The Ubiquitin–Proteasome System (UPS) and Viral Infection in Plants
Abstract
:1. Introduction
2. The 26S Proteasome
3. Ubiquitinylating Enzyme Cascade
4. The Ubiquitin (Ub)
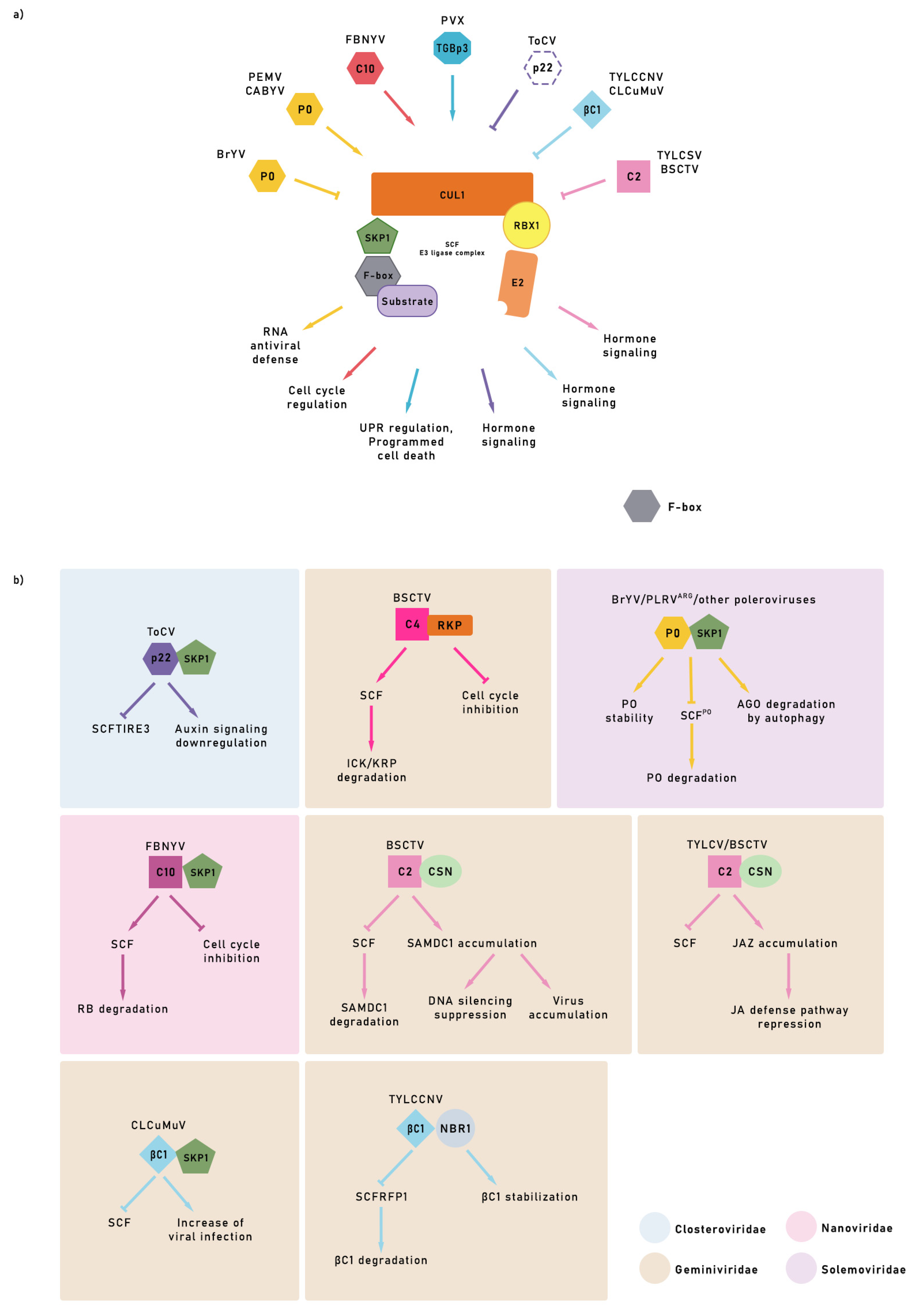
5. Ubiquitin-Proteasome System (UPS) Role under Viral Infection
6. Viral Movement Proteins and UPS
7. Unfolded Protein Response and UPS
8. The Role of the UPS in Plant Immunity
9. Viral Protein Degradation by the UPS
10. Viral Strategy Using E3 to Protect Unstable Proteins
11. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Glickman, M.H.; Ciechanover, A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Ling, Q.; Jarvis, P. Plant Signaling: Ubiquitin Pulls the Trigger on Chloroplast Degradation. Curr. Biol. 2016, 26, R38–R40. [Google Scholar] [CrossRef]
- Oh, E.; Akopian, D.; Rape, M. Principles of Ubiquitin-Dependent Signaling. Annu. Rev. Cell Dev. Biol. 2018, 34, 137–162. [Google Scholar] [CrossRef]
- He, D.; Damaris, R.; Li, M.; Khan, I.; Yang, P. Advances on Plant Ubiquitylome—From Mechanism to Application. Int. J. Mol. Sci. 2020, 21, 7909. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef]
- Ciechanover, A.; Schwartz, A.L. The ubiquitin-proteasome pathway: The complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. USA 1998, 95, 2727–2730. [Google Scholar] [CrossRef]
- Varshavsky, A. The early history of the ubiquitin field. Protein Sci. 2006, 15, 647–654. [Google Scholar] [CrossRef]
- Vogel, F.; Hofius, D.; Sonnewald, U. Intracellular Trafficking of Potato Leafroll VirusMovement Protein in Transgenic Arabidopsis. Traffic 2007, 8, 1205–1214. [Google Scholar] [CrossRef]
- Moon, J.; Parry, G.; Estelle, M. The Ubiquitin-Proteasome Pathway and Plant Development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Fu, H.; Walker, J.; Papa, C.M.; Smalle, J.; Ju, Y.M.; Vierstra, R.D. Purification of the Arabidopsis 26S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 2004, 279, 6401–6413. [Google Scholar] [CrossRef] [PubMed]
- Voges, D.; Zwickl, P.; Baumeister, W. The 26S Proteasome: A Molecular Machine Designed for Controlled Proteolysis. Annu. Rev. Biochem. 1999, 68, 1015–1068. [Google Scholar] [CrossRef]
- Groll, M.; Heinemeyer, W.; Jäger, S.; Ullrich, T.; Bochtler, M.; Wolf, D.H.; Huber, R. The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. Proc. Natl. Acad. Sci. USA 1999, 96, 10976–10983. [Google Scholar] [CrossRef]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Pathare, G.R.; Nagy, I.; Śledź, P.; Anderson, D.J.; Zhou, H.-J.; Pardon, E.; Steyaert, J.; Förster, F.; Bracher, A.; Baumeister, W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA 2014, 111, 2984–2989. [Google Scholar] [CrossRef] [PubMed]
- Dong, O.X.; Ronald, P.C. Genetic Engineering for Disease Resistance in Plants: Recent Progress and Future Perspectives. Plant Physiol. 2019, 180, 26–38. [Google Scholar] [CrossRef]
- Downes, B.; Vierstra, R. Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem. Soc. Trans. 2005, 33, 393–399. [Google Scholar] [CrossRef]
- Dielen, A.-S.; Badaoui, S.; Candresse, T.; German-Retana, S. The ubiquitin/26S proteasome system in plant-pathogen interactions: A never-ending hide-and-seek game. Mol. Plant Pathol. 2010, 11, 293–308. [Google Scholar] [CrossRef]
- Kosarev, P.; Mayer, K.F.; Hardtke, C.S. Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 2002, 3, research0016.1. [Google Scholar] [CrossRef]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional Analysis of the RING-Type Ubiquitin Ligase Family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 Ligase Essential for Arabidopsis Growth and Development, Is Involved in Abscisic Acid Signaling. Plant Cell 2006, 18, 3415–3428. [Google Scholar] [CrossRef] [PubMed]
- Ban, Z.; Estelle, M. CUL3 E3 ligases in plant development and environmental response. Nat. Plants 2021, 7, 6–16. [Google Scholar] [CrossRef]
- Thomann, A.; Dieterle, M.; Genschik, P. Plant CULLIN-based E3s: Phytohormones come first. FEBS Lett. 2005, 579, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Puig, J.; Pauluzzi, G.; Guiderdoni, E.; Gantet, P. Regulation of Shoot and Root Development through Mutual Signaling. Mol. Plant 2012, 5, 974–983. [Google Scholar] [CrossRef]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef]
- Zhu, T.; Zhou, X.; Zhang, J.; Zhang, W.; Zhang, L.; You, C.; Jameson, P.E.; Ma, P.; Guo, S. Ethylene-induced NbMYB4L is involved in resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant Pathol. 2021, 23, 16–31. [Google Scholar] [CrossRef]
- Villalobos, L.I.A.C.; Lee, S.; De Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef]
- Qin, Q.; Li, G.; Jin, L.; Huang, Y.; Wang, Y.; Wei, C.; Xu, Z.; Yang, Z.; Wang, H.; Li, Y. Auxin response factors (ARFs) differentially regulate rice antiviral immune response against rice dwarf virus. PLoS Pathog. 2020, 16, e1009118. [Google Scholar] [CrossRef]
- Wager, A.; Browse, J. Social Network: JAZ Protein Interactions Expand Our Knowledge of Jasmonate Signaling. Front. Plant Sci. 2012, 3, 41. [Google Scholar] [CrossRef]
- Major, I.; Yoshida, Y.; Campos, M.; Kapali, G.; Xin, X.; Sugimoto, K.; Ferreira, D.D.O.; He, S.Y.; Howe, G.A. Regulation of growth–defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [Green Version]
- Gagne, J.M.; Downes, B.P.; Shiu, S.-H.; Durski, A.M.; Vierstra, R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Shabek, N.; Zheng, N. Plant ubiquitin ligases as signaling hubs. Nat. Struct. Mol. Biol. 2014, 21, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.R. E3 Ubiquitin Ligases: Key Regulators of Hormone Signaling in Plants. Mol. Cell. Proteom. 2018, 17, 1047–1054. [Google Scholar] [CrossRef]
- Ryu, M.Y.; Cho, S.K.; Hong, Y.; Kim, J.; Kim, J.H.; Kim, G.M.; Chen, Y.-J.; Knoch, E.; Møller, B.L.; Kim, W.T.; et al. Classification of barley U-box E3 ligases and their expression patterns in response to drought and pathogen stresses. BMC Genom. 2019, 20, 326. [Google Scholar] [CrossRef]
- Sharma, B.; Taganna, J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 2020, 10, 9581. [Google Scholar] [CrossRef]
- Jiménez-López, D.; Muñóz-Belman, F.; González-Prieto, J.M.; Aguilar-Hernández, V.; Guzmán, P. Repertoire of plant RING E3 ubiquitin ligases revisited: New groups counting gene families and single genes. PLoS ONE 2018, 13, e0203442. [Google Scholar] [CrossRef] [PubMed]
- Parveen, A.; Rahim, M.S.; Sharma, A.; Mishra, A.; Kumar, P.; Fandade, V.; Kumar, P.; Bhandawat, A.; Verma, S.K.; Roy, J. Genome-wide analysis of RING-type E3 ligase family identifies potential candidates regulating high amylose starch biosynthesis in wheat (Triticum aestivum L.). Sci. Rep. 2021, 11, 11461. [Google Scholar] [CrossRef]
- Meng, X.; Liu, J.; Zhao, M. Genome-wide identification of RING finger genes in flax (Linum usitatissimum) and analyses of their evolution. PeerJ 2021, 9, e12491. [Google Scholar] [CrossRef]
- Callis, J.; Pöllmann, L.; Shanklin, J.; Wettern, M.; Vierstra, R. Sequence of a cDNA fromChlamydomonas reinhardiiencoding a ubiquitin 52 amino acid extension protein. Nucleic Acids Res. 1989, 17, 8377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doroodian, P.; Hua, Z. The Ubiquitin Switch in Plant Stress Response. Plants 2021, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin–proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef]
- Lam, Y.A.; Xu, W.; DeMartino, G.N.; Cohen, R.E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 1997, 385, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Luo, H. Interplay between the virus and the ubiquitin–proteasome system: Molecular mechanism of viral pathogenesis. Curr. Opin. Virol. 2015, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chenon, M.; Camborde, L.; Cheminant, S.; Jupin, I. A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity. EMBO J. 2011, 31, 741–753. [Google Scholar] [CrossRef]
- Patel, A.; McBride, J.A.; Mark, B.L. The endopeptidase of the maize-affecting Marafivirus type member maize rayado fino virus doubles as a deubiquitinase. J. Biol. Chem. 2021, 297, 100957. [Google Scholar] [CrossRef]
- Imura, Y.; Molho, M.; Chuang, C.; Nagy Peter, D. Cellular Ubc2/Rad6 E2 Ubiquitin-Conjugating Enzyme Facilitates Tombusvirus Replication in Yeast and Plants. Virology 2015, 484, 265–275. Available online: https://www.sciencedirect.com/science/article/pii/S0042682215002895 (accessed on 11 September 2022). [CrossRef]
- Nagy, P.D. The Roles of Host Factors in Tombusvirus RNA Recombination. Adv. Virus Res. 2011, 81, 63–84. [Google Scholar] [CrossRef]
- Sasvari, Z.; Gonzalez, P.A.; Nagy, D.P. Tombusvirus-Yeast Interactions Identify Conserved Cell-Intrinsic Viral Restriction Factors. Front. Plant Sci. 2014, 5, 383. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4127529/ (accessed on 11 September 2022). [CrossRef]
- Barajas, D.; Jiang, Y.; Nagy, P.D. A Unique Role for the Host ESCRT Proteins in Replication of Tomato bushy stunt virus. PLoS Pathog. 2009, 5, e1000705. [Google Scholar] [CrossRef] [Green Version]
- Barajas, D.; Nagy, P.D. Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology 2010, 397, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Li, Z.; Nagy, P.D. The Nedd4-Type Rsp5p Ubiquitin Ligase Inhibits Tombusvirus Replication by Regulating Degradation of the p92 Replication Protein and Decreasing the Activity of the Tombusvirus Replicase. J. Virol. 2009, 83, 11751–11764. [Google Scholar] [CrossRef]
- Prasanth, K.R.; Barajas, D.; Nagy, P.D. The Proteasomal Rpn11 Metalloprotease Suppresses Tombusvirus RNA Recombination and Promotes Viral Replication via Facilitating Assembly of the Viral Replicase Complex. J. Virol. 2015, 89, 2750–2763. [Google Scholar] [CrossRef]
- Zhang, C.; Wei, Y.; Xu, L.; Wu, K.-C.; Yang, L.; Shi, C.-N.; Yang, G.-Y.; Chen, D.; Yu, F.-F.; Xie, Q.; et al. A Bunyavirus-Inducible Ubiquitin Ligase Targets RNA Polymerase IV for Degradation during Viral Pathogenesis in Rice. Mol. Plant 2020, 13, 836–850. [Google Scholar] [CrossRef] [PubMed]
- Drugeon, G.; Jupin, I. Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 2002, 83, 3187–3197. [Google Scholar] [CrossRef] [PubMed]
- Reichel, C.; Beachy, R.N. Degradation of Tobacco Mosaic Virus Movement Protein by the 26S Proteasome. J. Virol. 2000, 74, 3330–3337. [Google Scholar] [CrossRef]
- Ye, C.; Verchot, J. Role of unfolded protein response in plant virus infection. Plant Signal. Behav. 2011, 6, 1212–1215. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Serino, G.; Deng, X.-W.; Dinesh-Kumar, S.P. Role of SCF Ubiquitin-Ligase and the COP9 Signalosome in the N Gene–Mediated Resistance Response to Tobacco mosaic virus. Plant Cell 2002, 14, 1483–1496. [Google Scholar] [CrossRef]
- Ao, K.; Tong, M.; Li, L.; Lüdke, D.; Lipka, V.; Chen, S.; Wiermer, M.; Li, X. SCF SNIPER7 controls protein turnover of unfoldase CDC48A to promote plant immunity. New Phytol. 2020, 229, 2795–2811. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, C.; Li, X. SCFSNIPER4 controls the turnover of two redundant TRAF proteins in plant immunity. Plant J. 2018, 95, 504–515. [Google Scholar] [CrossRef] [Green Version]
- Tong, M.; Kotur, T.; Liang, W.; Vogelmann, K.; Kleine, T.; Leister, D.; Brieske, C.; Yang, S.; Lüdke, D.; Wiermer, M.; et al. E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 2017, 215, 1516–1532. [Google Scholar] [CrossRef] [PubMed]
- Furniss, J.J.; Grey, H.; Wang, Z.; Nomoto, M.; Jackson, L.; Tada, Y.; Spoel, S.H. Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity. PLoS Pathog. 2018, 14, e1007447. [Google Scholar] [CrossRef]
- Rajsbaum, R.; García-Sastre, A. Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol. 2013, 21, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Mahon, C.; Krogan, N.J.; Craik, C.S.; Pick, E. Cullin E3 ligases and their rewiring by viral factors. Biomolecules 2014, 4, 897–930. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, C.; Liu, X.; Navas-Castillo, J.; Zang, L.; Fan, Z.; Zhu, X.; Zhou, T. Tomato chlorosis virus–encoded p22 suppresses auxin signalling to promote infection via interference with SKP1-Cullin-F-box TIR1 complex assembly. Plant Cell Environ. 2021, 44, 3155–3172. [Google Scholar] [CrossRef]
- Chen, S.; Sun, X.; Shi, Y.; Wei, Y.; Han, X.; Li, H.; Chen, L.; Sun, B.; Sun, H.; Shi, Y. Cucurbit Chlorotic Yellows Virus p22 Protein Interacts with Cucumber SKP1LB1 and Its F-Box-Like Motif Is Crucial for Silencing Suppressor Activity. Viruses 2019, 11, 818. [Google Scholar] [CrossRef]
- Lai, J.; Chen, H.; Teng, K.; Zhao, Q.; Zhang, Z.; Li, Y.; Liang, L.; Xia, R.; Wu, Y.; Guo, H.; et al. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 2008, 57, 905–917. [Google Scholar] [CrossRef]
- Kelley, D.R.; Estelle, M. Ubiquitin-Mediated Control of Plant Hormone Signaling. Plant Physiol. 2012, 160, 47–55. [Google Scholar] [CrossRef]
- Eini, O.; Dogra, S.; Selth, L.A.; Dry, I.B.; Randles, J.W.; Rezaian, M.A. Interaction with a Host Ubiquitin-Conjugating Enzyme Is Required for the Pathogenicity of a Geminiviral DNA β Satellite. Mol. Plant-Microbe Interact. 2009, 22, 737–746. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, R.; et al. CLCuMuB βC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, e1005668. [Google Scholar] [CrossRef] [Green Version]
- Bachmair, A.; Becker, F.; Masterson, R.V.; Schell, J. Perturbation of the ubiquitin system causes leaf curling, vascular tissue alterations and necrotic lesions in a higher plant. EMBO J. 1990, 9, 4543–4549. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ismayil, A.; Liu, Y. Autophagy in Plant-Virus Interactions. Annu. Rev. Virol. 2020, 7, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, Q.; Zhao, T.; Xiang, H.; Zhang, X.; Wu, Z.; Zhou, C.; Zhang, X.; Wang, Y.; Zhang, Y.; et al. Interaction between Brassica yellows virus silencing suppressor P0 and plant SKP1 facilitates stability of P0 in vivo against degradation by proteasome and autophagy pathways. New Phytol. 2019, 222, 1458–1473. [Google Scholar] [CrossRef]
- Barón, M.P.B.; Delfosse, V.C.; Agrofoglio, Y.C.; Nahirñak, V.; Almasia, N.I.; Rovere, C.V.; Distéfano, A.J. Argentinian potato leafroll virus P0 protein: Novel activities for a previously known suppressor. Plant Pathol. 2020, 70, 259–274. [Google Scholar] [CrossRef]
- Baumberger, N.; Tsai, C.-H.; Lie, M.; Havecker, E.; Baulcombe, D.C. The Polerovirus Silencing Suppressor P0 Targets ARGONAUTE Proteins for Degradation. Curr. Biol. 2007, 17, 1609–1614. [Google Scholar] [CrossRef]
- Csorba, T.; Lózsa, R.; Hutvágner, G.; Burgyán, J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010, 62, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Bortolamiol, D.; Pazhouhandeh, M.; Marrocco, K.; Genschik, P.; Ziegler-Graff, V. The Polerovirus F-Box Protein P0 Targets ARGONAUTE1 to Suppress RNA Silencing. Curr. Biol. 2007, 17, 1615–1621. Available online: https://www.cell.com/current-biology/abstract/S0960-9822(07)01778-2 (accessed on 11 September 2022). [CrossRef]
- Aronson, M.N.; Meyer, A.D.; Györgyey, J.; Katul, L.; Vetten, H.J.; Gronenborn, B.; Timchenko, T. Clink, a Nanovirus-Encoded Protein, Binds both pRB and SKP1. J. Virol. 2000, 74, 2967–2972. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, H.; Huang, X.; Xia, R.; Zhao, Q.; Lai, J.; Teng, K.; Li, Y.; Liang, L.; Du, Q.; et al. BSCTV C2 Attenuates the Degradation of SAMDC1 to Suppress DNA Methylation-Mediated Gene Silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef]
- Lozano-Duran, R.; Bejarano, E.R. Geminivirus C2 protein might be the key player for geminiviral co-option of SCF-mediated ubiquitination. Plant Signal. Behav. 2011, 6, 999–1001. [Google Scholar] [CrossRef] [Green Version]
- Rosas-Díaz, T.; Macho, A.P.; Beuzón, C.R.; Lozano-Durán, R.; Bejarano, E.R. The C2 Protein from the Geminivirus Tomato Yellow Leaf Curl Sardinia Virus Decreases Sensitivity to Jasmonates and Suppresses Jasmonate-Mediated Defences. Plants 2016, 5, 8. [Google Scholar] [CrossRef]
- Hotton, S.K.; Callis, J. Regulation of Cullin RING Ligases. Annu. Rev. Plant Biol. 2008, 59, 467–489. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Isono, E. The COP9 signalosome and its role in plant development. Eur. J. Cell Biol. 2010, 89, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. The Cullin-RING Ubiquitin-Protein Ligases. Annu. Rev. Plant Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Rosas-Díaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Deng, X.W.; Bejarano, E.R. Geminiviruses Subvert Ubiquitination by Altering CSN-Mediated Derubylation of SCF E3 Ligase Complexes and Inhibit Jasmonate Signaling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, M.; Gong, P.; Li, F.; Zhou, X. Selective autophagic receptor NbNBR1 prevents NbRFP1-mediated UPS-dependent degradation of βC1 to promote geminivirus infection. PLoS Pathog. 2021, 17, e1009956. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobaina, D.P.; Tarazi, R.; Castorino, T.; Vaslin, M.F.S. The Ubiquitin–Proteasome System (UPS) and Viral Infection in Plants. Plants 2022, 11, 2476. https://doi.org/10.3390/plants11192476
Lobaina DP, Tarazi R, Castorino T, Vaslin MFS. The Ubiquitin–Proteasome System (UPS) and Viral Infection in Plants. Plants. 2022; 11(19):2476. https://doi.org/10.3390/plants11192476
Chicago/Turabian StyleLobaina, Dania P., Roberto Tarazi, Tamara Castorino, and Maite F. S. Vaslin. 2022. "The Ubiquitin–Proteasome System (UPS) and Viral Infection in Plants" Plants 11, no. 19: 2476. https://doi.org/10.3390/plants11192476
APA StyleLobaina, D. P., Tarazi, R., Castorino, T., & Vaslin, M. F. S. (2022). The Ubiquitin–Proteasome System (UPS) and Viral Infection in Plants. Plants, 11(19), 2476. https://doi.org/10.3390/plants11192476





