Identification of Key Genes Related to Dormancy Control in Prunus Species by Meta-Analysis of RNAseq Data
Abstract
:1. Introduction
2. Results
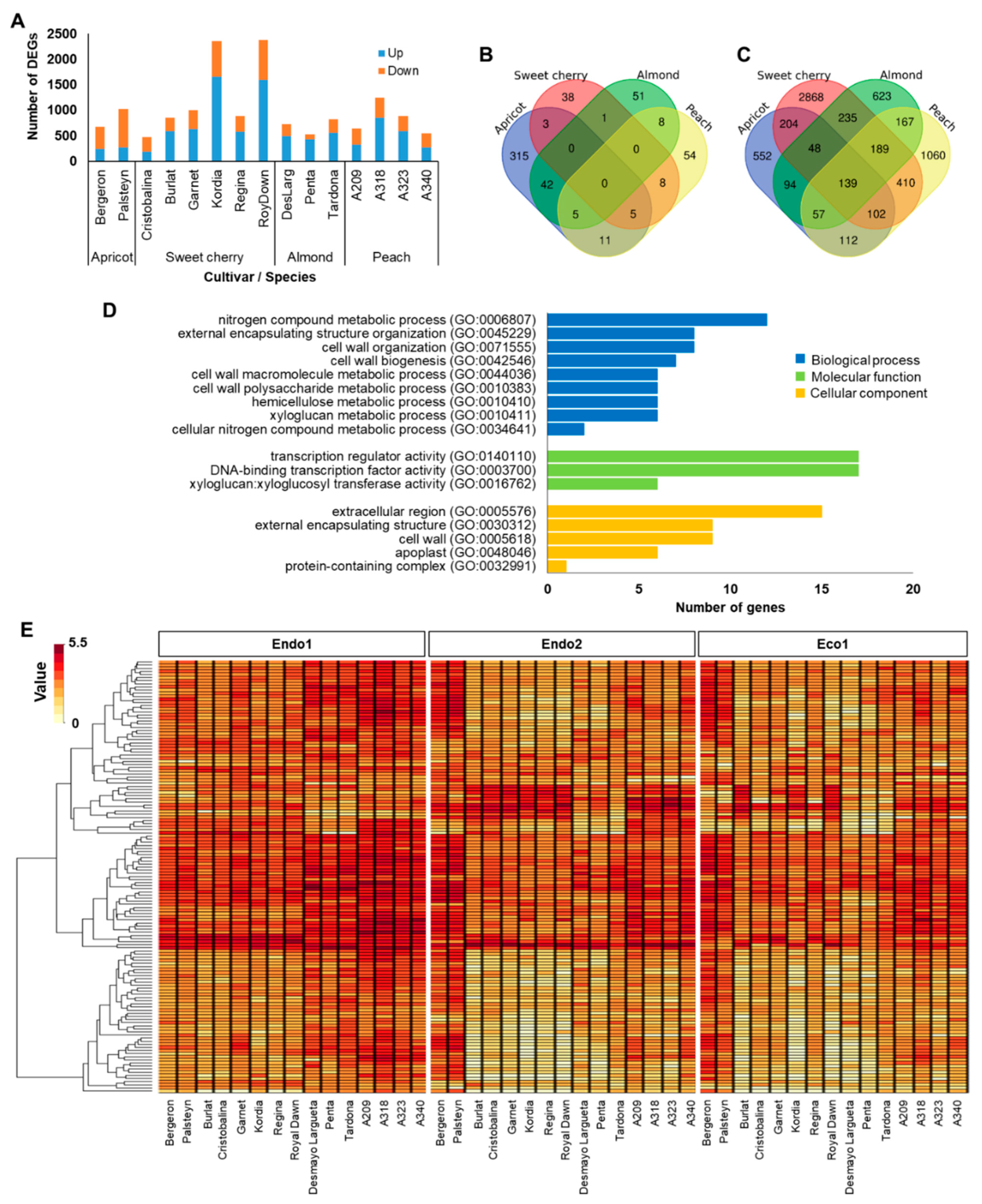
2.1. Summary of Meta-Analysis of RNAseq Data
2.2. Endodormancy-Related Genes
2.3. Ecodormancy-Related Genes
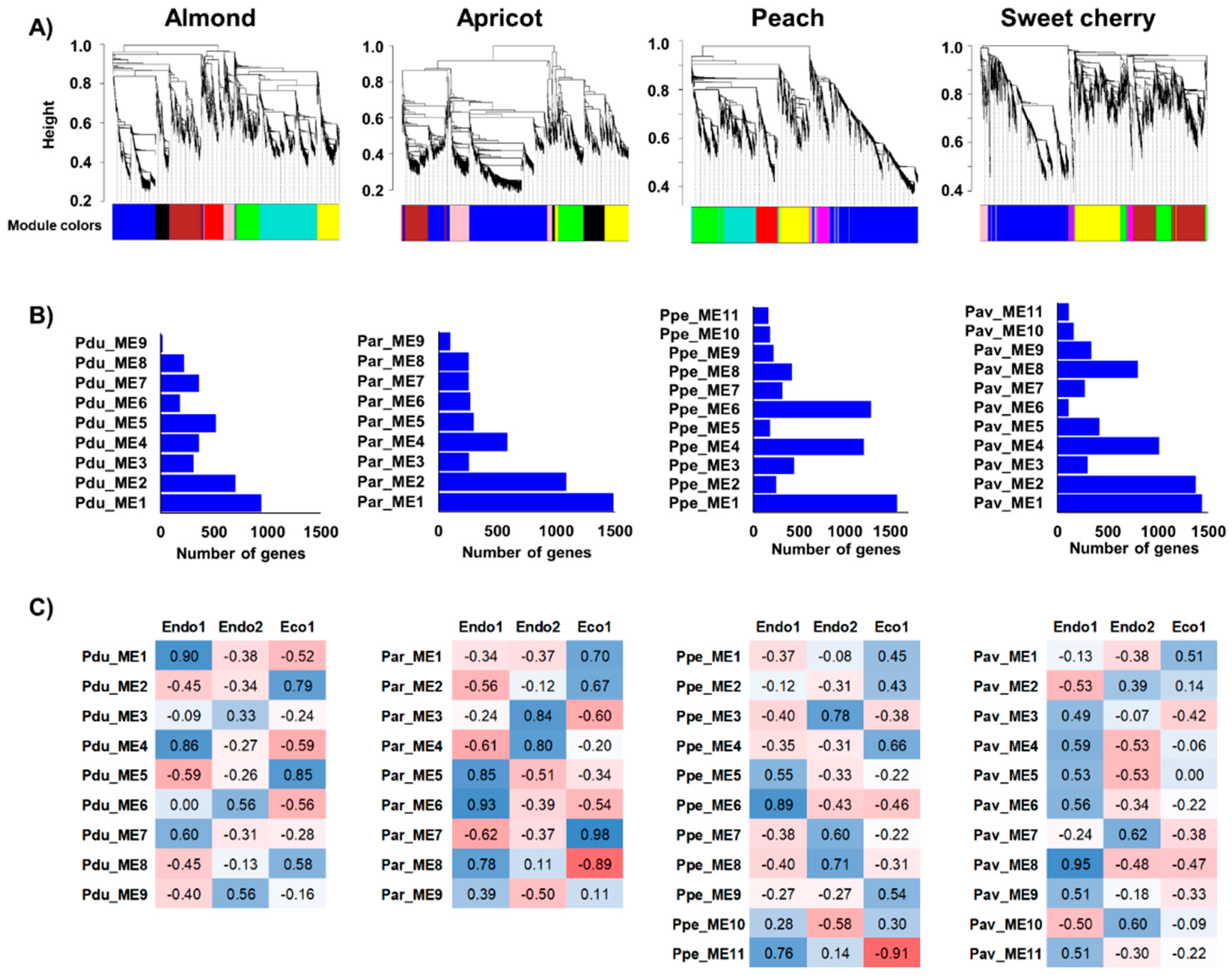
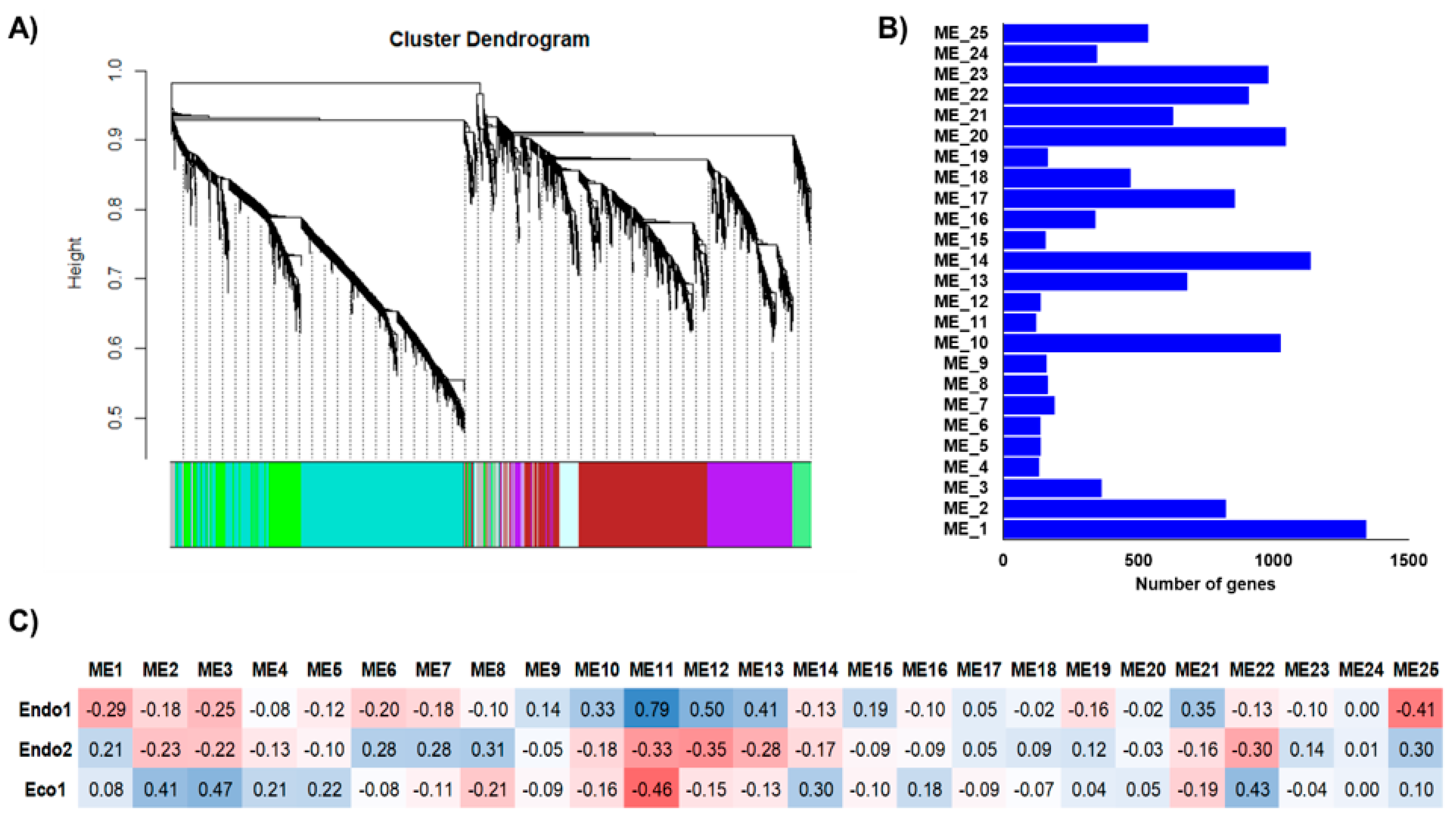
2.4. Individual and Integrated Prunus Co-Expression Networks
2.5. Candidate Genes in Major Prunus Bloom Time and Dormancy-Related QTLs
3. Discussion
4. Materials and Methods
4.1. Selection of RNAseq Studies and Sample Points Normalization
4.2. Read Alignment and Differentially Expressed Gene Analysis
4.3. GO Enrichment and Expression Profile Analysis
4.4. Gene Co-Expression Network Analysis
4.5. Candidate Gene Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Olsen, J.E. Light and temperature sensing and signaling in induction of bud dormancy in woody plants. Plant Mol. Biol. 2010, 73, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endo-, para-and ecodormancy: Physiological terminology and classification for dormancy research. Hortic. Sci. 1987, 22, 271–277. [Google Scholar]
- Rohde, A.; Bhalerao, R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007, 12, 217–223. [Google Scholar] [CrossRef]
- Fadón, E.; Herrero, M.; Rodrigo, J. Flower bud dormancy in Prunus species. In Advances in Plant Dormancy; Springer: Berlin/Heidelberg, Germany, 2015; pp. 123–135. [Google Scholar] [CrossRef]
- Cooke, J.E.K.; Eriksson, M.E.; Junttila, O. The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant Cell Environ. 2012, 35, 1707–1728. [Google Scholar] [CrossRef]
- Luedeling, E. Climate change impacts on winter chill for temperate fruit and nut production: A review. Sci. Hortic. 2012, 144, 218–229. [Google Scholar] [CrossRef]
- Fadón, E.; Herrera, S.; Guerrero, B.I.; Guerra, M.R.; Rodrigo, J. Chilling and heat requirements of temperate stone fruit trees (Prunus sp.). Agronomy 2020, 10, 409. [Google Scholar] [CrossRef]
- Fan, S.; Bielenberg, D.G.; Zhebentyayeva, T.N.; Reighard, G.L.; Okie, W.R.; Holland, D.; Abbott, A.G. Mapping quantitative trait loci associated with chilling requirement, heat requirement and bloom date in peach (Prunus persica). New Phytol. 2010, 185, 917–930. [Google Scholar] [CrossRef]
- Campoy, J.A.; Ruiz, D.; Allderman, L.; Cook, N.; Egea, J. The fulfilment of chilling requirements and the adaptation of apricot (Prunus armeniaca L) in warm winter climates: An approach in Murcia (Spain) and the Western Cape (South Africa). Eur. J. Agron. 2011, 37, 43–55. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Dicenta, F.; Martinez-Gomez, P. Inheritance of chilling and heat requirements for flowering in almond and QTL analysis. Tree Genet. Genomes 2012, 8, 379–389. [Google Scholar] [CrossRef]
- Castède, S.; Campoy, J.A.; Quero-García, J.; Le Dantec, L.; Lafargue, M.; Barreneche, T.; Weden, B.; Dirlewanger, E. Genetic determinism of phenological traits highly affected by climate change in Prunus avium: Flowering date dissection into chilling and heat requirements. New Phytol. 2014, 202, 703–715. [Google Scholar] [CrossRef]
- Guo, L.; Dai, J.H.; Wang, M.C.; Xu, J.C.; Luedeling, E. Responses of spring phenology in temperate zone trees to climate warming: A case study of apricot flowering in China. Agric. For. Meteorol. 2015, 201, 1–7. [Google Scholar] [CrossRef]
- Atagul, O.; Calle, A.; Demirel, G.; Lawton, J.M.; Bridges, W.C.; Gasic, K. Estimating heat requirement for flowering in peach germplasm. Agronomy 2022, 12, 1002. [Google Scholar] [CrossRef]
- Badeck, F.W.; Bondeau, A.; Böttcher, K.; Doktor, D.; Lucht, W.; Schaber, J.J.; Sitch, S. Responses of spring phenology to climate change. New Phytol. 2004, 162, 295–309. [Google Scholar] [CrossRef]
- Atkinson, C.J.; Brennan, R.M.; Jones, H.G. Declining chilling and its impact on temperate perennial crops. Environ. Exp. Bot. 2013, 91, 48–62. [Google Scholar] [CrossRef]
- Vitasse, Y.; Lenz, A.; Körner, C. The interaction between freezing tolerance and phenology in temperate deciduous trees. Front. Plant Sci. 2014, 5, 541. [Google Scholar] [CrossRef]
- Lloret, A.; Badenes, M.L.; Ríos, G. Modulation of dormancy and growth responses in reproductive buds of temperate trees. Front. Plant Sci. 2018, 9, 1368. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Quero-García, J.; Le Dantec, L.; Lambert, P.; Ruiz, D.; Dondini, L.; Illa, E.; Quilot-Turion, B.; Audergon, J.M.; Tartarini, S.; et al. Comparison of the genetic determinism of two key phenologycal traits, flowering and maturity dates, in three Prunus species: Peach, apricot and sweet cherry. Heredity 2012, 109, 280–292. [Google Scholar] [CrossRef]
- Zhebentyayeva, T.N.; Fan, S.; Chandra, A.; Bielenberg, D.G.; Reighard, G.L.; Okie, W.R.; Abbott, A.G. Dissection of chilling requirement and bloom date QTLs in peach using a whole genome sequencing of sibling trees from an F2 mapping population. Tree Genet. Genomes 2014, 10, 35–51. [Google Scholar] [CrossRef]
- Bielenberg, D.G.; Rauh, B.; Fan, S.; Gasic, K.; Abbott, A.G.; Reighard, G.L.; Okie, W.R.; Wells, C.E. Genotyping by sequencing for SNP-based linkage map construction and QTL analysis of chilling requirement and bloom date in peach [Prunus persica (L.) Batsch]. PLoS ONE 2015, 10, e0139406. [Google Scholar] [CrossRef]
- Salazar, J.A.; Ruiz, D.; Campoy, J.A.; Tartarini, S.; Dondini, L.; Martínez-Gómez, P. Inheritance of reproductive phenology traits and relates QTL identification in apricot. Tree Genet. Genomes 2016, 12, 71. [Google Scholar] [CrossRef]
- Calle, A.; Cai, L.; Iezzoni, A.; Wünsch, A. Genetic dissection of bloom time in low chilling sweet cherry (Prunus avium L.) using a multi-family QTL approach. Front. Plant Sci. 2020, 10, 1647. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, S.; Lawton-Rauh, A.L.; Reighard, G.L.; Abbott, A.G.; Bielenberd, D.G. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 2009, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, R.; Yamane, H.; Ooka, T.; Jotatsu, H.; Kitamura, Y.; Akagi, T.; Tao, R. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 2011, 157, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Traver, C.; Guerrero, B.I.; Badenes, M.L.; Rodrigo, J.; Ríos, G.; Lloret, A. Structure and expression of bud dormancy-associated MADS-box genes (DAM) in European plum. Front. Plant Sci. 2020, 11, 1288. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Z.; Li, H.; Jiu, S.; Qu, Y.; Wang, L.; Ma, C.; Xu, W.; Wang, S.; Zhang, C. Dormancy-Associated MADS-Box (DAM) genes influence chilling requirement of sweet cherries and co-regulate flower development with SOC1 gene. Int. J. Mol. Sci. 2020, 21, 921. [Google Scholar] [CrossRef] [PubMed]
- Calle, A.; Grimplet, J.; Le Dantec, L.; Wünsch, A. Identification and characterization of DAMs mutations associated with early blooming in sweet cherry, and validation of DNA-based markers for selection. Front. Plant Sci. 2021, 12, 621491. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Chhajed, S.; Vashisth, T.; Olmstead, M.A.; Olmstead, J.W.; Colquhoun, T.A. Transcriptomic study of early responses to the bud dormancy-breaking agent hydrogen cyanamide in ‘TropicBeauty’ peach. J. Am. Soc. Hortic. Sci. 2019, 144, 244–256. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, P.Y.; Zhong, S.; Dardick, C.; Callahan, A.; An, Y.Q.; van Knocker, S.; Yang, Y.; Zhong, G.Y.; Abbott, A.; et al. Thermal-responsive genetic and epigenetic regulation of DAM cluster controlling dormancy and chilling requirement in peach floral buds. Hortic. Res. 2020, 7, 114. [Google Scholar] [CrossRef]
- Yu, J.; Conrad, A.O.; Decroocq, V.; Zhebentyayeva, T.; Williams, D.E.; Bennett, D.; Roch, G.; Audergon, J.-M.; Dardick, C.; Liu, Z.; et al. Distinctive gene expression patterns define endodormancy to ecodormancy transition in apricot and peach. Front. Plant Sci. 2020, 11, 180. [Google Scholar] [CrossRef]
- Prudencio, Á.S.; Hoeberichts, F.A.; Dicenta, F.; Martínez-Gómez, P.; Sánchez-Pérez, R. Identification of early and late flowering time candidate genes in endodormant and ecodormant almond flower buds. Tree Physiol. 2021, 41, 589–605. [Google Scholar] [CrossRef]
- Ionescu, I.A.; López-Ortega, G.; Burow, M.; Bayo-Canha, A.; Junge, A.; Gericke, O.; Møller, B.L.; Sánchez-Pérez, R. Transcriptome and metabolite changes during hydrogen cyanamide-induced floral bud break in sweet cherry. Front. Plant Sci. 2017, 8, 1233. [Google Scholar] [CrossRef] [PubMed]
- Vimont, N.; Fouché, M.; Campoy, J.A.; Tong, M.; Arkoun, M.; Yvin, J.C.; Wigge, P.A.; Dirlewanger, E.; Cortijo, S.; Wenden, B. From bud formation to flowering: Transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy. BMC Genom. 2019, 20, 974. [Google Scholar] [CrossRef]
- Vimont, N.; Quah, F.X.; Schöepfer, D.G.; Roudier, F.; Dirlewanger, E.; Wigge, P.A.; Wenden, B.; Cortijo, S. ChIP-seq and RNA-seq for complex and low-abundance tree buds reveal chromatin and expression co-dynamics during sweet cherry bud dormancy. Tree Genet. Genomes 2020, 16, 9. [Google Scholar] [CrossRef]
- Villar, L.; Lienqueo, I.; Llanes, A.; Rojas, P.; Perez, J.; Correa, F.; Sagredo, B.; Masciarelli, O.; Luna, V.; Almada, R. Comparative transcriptomic analysis reveals novel roles of transcription factors and hormones during the flowering induction and floral bud differentiation in sweet cherry trees (Prunus avium L. cv. Bing). PLoS ONE 2020, 15, e0230110. [Google Scholar] [CrossRef]
- Rothkegel, K.; Sandoval, P.; Soto, E.; Ulloa, L.; Riveros, A.; Lillo-Carmona, V.; Cáceres-Molina, J.; Almeida, A.M.; Meneses, C. Dormant but active: Chilling accumulation modulates the epigenome and transcriptome of Prunus avium during bud dormancy. Front. Plant Sci. 2020, 11, 1115. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, Y.; Xin, D.; Chen, W.; Shao, X.; Wang, Y.; Guo, W. RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus). Gene 2015, 555, 362–376. [Google Scholar] [CrossRef] [PubMed]
- Habu, T.; Yamane, H.; Igarashi, K.; Hamada, K.; Yano, K.; Tao, R. 454-pyrosequencing of the transcriptome in leaf and flower buds of Japanese apricot (Prunus mume Sieb. et Zucc.) at different dormant stages. J. Jpn. Soc. Hortic. Sci. 2012, 81, 239–250. [Google Scholar] [CrossRef]
- Zhong, W.; Gao, Z.; Zhuang, W.; Shi, T.; Zhang, Z.; Ni, Z. Genome-wide expression profiles of seasonal bud dormancy at four critical stages in Japanese apricot. Plant Mol. Biol. 2013, 83, 247–264. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhuo, X.; Zhao, K.; Zheng, T.; Han, Y.; Yuan, C.; Zhang, Q. Transcriptome profiles reveal the crucial roles of hormone and sugar in the bud dormancy of Prunus mume. Sci. Rep. 2018, 8, 5090. [Google Scholar] [CrossRef]
- Wang, Z.; Song, S.; Sheng, S.; Tian, J.; Wu, R.; Pang, X. Comparative transcriptome analysis identifies differentially expressed genes between normal and late-blooming Siberian apricot. J. For. Res. 2019, 30, 2277–2288. [Google Scholar] [CrossRef]
- Barros, P.M.; Gonçalves, N.; Saibo, N.J.; Oliveira, M.M. Cold acclimation and floral development in almond bud break: Insights into the regulatory pathways. J. Exp. Bot. 2012, 63, 4585–4596. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhou, Y.; Ahmad, S.; Yong, X.; Xie, X.; Han, Y.; Li, Y.; Sun, L.; Zhang, Q. PmCBFs synthetically affect PmDAM6 by alternative promoter binding and protein complexes towards the dormancy of bud for Prunus mume. Sci. Rep. 2018, 8, 4527. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Sun, W.; Xu, Y.; Sabir, I.A.; Abdullah, M.; Wang, S.; Jiu, S.; Zhang, C. Cold induced genes (CIGs) regulate flower development and dormancy in Prunus avium L. Plant Sci. 2021, 313, 111061. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Han, X.; Wang, Q.; Wang, X.; Chen, X.; Li, L.; Fu, X.; Gao, D. EARLY BUD BREAK 1 triggers bud break in peach trees by regulating hormone metabolism, the cell cycle, and cell wall modifications. J. Exp. Bot. 2020, 71, 3512–3523. [Google Scholar] [CrossRef]
- Beauvieux, R.; Wenden, B.; Dirlewanger, E. Bud dormancy in perennial fruit tree species: A pivotal role for oxidative cues. Front. Plant Sci. 2018, 9, 657. [Google Scholar] [CrossRef]
- Hernández, J.A.; Díaz-Vivancos, P.; Acosta-Motos, J.R.; Alburquerque, N.; Martínez, D.; Carrera, E.; García-Bruntón, J.; Barba-Espín, G. Interplay among antioxidant system, hormone profile and carbohydrate metabolism during bud dormancy breaking in a high-chill peach variety. Antioxidants 2021, 10, 560. [Google Scholar] [CrossRef]
- Liu, J.; Sherif, S.M. Hormonal orchestration of bud dormancy cycle in deciduous woody perennials. Front. Plant Sci. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Vimont, N.; Schwarzenberg, A.; Domijan, M.; Donkpegan, A.S.L.; Beauvieux, R.; le Dantec, L.; Arkoun, M.; Jamois, F.; Yvin, J.C.; Wigge, P.A.; et al. Fine tuning of hormonal signaling is linked to dormancy status in sweet cherry flower buds. Tree Physiol. 2021, 41, 544–561. [Google Scholar] [CrossRef]
- Li, P.; Zheng, T.; Zhang, Z.; Liu, W.; Qiu, L.; Wang, J.; Cheng, T.; Zhang, Q. Integrative identification of crucial genes associated with plant hormone-mediated bud dormancy in Prunus mume. Front. Genet. 2021, 12, 698598. [Google Scholar] [CrossRef]
- Benny, J.; Pisciotta, A.; Caruso, T.; Martinelli, F. Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of R.NA-Seq data. BMC Plant Biol. 2019, 19, 194. [Google Scholar] [CrossRef] [Green Version]
- Balan, B.; Caruso, T.; Martinelli, F. Gaining insight into exclusive and common transcriptomic features linked with biotic stress responses in Malus. Front. Plant Sci. 2017, 8, 1569. [Google Scholar] [CrossRef]
- Canton, M.; Forestan, C.; Bonghi, C.; Varotto, S. Meta-analysis of RNA-Seq studies reveals genes with dominant functions during flower bud endo- to eco-dormancy transition in Prunus species. Sci. Rep. 2021, 11, 13173. [Google Scholar] [CrossRef] [PubMed]
- Olukolu, B.A.; Trainin, T.; Fan, S.; Kole, C.; Bielenberg, D.G.; Reighard, G.L.; Abbott, A.G.; Holland, D. Genetic linkage mapping for molecular dissection of chilling requirements and budbreak in apricot (Prunus armeniaca L.). Genome 2009, 52, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Romeu, J.F.; Monforte, A.; Sánchez, G.; Granell, A.; García-Brutón, J.; Badenes, M.L.; Ríos, G. Quantitative trait loci affecting reproductive phenology in peach. BMC Plant Biol. 2014, 14, 52. [Google Scholar] [CrossRef]
- Kitamura, Y.; Habu, T.; Yamane, H.; Nishiyama, S.; Kajita, K.; Soube, T.; Kawai, T.; Numaguchi, K.; Nakazaki, T.; Kitajima, A.; et al. Identification of QTLs controlling chilling and heat requirements for dormancy release and bud break in Japanese apricot (Prunus mume). Tree Genet. Genomes 2018, 14, 33. [Google Scholar] [CrossRef]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 1–18. [Google Scholar] [CrossRef]
- Wang, D.; Gao, Z.; Du, P.; Xiao, W.; Tan, Q.; Chen, X.; Li, L.; Gao, D. Expression of ABA metabolism-related genes suggests similarities and differences between seed dormancy and bud dormancy of peach (Prunus persica). Front. Plant Sci. 2015, 6, 1248. [Google Scholar] [CrossRef]
- Chmielewski, F.M.; Gotz, K.P.; Homann, T.; Huschek, G.; Rawel, H. M Identification of endodormancy release for cherries (Prunus avium L.) by abscisic acid and sugars. J. Hortic. 2017, 4, 3. [Google Scholar] [CrossRef]
- Zheng, C.; Halaly, T.; Acheampong, A.K.; Takebayashi, Y.; Jikumaru, Y.; Kamiya, Y.; Or, E. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 2015, 66, 1527–1542. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Niu, Q.; He, L.; Teng, Y.; Bai, S. Abscisic acid (ABA) promotes the induction and maintenance of pear (Pyrus pyrifolia white pear group) flower bud endodormancy. Int. J. Mol. Sci. 2018, 19, 310. [Google Scholar] [CrossRef] [Green Version]
- Cai, L.; Stegmeir, T.; Sebolt, A.; Zheng, C.; Bink, M.C.A.M.; Iezzoni, A. Identification of bloom date QTLs and haplotype analysis in tetraploid sour cherry (Prunus cerasus). Tree Genet. Genomes 2018, 14, 22. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Espasandin, F.D.; Maiale, S.J.; Calzadilla, P.; Ruiz, O.A.; Sansberro, P.A. Transcriptional regulation of 9-cis-epoxycarotenoid dioxygenase (NCED) gene by putrescine accumulation positively modulates ABA synthesis and drought tolerance in Lotus tenuis plants. Plant Physiol. Biochem. 2014, 76, 29–35. [Google Scholar] [CrossRef]
- Wang, P.; Lu, S.; Zhang, X.; Hyden, B.; Qin, L.; Liu, L.; Bai, Y.; Han, Y.; Wen, Z.; Xu, J.; et al. Double NCED isozyme control ABA biosynthesis for ripening and senescent regulation in peach fruits. Plant Sci. 2021, 304, 110739. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Bai, S.; Saito, T.; Ito, A.; Moriguchi, T. Dormancy-associated MADS-Box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 2017, 58, 1378–1390. [Google Scholar] [CrossRef]
- Duan, C.; Li, X.; Gao, D.; Liu, H.; Li, M. Studies on regulations of endogenous ABA and GA3 in sweet cherry flower buds on dormancy. Acta Hortic. Sin. 2004, 31, 149–154. [Google Scholar]
- Zhuang, W.; Gao, Z.; Wang, L.; Zhong, W.; Ni, Z.; Zhang, Z. Comparative proteomic and transcriptomic approaches to address the active role of GA4 in Japanese apricot flower bud dormancy release. J. Exp. Bot. 2013, 64, 4953–4966. [Google Scholar] [CrossRef]
- Zhuang, W.; Gao, Z.; Wen, L.; Huo, X.; Cai, B.; Zhang, Z. Metabolic changes upon flower bud break in Japanese apricot are enhanced by exogenous GA4. Hortic. Res. 2015, 2, 15046. [Google Scholar] [CrossRef]
- Choubane, D.; Rabot, A.; Mortreau, E.; Legourrierec, J.; Péron, T.; Foucher, F.; Ahcène, Y.; Pelleschi-Travier, S.; Leduc, N.; Hamama, L.; et al. Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp. J. Plant Physiol. 2012, 169, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.H.; Zhong, W.J.; Huo, X.M.; Zhuang, W.B.; Ni, Z.J.; Gao, Z.H. Expression analysis of ABA- and GA-related genes during four stages of bud dormancy in Japanese apricot (Prunus mume Sieb. et Zucc). J. Hortic. Sci. Biotechnol. 2016, 91, 362–369. [Google Scholar] [CrossRef]
- Zawaski, C.; Kadmiel, M.; Pickens, J.; Ma, C.; Strauss, S.; Busov, V. Repression of gibberellin biosynthesis or signaling produces striking alterations in poplar growth, morphology, and flowering. Planta 2011, 234, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Cao, H.; Hao, X.; Zeng, J.; Qian, W.; Guo, Y.; Ye, N.; Yang, Y.; Wang, X. Differential expression of gibberellin- and abscisic acid-related genes implies their roles in the bud activity-dormancy transition of tea plants. Plant Cell Rep. 2018, 37, 425–441. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Wisniewski, M.; Norelli, J.; Artlip, T. Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 2015, 6, 85. [Google Scholar] [CrossRef]
- Saito, T.; Bai, S.; Imai, T.; Ito, A.; Nakajima, I.; Moriguchi, T. Histone modification and signaling cascade of the dormancy-associated MADS-box gene, PpMADS 13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ. 2015, 38, 1157–1166. [Google Scholar] [CrossRef]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Z.; Kang, J.; Kang, D.; Gu, H.; Qin, G. AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2013, 31, 87–97. [Google Scholar] [CrossRef]
- Fennell, A.Y.; Schlauch, K.A.; Gouthu, S.; Deluc, L.G.; Khadka, V.; Sreekantan, L.; Grimplet, J.; Cramer, G.R.; Mathiason, K.L. Short day transcriptomic programming during induction of dormancy in grapevine. Front. Plant Sci. 2015, 6, 834. [Google Scholar] [CrossRef]
- Guy, C.L.; Haskell, D. Induction of freezing tolerance in spinach is associated with the synthesis of cold acclimation induced proteins. Plant Physiol. 1987, 84, 872–878. [Google Scholar] [CrossRef] [Green Version]
- Guy, C.L.; Haskell, D. Preliminary characterization of high molecular mass proteins associated with cold acclimation in spinach. Plant Physiol. Biochem. 1989, 27, 777–784. [Google Scholar]
- Kaye, C.; Neven, L.; Hofig, A.; Li, Q.B.; Haskell, D.; Guy, C. Characterization of a gene for spinach CAP160 and expression of two spinach cold-acclimation proteins in tobacco. Plant Physiol. 1998, 116, 1367–1377. [Google Scholar] [CrossRef]
- Yu, D.J.; Jun, S.H.; Park, J.; Kwon, J.H.; Lee, H.J. Transcriptome analysis of genes involved in cold hardiness of peach tree (Prunus persica) shoots during cold acclimation and deacclimation. Genes 2020, 11, 611. [Google Scholar] [CrossRef]
- Kim, M.H.; Sasaki, K.; Imai, R. Cold shock domain protein 3 regulates freezing tolerance in Arabidopsis thaliana. J. Biol. Chem. 2009, 284, 23454–23460. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Bocca, S.N.; Ramos, R.L.; Barroco, R.M.; Magioli, C.; Jorge, V.C.; Coutinho, T.C.; Rangel-Lima, C.M.; De Rycke, R.; Inze, D.; et al. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 2007, 225, 1339–1351. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, K.; Hill, K.; Perry, S.E.; Sentoku, N.; Long, J.A.; Karlson, D.T. Arabidopsis cold shock domain proteins: Relationships to floral and silique development. J. Exp. Bot. 2009, 60, 1047–1062. [Google Scholar] [CrossRef]
- Rinne, P.L.; van der Schoot, C. Plasmodesmata at the crossroads between development, dormancy, and defense. Can. J. Bot. 2003, 81, 1182–1197. [Google Scholar] [CrossRef]
- Lee, Y.; Karunakaran, C.; Lahlali, R.; Liu, X.; Tanino, K.K.; Olsen, J.E. Photoperiodic regulation of growth-dormancy cycling through induction of multiple bud–shoot barriers preventing water transport into the winter buds of norway spruce. Front. Plant Sci. 2017, 8, 2109. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef]
- Rinne, P.L.; Welling, A.; Vahala, J.; Ripel, L.; Ruonala, R.; Kangasjärvi, J.; van der Schoot, C. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible, 1, 3-β-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 2011, 23, 130–146. [Google Scholar] [CrossRef]
- Rinne, P.L.; Kaikuranta, P.M.; van der Schoot, C. The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 2001, 26, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Tylewicz, S.; Petterle, A.; Marttila, S.; Miskolczi, P.; Azeez, A.; Singh, R.K.; Immanen, J.; Mähler, N.; Hvidsten, T.R.; Eklund, D.M.; et al. Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science 2018, 360, 212–215. [Google Scholar] [CrossRef]
- Díaz-Riquelme, J.; Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M.J. Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 2012, 12, 181. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef]
- Fadón, E.; Herrero, M.; Rodrigo, J. Dormant flower buds actively accumulate starch over winter in sweet cherry. Front. Plant Sci. 2018, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- El Kayal, W.; Allen, C.C.; Ju, C.J.; Adams, E.; King-Jones, S.; Zaharia, L.I.; Abrams, S.R.; Cooke, J.E. Molecular events of apical bud formation in white spruce, Picea glauca. Plant Cell Environ. 2011, 34, 480–500. [Google Scholar] [CrossRef] [PubMed]
- Ríos, G.; Tadeo, F.R.; Leida, C.; Badenes, M. Prediction of components of the sporopollenin synthesis pathway in peach by genomic and expression analyses. BMC Genom. 2013, 14, 40. [Google Scholar] [CrossRef]
- Zou, T.; Li, S.; Liu, M.; Wang, T.; Xiao, Q.; Chen, D.; Li, Q.; Liang, Y.; Zhu, J.; Liang, Y.; et al. An atypical strictosidine synthase, OsSTRL2, plays key roles in anther development and pollen wall formation in rice. Sci. Rep. 2017, 7, 6863. [Google Scholar] [CrossRef]
- Phan, H.A.; Iacuone, S.; Li, S.F.; Parish, R.W. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 2011, 23, 2209–2224. [Google Scholar] [CrossRef]
- Sorensen, A.M.; Kröber, S.; Unte, U.S.; Huijser, P.; Dekker, K.; Saedler, H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003, 33, 413–423. [Google Scholar] [CrossRef]
- Xu, J.; Yang, C.; Yuan, Z.; Zhang, D.; Gondwe, M.Y.; Ding, Z.; Liang, W.; Zhang, D.; Wilson, Z.A. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 2010, 22, 91–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadón, E.; Herrero, M.; Rodrigo, J. Anther and pollen development in sweet cherry (Prunus avium L.) in relation to winter dormancy. Protoplasma 2019, 256, 733–744. [Google Scholar] [CrossRef]
- Julián, C.; Rodrigo, J.; Herrero, M. Stamen development and winter dormancy in apricot (Prunus armeniaca). Ann. Bot. 2011, 108, 617–625. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information: Sequence Read Archive (NCBI SRA). Available online: https:www.ncbi.nlm.nih.gov/sra (accessed on 20 September 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Dirlewanger, E.; Graziano, E.; Joobeur, T.; Garriga-Calderé, F.; Cosson, P.; Howad, W.; Arús, P. Comparative mapping and marker-assisted selection in Rosaceae fruit crop. Proc. Natl. Acad. Sci. USA 2004, 101, 9891–9896. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots; Various R Programming Tools for Plotting Data. R Package Version 2.17.0. Available online: http://CRA.N.R-project.org/package=gplots (accessed on 27 September 2021).
- Gene Ontology, C. The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, 325–334. [Google Scholar] [CrossRef]
- Gene Ontology Database. Available online: http://www.geneontology.org (accessed on 14 December 2021).
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, 1137–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Species | Source | Cultivar | Blooming Phenotype | Location | SRA Access |
|---|---|---|---|---|---|
| P. armeniaca | [30] | Palsteyn | Early | Ain, France | PRJNA567655 |
| Bergeron | Late | Ain, France | PRJNA567655 | ||
| P. avium | [33] | Cristobalina | Extra early | Lot-et-Garonne, France | PRJNA540235 |
| Garnet | Mid-season | Lot-et-Garonne, France | PRJNA540235 | ||
| Regina | Late | Lot-et-Garonne, France | PRJNA540235 | ||
| [34] | Burlat | Early | Nouvelle-Aquitaine, France | PRJNA595502 | |
| [36] | Royal Dawn | Early | O’Higgins, Chile | PRJNA611731 | |
| Kordia | Extra late | Valparaiso, Chile | PRJNA611733 | ||
| P. dulcis | [31] | Desmayo largueta | Extra early | Murcia, Spain | PRJNA610711 |
| Penta | Late | Murcia, Spain | PRJNA610711 | ||
| Tardona | Extra late | Murcia, Spain | PRJNA610711 | ||
| P. persica | [30] | A209 | Early | Clemson (SC), US | PRJNA567655 |
| A340 | Early | Clemson (SC), US | PRJNA567655 | ||
| A318 | Late | Clemson (SC), US | PRJNA567655 | ||
| A323 | Late | Clemson (SC), US | PRJNA567655 |
| Region | Gene ID | Position (Mbp) | Annotation |
|---|---|---|---|
| Chr1: 0–6.0 Mbp | Prupe.1G030500.1 | 2.2 | transcription factor bHLH92 |
| (Peach, sweet cherry) | Prupe.1G069800.1 | 5.0 | probable xyloglucan endotransglucosylase/hydrolase protein |
| Prupe.1G074400.1 | 5.3 | transcription factor bHLH35 | |
| Chr1: 43–43.5 Mbp (Almond, apricot, peach, and sweet cherry) | Prupe.1G531100.1 | 43.4 | MADS-box protein JOINTLESS |
| Prupe.1G531600.1 | 43.5 | MADS-box protein JOINTLESS | |
| Prupe.1G531700.1 | 43.5 | MADS-box protein JOINTLESS | |
| Chr2: 12–18 Mbp | Prupe.2G106300.3 | 16.4 | glycine-rich protein A3 |
| (Apricot, peach) | Prupe.2G113500.1 | 17.2 | protein ECERIFERUM 1 |
| Prupe.2G122600.1 | 17.9 | protein NRT1/PTR FAMILY 7.3 | |
| Chr2: 27–30 Mbp | Prupe.2G266100.1 | 27.3 | methylesterase 17 |
| (Almond, sweet cherry) | Prupe.2G288400.1 | 28.3 | protein HOTHEAD-like |
| Prupe.2G289500.1 | 28.4 | dehydration-responsive element-binding protein 1A | |
| Prupe.2G289600.1 | 28.4 | ethylene-responsive transcription factor ERF027 | |
| Prupe.2G294400.4 | 28.6 | low-temperature-induced 65 kDa protein | |
| Prupe.2G294400.2 | 28.6 | low-temperature-induced 65 kDa protein | |
| Prupe.2G318900.1 | 29.8 | two-component response regulator-like APRR5 | |
| Chr4: 1.5–7 Mbp | Prupe.4G036800.1 | 1.7 | F-box protein At1g61340 |
| (Almond, apricot, Peach, and sweet cherry) | Prupe.4G040900.1 | 1.9 | NAC transcription factor 25 |
| Prupe.4G046800.1 | 2.2 | CASP-like protein 1C3 | |
| Prupe.4G070500.1 | 3.5 | floral homeotic protein AGAMOUS | |
| Prupe.4G080700.1 | 3.9 | gibberellin 2-beta-dioxygenase | |
| Prupe.4G082000.1 | 4.0 | probable 9-cis-epoxycarotenoid dioxygenase NCED5 | |
| Prupe.4G101900.1 | 5.2 | CASP-like protein 1B1 | |
| Chr4: 10–15 Mbp | Prupe.4G192000.1 | 11.5 | myb-related protein 308 |
| (Peach, sweet cherry) | Prupe.4G200200.2 | 12.4 | scarecrow-like protein 21 |
| Prupe.4G204600.2 | 12.7 | gibberellin 2-beta-dioxygenase 8 | |
| Prupe.4G204600.3 | 12.7 | gibberellin 2-beta-dioxygenase 8 | |
| Chr5: 8–14 Mbp | Prupe.5G072800.1 | 8.7 | 14 kDa proline-rich protein DC2.15 |
| (Apricot, peach) | Prupe.5G073500.1 | 8.8 | 4 kDa proline-rich protein DC2.15-like |
| Prupe.5G083900.1 | 9.6 | sugar carrier protein C | |
| Prupe.5G089900.1 | 10.0 | dehydration-responsive element-binding protein 1A | |
| Prupe.5G090000.1 | 10.1 | dehydration-responsive element-binding protein 1E | |
| Prupe.5G090100.1 | 10.1 | dehydration-responsive element-binding protein 1E | |
| Prupe.5G090200.1 | 10.1 | dehydration-responsive element-binding protein 1A | |
| Prupe.5G090300.1 | 10.1 | ethylene-responsive transcription factor ERF027 | |
| Prupe.5G090500.1 | 10.1 | dehydration-responsive element-binding protein 1F | |
| Prupe.5G090600.1 | 10.1 | ethylene-responsive transcription factor ERF027 | |
| Prupe.5G090800.1 | 10.1 | ethylene-responsive transcription factor ERF027 | |
| Prupe.5G090900.1 | 10.1 | sugar carrier protein C | |
| Chr6: 25–29 Mbp | Prupe.6G284000.1 | 26.4 | abscisic acid receptor PYL4 |
| (Peach) | Prupe.6G315700.2 | 28.1 | calmodulin-binding protein 60 G |
| Chr7: 11–19 Mbp | Prupe.7G142500.1 | 15.7 | dof zinc finger protein DOF3.4 |
| (Almond, apricot, peach) | Prupe.7G161100.1 | 16.7 | cold shock protein CS66 |
| Prupe.7G168200.1 | 17.1 | gibberellin-regulated protein 11 | |
| Chr8: 10–13 | Prupe.8G082100.1 | 11.5 | auxin-responsive protein SAUR50 |
| (Peach) | Prupe.8G083400.1 | 11.7 | NADP-dependent D-sorbitol-6-phosphate dehydrogenase |
| Prupe.8G083400.2 | 11.7 | NADP-dependent D-sorbitol-6-phosphate dehydrogenase | |
| Prupe.8G095800.1 | 12.8 | protein RADIALIS-like 3 | |
| Prupe.8G096400.1 | 12.9 | glycine-rich RNA-binding protein-like |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calle, A.; Saski, C.; Wünsch, A.; Grimplet, J.; Gasic, K. Identification of Key Genes Related to Dormancy Control in Prunus Species by Meta-Analysis of RNAseq Data. Plants 2022, 11, 2469. https://doi.org/10.3390/plants11192469
Calle A, Saski C, Wünsch A, Grimplet J, Gasic K. Identification of Key Genes Related to Dormancy Control in Prunus Species by Meta-Analysis of RNAseq Data. Plants. 2022; 11(19):2469. https://doi.org/10.3390/plants11192469
Chicago/Turabian StyleCalle, Alejandro, Christopher Saski, Ana Wünsch, Jérôme Grimplet, and Ksenija Gasic. 2022. "Identification of Key Genes Related to Dormancy Control in Prunus Species by Meta-Analysis of RNAseq Data" Plants 11, no. 19: 2469. https://doi.org/10.3390/plants11192469





