | −8.3 | −8.3 | Chloroplast |
| 3 | P12332 | Chlorophyll a–b binding protein (fragment) | 22.029 | 19.0243902 | Silene latifolia subsp. alba | −9.1 | −9.1 | Chloroplast |
| 4 | P15194 | Chlorophyll a–b binding protein type 2 member 1B | 29.014 | 31.3868613 | Pinus sylvestris | −12.2 | −12.2 | Chloroplast |
| 5 | P12328 | Chlorophyll a–b binding protein of LHCII type I | 28.359 | 15.1515152 | Lemna gibba | −12.5 | −12.5 | Chloroplast |
| 6 | A1E9K3 | Cytochrome f | 35.341 | 49.0625 | Hordeum vulgare | −5.9 | −5.9 | Chloroplast |
| 7 | P69390 | Cytochrome b559 subunit alpha | 9.439 | 39.7590361 | Hordeum vulgare | −3.9 | −2.6 | Chloroplast |
| 8 | O64422 | Fructose-1,6-bisphosphatase | 43.577 | 26.8472906 | Oryza sativa subsp. japonica | −0.3 | −1.9 | Chloroplast |
| 9 | Q01516 | Fructose-bisphosphate aldolase 1 (fragment) | 38.633 | 15.7303370 | Pisum sativum | −4.2 | −4.2 | Chloroplast |
| 10 | P17606 | Malate dehydrogenase (NADP) 1 | 46.426 | 17.4825174 | Sorghum bicolor | 0.1 | −8.1 | Chloroplast |
| 11 | B3TNA5 | NAD(P)H-quinone oxidoreductase subunit H | 45.795 | 30.2798982 | Brachypodium distachyon | −7.4 | −7.4 | Chloroplast |
| 12 | P46722 | NAD(P)H-quinone oxidoreductase subunit I | 21.143 | 51.1111111 | Zea mays | −10.0 | −10.0 | Chloroplast |
| 13 | O49079 | Oxygen-evolving enhancer protein 1 | 34.848 | 21.8844984 | Fritillaria agrestis | −0.3 | −1.6 | Chloroplast |
| 14 | Q09ME8 | Photosystem II reaction center protein H | 7.664 | 23.2876712 | Citrus sinensis | −9.4 | −9.4 | Chloroplast |
| 15 | Q40073 | Ribulose bisphosphate carboxylase/oxygenase activase A | 51.041 | 20.4741379 | Hordeum vulgare | −0.5 | −1.5 | Chloroplast |
| 16 | Q37227 | Ribulose bisphosphate carboxylase large chain (fragment) | 49.103 | 28.8939051 | Iris germanica | −7.2 | −7.2 | Chloroplast |
| 17 | O23813 | Sulfite reductase (ferredoxin) | 69.971 | 19.0551181 | Zea mays | −4.3 | −12.5 | Chloroplast |
| 18 | Q0P3P2 | ATP synthase subunit beta | 51.83 | 21.9461697 | Ostreococcus tauri | 0.3 | 3.5 | Chloroplast |
| 19 | P55240 | Glucose-1-phosphate adenylyltransferase small subunit (fragment) | 13.238 | 20 | Zea mays | 0 | 8.1 | Chloroplast, Amyloplast |
| 20 | Q9SQL2 | Chlorophyll a–b binding protein P4 | 27.212 | 18.2539683 | Pisum sativum | 10.7 | 2.5 | Chloroplast |
| 21 | Q9LD57 | Phosphoglycerate kinase 1 | 50.081 | 25.3638253 | Arabidopsis thaliana | 0 | 12.0 | Chloroplast |
| 22 | A6YGB8 | Photosystem II protein D1 | 38.125 | 18.3139535 | Pleurastrum terricola | 14.0 | 13.9 | Chloroplast |
| 23 | P08927 | RuBisCO large subunit-binding protein subunit beta | 62.945 | 21.3445378 | Pisum sativum | 2.7 | 7.3 | Chloroplast |
| Protein metabolism | ||||||||
| 24 | O81154 | Cysteine synthase | 34.32 | 19.3846154 | Solanum tuberosum | −7.2 | −7.2 | Cytoplasm |
| 25 | Q9LST6 | Proteasome subunit beta type-2 | 23.463 | 19.8113207 | Oryza sativa subsp. japonica | −5.7 | −5.7 | Nucleus, cytoplasm |
| 26 | A2YXU2 | Proteasome subunit alpha type-7-A | 27.279 | 35.3413655 | Oryza sativa subsp. indica | −7.3 | 0.7 | Nucleus, cytoplasm |
| 27 | A1E9M6 | 30S ribosomal protein S8 | 15.7 | 19.1176470 | Hordeum vulgare | −7.4 | −7.4 | Chloroplast |
| 28 | P51427 | 40S ribosomal protein S5-2 | 22.907 | 25.6038647 | Arabidopsis thaliana | −10.4 | −10.4 | Cell Wall, cytosol, plasma membrane, ribosomes |
| 29 | Q949H0 | 40S ribosomal protein S7 | 22.064 | 16.7539267 | Hordeum vulgare | −8.3 | −8.3 | Ribosome |
| 30 | P0DKK8 | 40S ribosomal protein S10-1 | 20.25 | 22.4043715 | Oryza sativa subsp. japonica | −0.9 | −8.6 | Cytoplasm |
| 31 | P17093 | 40S ribosomal protein S11 | 17.822 | 16.9811321 | Glycine max | −6.8 | −6.8 | Cytosol |
| 32 | B7F845 | 60S ribosomal protein L10a | 24.467 | 20.8333333 | Oryza sativa subsp. japonica | −8.1 | −8.1 | Cytosol |
| 33 | P42794 | 60S ribosomal protein L11-2 | 20.848 | 17.5824175 | Arabidopsis thaliana | −7.3 | −7.3 | Nucleus, cytoplasm |
| 34 | O48557 | 60S ribosomal protein L17 | 19.494 | 31.5789473 | Zea mays | −6.4 | −6.4 | Cytosol |
| 35 | P49690 | 60S ribosomal protein L23 | 15.017 | 48.5714285 | Arabidopsis thaliana | −2.1 | −9.5 | Cytosol, endoplasmic reticulum, extracellular region or secreted, nucleus |
| 36 | P43643 | Elongation factor 1-alpha | 49.251 | 27.0693512 | Nicotiana tabacum | 1.5 | −8.3 | Cytoplasm |
| 37 | Q6EN80 | 30S ribosomal protein S19. | 10.689 | 19.3548387 | Oryza nivara | −0.2 | 1.7 | Chloroplast |
| 38 | P50300 | S-adenosylmethionine synthase | 43.141 | 16.2849872 | Pinus banksiana | 10.1 | 10.0 | Cytoplasm |
| Carbohydrate and energy metabolism | ||||||||
| 39 | P12863 | Triosephosphate isomerase | 27.008 | 38.3399209 | Zea mays | −1.5 | −1.2 | Cytoplasm |
| 40 | P92549 | ATP synthase subunit alpha | 55.011 | 26.035503 | Arabidopsis thaliana | 0.8 | −9.6 | Mitochondrion |
| 41 | P00056 | Cytochrome c | 12.005 | 39.6396396 | Zea mays | 1.0 | −9.8 | Mitochondrion |
| 42 | P42895 | Enolase 2 | 48.132 | 45.0672646 | Zea mays | −6.4 | −6.4 | Cytoplasm |
| 43 | Q6XZ79 | Fructokinase-1 | 34.669 | 26.0061919 | Zea mays | −2.1 | 0.2 | Cytosol |
| 44 | P26518 | Glyceraldehyde-3-phosphate dehydrogenase | 36.959 | 20.5278592 | Magnolia liliiflora | 1.4 | −8.5 | Cytoplasm |
| 45 | P26517 | Glyceraldehyde-3-phosphate dehydrogenase 1 | 36.491 | 44.2136499 | Hordeum vulgare | −10.7 | −10.7 | Cytoplasm |
| 46 | Q09054 | Glyceraldehyde-3-phosphate dehydrogenase 2 | 36.519 | 39.7626112 | Zea mays | 1.2 | −8.1 | Cytoplasm |
| 47 | P93805 | Phosphoglucomutase, cytoplasmic 2 | 63.002 | 39.4511149 | Zea mays | −0.3 | −1.6 | Cytoplasm |
| 48 | O24047 | Malate dehydrogenase | 35.475 | 25.6024096 | Mesembryanthemum crystallinum | −7.7 | −7.7 | Cytoplasm |
| 49 | Q9ZP06 | Malate dehydrogenase 1. | 35.782 | 18.4750733 | Arabidopsis thaliana | −8.4 | −8.4 | Mitochondrion |
| 50 | P80269 | NADH dehydrogenase (ubiquinone) iron–sulfur protein 8. | 26.361 | 23.1441048 | Solanum tuberosum | −8.8 | −8.8 | Mitochondrion |
| 51 | P42066 | Phosphoenolpyruvate carboxykinase (ATP) | 74.35 | 23.2835820 | Cucumis sativus | 0.8 | −2.0 | Cytoplasm |
| 52 | Q9XFA2 | Phosphoenolpyruvate carboxykinase (ATP) 2 | 68.59 | 22.5239617 | Urochloa panicoides | −0.7 | −9.4 | Cytoplasm |
| 53 | Q6YZX6 | Putative aconitate hydratase | 98.021 | 15.0334075 | Oryza sativa subsp. japonica | 1.5 | −6.4 | Cytoplasm |
| 54 | Q6ZFT5 | Ribose-phosphate pyrophosphokinase 4 | 36.126 | 17.8461538 | Oryza sativa subsp. japonica | 1.1 | −6.6 | Cytosol, plasma membrane, cytoplasm, |
| 55 | Q42971 | Enolase | 47.942 | 39.6860986 | Oryza sativa subsp. japonica | 1.5 | 0 | Cytoplasm |
| 56 | Q6AVT2 | Glucose-1-phosphate adenylyltransferase large subunit 1 | 55.392 | 17.4168297 | Oryza sativa subsp. japonica | 1.1 | −8.3 | Chloroplast, amyloplast |
| 57 | Q6Z1G7 | Pyruvate dehydrogenase E1 component subunit beta-1 | 39.919 | 15.2406417 | Oryza sativa subsp. japonic | 8.9 | 0 | Mitochondrion |
| Antioxidant defense and stress response | ||||||||
| 58 | P38559 | Glutamine synthetase root isozyme 1 | 39.226 | 24.6498599 | Zea mays | −6.7 | −6.7 | Cytoplasm |
| 59 | O23877 | Ferredoxin-NADP reductase, embryo isozyme | 41.788 | 16.9312169 | Oryza sativa subsp. japonica | 0.3 | −7.7 | Chloroplast |
| 60 | Q6QPJ6 | Peroxiredoxin Q | 23.402 | 22.5352112 | Populus jackii | −7.4 | −7.4 | Chloroplast |
| 61 | Q69SV0 | Probable L-ascorbate peroxidase 8 | 51.156 | 19.2468619 | Oryza sativa subsp. japonica | −0.3 | −8.3 | Chloroplast |
| 62 | Q02028 | Stromal 70 kDa heat-shock-related protein. | 75.469 | 24.5042493 | Pisum sativum | 1.5 | −9.0 | Chloroplast |
| 63 | Q6ZFU6 | Thioredoxin reductase NTRB | 34.655 | 17.2205438 | Oryza sativa subsp. japonica | 0.6 | −8.0 | Cytoplasm |
| 64 | Q9BAE0 | ATP-dependent zinc metalloprotease FTSH | 75.633 | 19.8300283 | Medicago sativa | 7.8 | 8.2 | Chloroplast |
| 65 | P14655 | Glutamine synthetase | 46.613 | 18.6915888 | Oryza sativa subsp. japonica | 7.8 | 0 | Chloroplast |
| 66 | Q01899 | Heat shock 70 kDa protein | 72.493 | 15.7037037 | Phaseolus vulgaris | 1.9 | −9.6 | Mitochondrion |
| 67 | P27322 | Heat shock cognate 70 kDa protein 2 | 70.663 | 30.7453416 | Solanum lycopersicum | 9.5 | 8.4 | Cytoplasm |
| 68 | Q08275 | 17.0 kDa class II heat shock protein | 17.036 | 18.1818181 | Zea mays | 1.8 | 2.3 | Cytoplasm |
| 69 | P27880 | 18.2 kDa class I heat shock protein | 18.154 | 19.6202531 | Medicago sativa | 6.6 | 0 | Cytoplasm |
| 70 | A5H8G4 | Peroxidase 1 | 38.33 | 17.7111717 | Zea mays | 2.2 | 2.4 | Vacuoles |
| 71 | Q9SMB1 | Spermidine synthase 1 | 35.124 | 15.7894737 | Oryza sativa subsp. japonica | 1.8 | 1.3 | |
| Proteins involved in RNA and DNA Processing | ||||||||
| 72 | O48556 | Soluble inorganic pyrophosphatase | 24.354 | 30.3738318 | Zea mays | −7.7 | 0.3 | Cytoplasm |
| 73 | Q99070 | Glycine-rich RNA-binding protein 2 | 16.35 | 23.8095238 | Sorghum bicolor | 1.4 | 2.2 | |
| Proteins involved in Growth, Development and Cellular Organization | ||||||||
| 74 | P20904 | Actin | 41.732 | 28.3819629 | Volvox carteri | 1.4 | 3.0 | Cytoskeleton |
| 75 | P02582 | Actin-1 | 41.591 | 20.2666667 | Zea mays | 8.4 | 9.2 | Cytoskeleton |
| 76 | Q9AY76 | Actin-depolymerizing factor 2 | 16.783 | 22.7586206 | Oryza sativa subsp. japonica | 4.3 | 0 | Cytoskeleton, Cytoplasm |
| 77 | Q41764 | Actin-depolymerizing factor 3 | 15.89 | 18.7050359 | Zea mays | 0 | 8.3 | Cytoplasm |
| 78 | P53492 | Actin-7 | 41.709 | 44.0318302 | Arabidopsis thaliana | 1.5 | 7.6 | Cytoskeleton |
| 79 | Q41738 | Thiamine thiazole synthase 1 | 37.081 | 19.7740113 | Zea mays | −0.5 | 1.7 | Chloroplast |
| Signal Transduction | ||||||||
| 80 | Q9SP07 | 14-3-3-Like protein | 29.235 | 39.7683398 | Lilium longiflorum | 0.1 | 2.1 | |
| 81 | Q7XTE8 | 14-3-3-Like protein GF14-B | 29.845 | 64.5038167 | Oryza sativa subsp. japonica | 1.0 | 2.2 | Nucleus, cytoplasm |
| 82 | Q2R2W2 | 14-3-3-Like protein GF14-D | 29.244 | 39.6226415 | Oryza sativa subsp. japonica | 2.7 | 2.4 | |
| 83 | P49104 | Ras-related protein Rab-2-B | 23.046 | 35.7142857 | Zea mays | 6.4 | 7.3 | Golgi apparatus, endoplasmic reticulum |
| 84 | P28186 | Ras-related protein RABE1c | 23.82 | 22.2222222 | Arabidopsis thaliana | −7.2 | 1.6 | Plasma membrane, Golgi apparatus |
| 85 | O49513 | Ras-related protein RABA1e | 24.315 | 23.0414747 | Arabidopsis thaliana | 8.6 | 7.6 | Plasma membrane |
| 86 | O24461 | Ras-related protein Rab7 | 23.212 | 19.8067632 | Prunus armeniaca | 0 | 6.3 | Plasma membrane |
| Membrane transport and cell wall metabolism | ||||||||
| 87 | P27080 | ADP, ATP carrier protein | 33.506 | 19.4805194 | Chlamydomonas reinhardtii | 0 | 6.0 | |
| 88 | P27081 | ADP/ATP carrier protein, mitochondrial (fragment) | 41.802 | 17.8756477 | Solanum tuberosum | 0 | 7.4 | Mitochondrion |
| 89 | P29036 | Ferritin-1 | 28.007 | 18.1102362 | Zea mays | 1.7 | 0.4 | Chloroplast, plastid |
| 90 | Q05737 | GTP-binding protein YPTM2 | 22.461 | 46.7980296 | Zea mays | 6.2 | 7.5 | Plasma membrane |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salas-Moreno, M.; Castillejo, M.Á.; Rodríguez-Cavallo, E.; Marrugo-Negrete, J.; Méndez-Cuadro, D.; Jorrín-Novo, J. Proteomic Changes in Paspalum fasciculatum Leaves Exposed to Cd Stress. Plants 2022, 11, 2455. https://doi.org/10.3390/plants11192455
Salas-Moreno M, Castillejo MÁ, Rodríguez-Cavallo E, Marrugo-Negrete J, Méndez-Cuadro D, Jorrín-Novo J. Proteomic Changes in Paspalum fasciculatum Leaves Exposed to Cd Stress. Plants. 2022; 11(19):2455. https://doi.org/10.3390/plants11192455
Chicago/Turabian StyleSalas-Moreno, Manuel, María Ángeles Castillejo, Erika Rodríguez-Cavallo, José Marrugo-Negrete, Darío Méndez-Cuadro, and Jesús Jorrín-Novo. 2022. "Proteomic Changes in Paspalum fasciculatum Leaves Exposed to Cd Stress" Plants 11, no. 19: 2455. https://doi.org/10.3390/plants11192455
APA StyleSalas-Moreno, M., Castillejo, M. Á., Rodríguez-Cavallo, E., Marrugo-Negrete, J., Méndez-Cuadro, D., & Jorrín-Novo, J. (2022). Proteomic Changes in Paspalum fasciculatum Leaves Exposed to Cd Stress. Plants, 11(19), 2455. https://doi.org/10.3390/plants11192455







