Control of Grain Shape and Size in Rice by Two Functional Alleles of OsPUB3 in Varied Genetic Background
Abstract
1. Introduction
2. Results
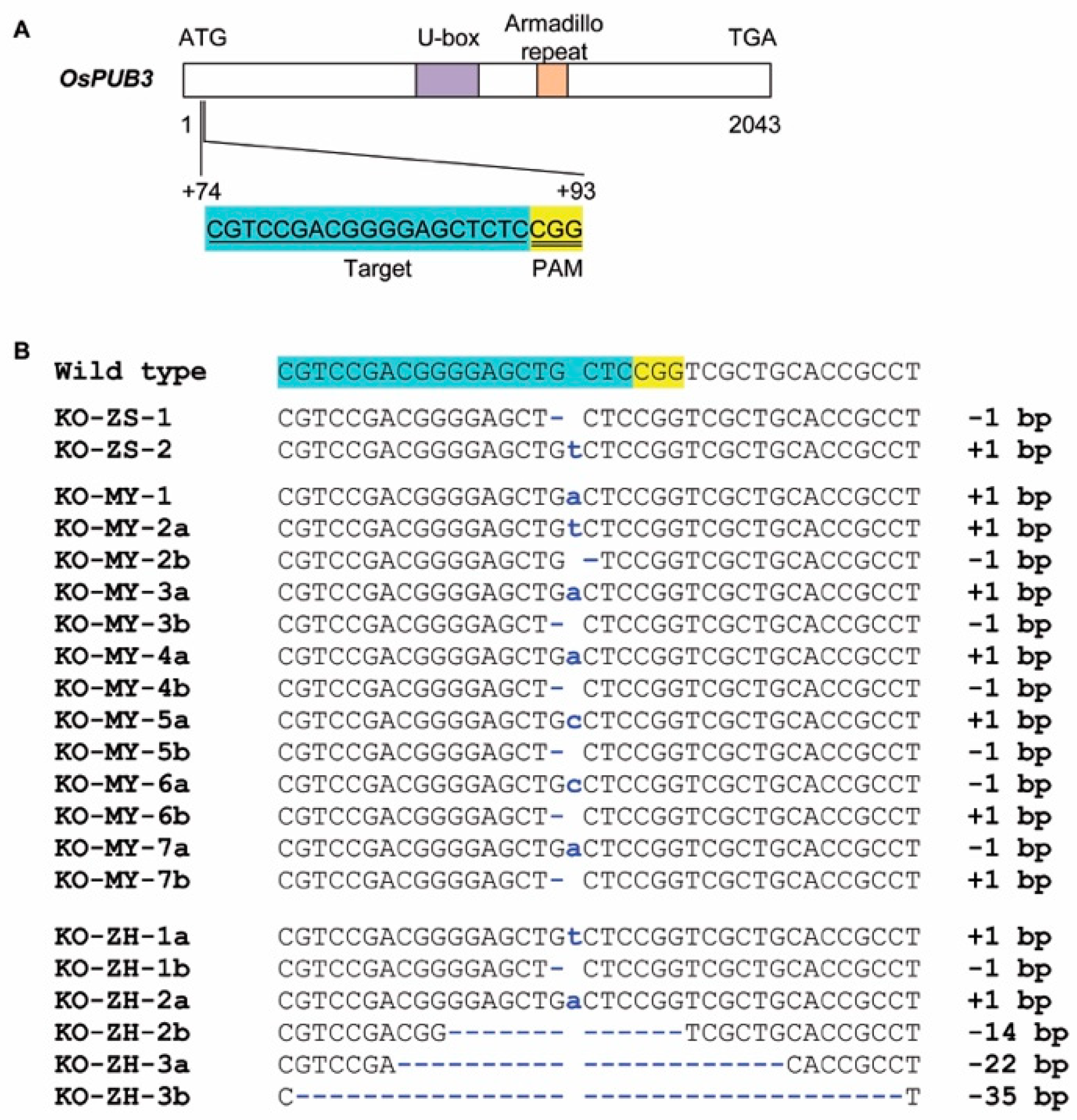
2.1. Knockout Mutants of OsPUB3 Produced from Three Rice Cultivars
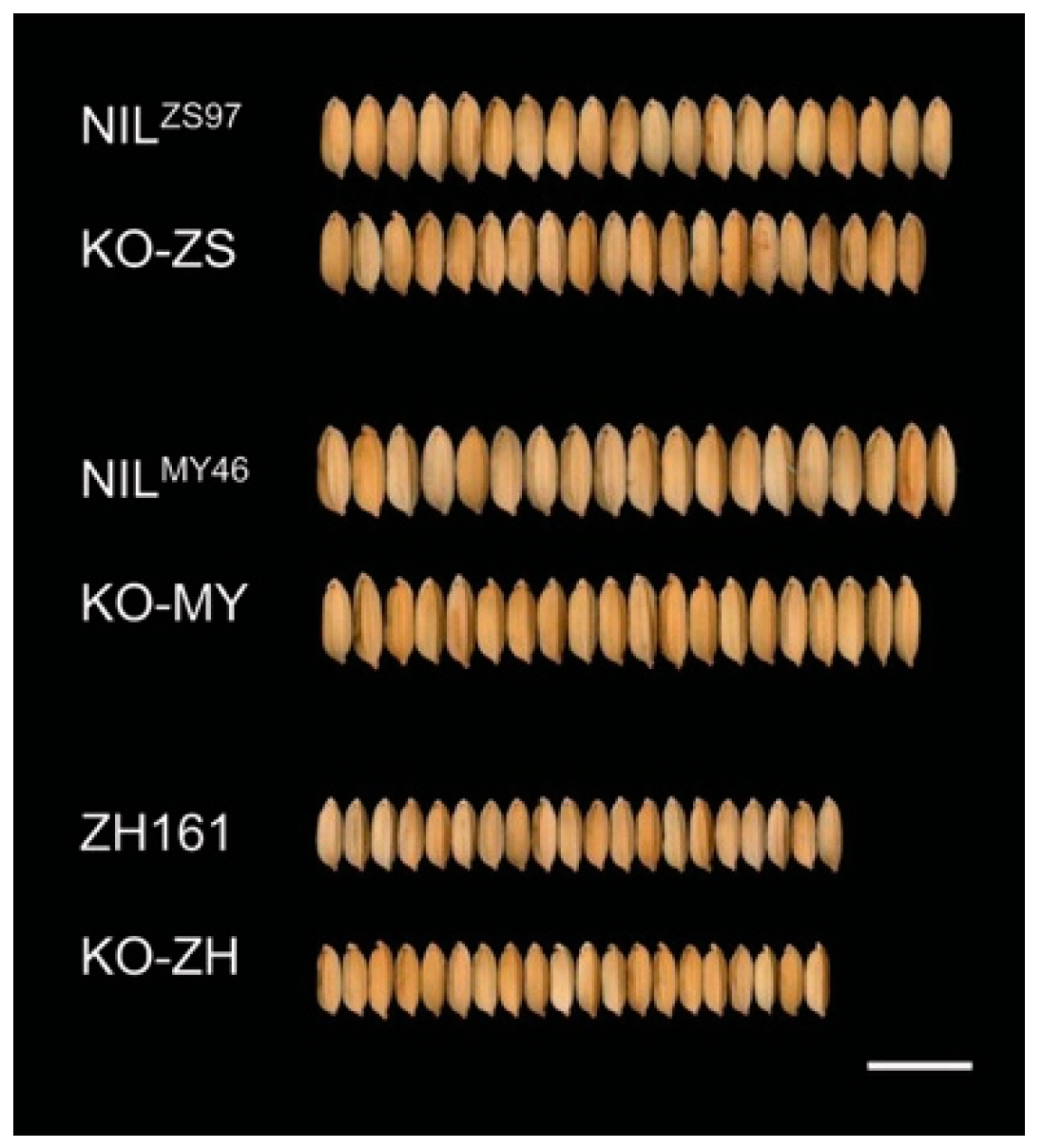
2.2. Phenotypic Change Due to OsPUB3 Knockout
2.3. Effects of Expressing OsPUB3 with Rice Actin 1 Promoter
3. Discussion
4. Materials and Methods
4.1. Knockout OsPUB3 in Three Rice Cultivars
4.2. Expressing OsPUB3 with Rice Actin 1 Promoter
4.3. Field Experiments and Phenotyping
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Calingacion, M.; Laborte, A.; Nelson, A.; Resurreccion, A.; Concepcin, J.C.; Daygon, V.D.; Mumm, R.; Reinke, R.; Dipti, S.; Bassinello, P.Z.; et al. Diversity of global rice markets and the science required for consumer-targeted rice breeding. PLoS ONE 2014, 9, e85106. [Google Scholar] [CrossRef]
- Huang, H.X.; Qian, Q. Progress in genetic research of rice grain shape and breeding achievements of long-grain shape and good quality japonica rice. Chin. J. Rice Sci. 2017, 31, 665–672. [Google Scholar]
- Mao, T.; Zhu, M.D.; Sheng, Z.H.; Shao, G.N.; Jiao, G.A.; Mawia, A.M.; Ahmad, S.; Xie, L.H.; Tang, S.Q.; Wei, X.J.; et al. Effects of grain shape genes editing on appearance quality of erect-panicle geng/japonica rice. Rice 2021, 14, 74. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, C.; Li, Q.; Liu, Q. Genetic control of grain appearance quality in rice. Biotechnol. Adv. 2022, 60, 108014. [Google Scholar] [CrossRef]
- Chan, A.N.; Wang, L.L.; Zhu, Y.J.; Fan, Y.Y.; Zhuang, J.Y.; Zhang, Z.H. Identification through fine mapping and verification using CRISPR/Cas9-targeted mutagenesis for a minor QTL controlling grain weight in rice. Theor. Appl. Genet. 2021, 134, 327–337. [Google Scholar] [CrossRef]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H.; et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S.; et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef]
- Yu, J.; Xiong, H.; Zhu, X.; Zhang, H.; Li, H.; Miao, J.; Wang, W.; Tang, Z.; Zhang, Z.; Yao, G.; et al. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 2017, 15, 28. [Google Scholar] [CrossRef]
- Liu, Q.; Han, R.; Wu, K.; Zhang, J.; Ye, Y.; Wang, S.; Chen, J.; Pan, Y.; Li, Q.; Xu, X.; et al. G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018, 9, 852. [Google Scholar] [CrossRef]
- Yu, J.; Miao, J.; Zhang, Z.; Xiong, H.; Zhu, X.; Sun, X.; Pan, Y.; Liang, Y.; Zhang, Q.; Abdul Rehman, R.M.; et al. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 2018, 16, 1667–1678. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Yu, H.X.; Ye, W.W.; Shan, J.X.; Dong, N.Q.; Guo, T.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; et al. A rice QTL GS3.1 regulates grain size through metabolic-flux distribution between flavonoid and lignin metabolons without affecting stress tolerance. Commun. Biol. 2021, 4, 1171–1185. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Li, Q.; Lu, L.; Liu, H.; Bai, X.; Zhou, X.; Wu, B.; Yuan, M.; Yang, L.; Xing, Y. A minor QTL, SG3, encoding an R2R3-MYB protein, negatively controls grain length in rice. Theor. Appl. Genet. 2020, 133, 2387–2399. [Google Scholar] [CrossRef]
- Qi, P.; Lin, Y.S.; Song, X.J.; Shen, J.B.; Huang, W.; Shan, J.X.; Zhu, M.Z.; Jiang, L.; Gao, J.P.; Lin, H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef]
- Hu, Z.; Lu, S.J.; Wang, M.J.; He, H.; Sun, L.; Wang, H.; Liu, X.H.; Jiang, L.; Sun, J.L.; Xin, X.; et al. A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 2018, 11, 736–749. [Google Scholar] [CrossRef]
- Qiao, J.; Jiang, H.; Lin, Y.; Shang, L.; Wang, M.; Li, D.; Fu, X.; Geisler, M.; Qi, Y.; Gao, Z.; et al. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 2021, 14, 1683–1698. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Song, X.J.; Kuroha, T.; Ayano, M.; Furuta, T.; Nagai, K.; Komeda, N.; Segami, S.; Miura, K.; Ogawa, D.; Kamura, T.; et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 76–81. [Google Scholar] [CrossRef]
- Wang, A.; Hou, Q.; Si, L.; Huang, X.; Luo, J.; Lu, D.; Zhu, J.; Shangguan, Y.; Miao, J.; Xie, Y.; et al. The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol. 2019, 180, 2077–2090. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.L.; Ma, S.P.; Xiao, Z.L.; Li, F.P.; Wei, X.; Lin, S.J.; Wang, X.L.; Ji, Z.; Fu, Y.; Pan, J.H.; et al. Natural variations in grain length 10 (GL10) regulate rice grain size. J. Genet. Genomics 2022, 49, 405–413. [Google Scholar] [CrossRef]
- Zuo, Z.W.; Zhang, Z.H.; Huang, D.R.; Fan, Y.Y.; Yu, S.B.; Zhuang, J.Y.; Zhu, Y.J. Control of thousand-grain weight by OsMADS56 in rice. Int. J. Mol. Sci. 2021, 23, 125. [Google Scholar] [CrossRef]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.; Shang, L.; Zhang, B.; Hu, J.; Wang, Y.; Lin, H.; Zhang, A.; Liu, C.; Peng, Y.; Zhu, L.; et al. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 2020, 227, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Xu, J.; Zeng, D.; Zhang, B.; Geng, M.; Zhang, G.; Huang, K.; Huang, L.; Xu, R.; Ge, S.; et al. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef]
- Shi, C.L.; Dong, N.Q.; Guo, T.; Ye, W.W.; Shan, J.X.; Lin, H.X. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Zhan, P.L.; Wei, X.; Xiao, Z.L.; Wang, X.L.; Ma, S.P.; Lin, S.J.; Li, F.P.; Bu, S.H.; Liu, Z.P.; Zhu, H.T.; et al. GW10, a member of P450 subfamily regulates grain size and grain number in rice. Theor. Appl. Genet. 2021, 134, 3941–3950. [Google Scholar] [CrossRef]
- Dong, N.Q.; Sun, Y.W.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629–2645. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xin, X.; He, Y.; Chen, H.; Li, Q.; Tang, X.; Zhong, Z.; Deng, K.; Zheng, X.; Akher, S.A.; et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 2019, 38, 475–485. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, J.; Ding, A.; Zhang, T.; Ren, X.; Zhang, L.; Li, Q.; Fan, X.; Zhang, C.; Liu, Q. Improving grain appearance of erect-panicle japonica rice cultivars by introgression of the null gs9 allele. J. Integr. Agric. 2021, 20, 2032–2042. [Google Scholar] [CrossRef]
- Huang, H.; Ye, Y.; Song, W.; Li, Q.; Han, R.; Wu, C.; Wang, S.; Yu, J.; Liu, X.; Fu, X.; et al. Modulating the C-terminus of DEP1 synergistically enhances grain quality and yield in rice. J. Genet. Genomics 2022, 49, 506–509. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Li, Y. Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, A.; Liu, X.; Chen, J. Grain size associated genes and the molecular regulatory mechanism in rice. Int. J. Mol. Sci. 2022, 23, 3169. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A.P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.H.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.Q.; Wang, D.K.; Wu, Y.B.; Huang, K.; Duan, P.G.; Li, N.; Xu, R.; Zeng, D.L.; Dong, G.J.; Zhang, B.L.; et al. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant 2021, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Wu, K.; Qian, Q.; Liu, Q.; Li, Q.; Pan, Y.J.; Ye, Y.F.; Liu, X.R.; Wang, J.; Zhang, J.Q.; et al. Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 2017, 27, 1142–1156. [Google Scholar] [CrossRef]
- Huang, K.; Wang, D.K.; Duan, P.G.; Zhang, B.L.; Xu, R.; Li, N.; Li, Y.H. WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 2017, 91, 849–860. [Google Scholar] [CrossRef]
- Dong, Q.; Zhang, Z.H.; Wang, L.L.; Zhu, Y.J.; Fan, Y.Y.; Mou, T.M.; Ma, L.Y.; Zhuang, J.Y. Dissection and fine-mapping of two QTL for grain size linked in a 460-kb region on chromosome 1 of rice. Rice 2018, 11, 44. [Google Scholar] [CrossRef]
- Zheng, K.L.; Huang, N.; Bennett, J.; Khush, G.S. PCR-Based Marker Assisted Selection in Rice Breeding: IRRI Discussion Paper Series No. 12; International Rice Research Institute: Los Banos, CA, USA, 1995. [Google Scholar]
- Hori, K.; Matsubara, K.; Yano, M. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor. Appl. Genet. 2016, 129, 2241–2252. [Google Scholar] [CrossRef]
- Chen, J.Y.; Zhang, H.W.; Zhang, H.L.; Ying, J.Z.; Ma, L.Y.; Zhuang, J.Y. Natural variation at qHD1 affects heading date acceleration at high temperatures with pleiotropism for yield traits in rice. BMC Plant Biol. 2018, 18, 112. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Yan, W.; Zhang, Z.; Lu, L.; Han, Z.; Zhao, H.; Liu, H.Y.; Song, P.; Hu, Y.; et al. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 2015, 208, 1056–1066. [Google Scholar] [CrossRef]
- Lin, S.J.; Liu, Z.P.; Zhang, K.; Yang, W.F.; Zhan, P.L.; Tan, Q.Y.; Gou, Y.J.; Ma, S.P.; Luan, X.; Huang, C.B.; et al. GL9 from Oryza glumaepatula controls grain size and chalkiness in rice. Crop J. 2022. [Google Scholar] [CrossRef]
- Pan, L.X.; Sun, Z.Z.; Zhang, C.Q.; Li, B.; Yang, Q.Q.; Chen, F.; Fan, X.L.; Zhao, D.S.; Lv, Q.M.; Yuan, D.Y.; et al. Allelic diversification of the Wx and ALK loci in indica restorer lines and their utilization in hybrid rice breeding in China over the last 50 years. Int. J. Mol. Sci. 2022, 23, 5941. [Google Scholar] [CrossRef] [PubMed]
- Byun, M.Y.; Cui, L.H.; Oh, T.K.; Jung, Y.J.; Lee1, A.; Park, K.Y.; Kang, B.G.; Kim, W.T. Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in rice (Oryza sativa L.). Plant Sci. 2017, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Ellis, B.E. Double jeopardy: Both overexpression and suppression of a redox-activated plant mitogen-activated protein kinase render tobacco plants ozone sensitive. Plant Cell 2022, 14, 2059–2069. [Google Scholar] [CrossRef]
- Hoppe, T.; Cassata, G.; Barral, J.M.; Springer, W.; Hutagalung, A.H.; Epstein, H.F.; Baumeister, R. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell 2004, 118, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.R.; Skinner, D.J.; Gasser, C.S. Roles of polarity determinants in ovule development. Plant J. 2009, 57, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Bernick, E.P.; Zhang, P.J.; Du, S.J. Knockdown and overexpression of Unc-45b result in defective myofibril organization in skeletal muscles of zebrafish embryos. BMC Cell Biol. 2010, 11, 70. [Google Scholar] [CrossRef] [PubMed]
- Schülein-Völk, C.; Wolf, E.; Zhu, J.; Xu, W.S.; Taranets, L.; Hellmann, A.; Jänicke, L.A.; Diefenbacher, M.E.; Behrens, A.; Eilers, M.; et al. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell Rep. 2014, 9, 1099–1109. [Google Scholar] [CrossRef]
- Landsverk, M.L.; Li, S.M.; Hutagalung, A.H.; Najafov, A.; Hoppe, T.; Barral, J.M.; Epstein, H.F. The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J. Cell Biol. 2007, 177, 205–210. [Google Scholar] [CrossRef]
- Wang, P.P.; Zhou, Z.H.; Hu, A.C.; Albuquerque, C.P.D.; Zhou, Y.; Hong, L.X.; Sierecki, E.; Ajiro, M.; Kruhlak, M.; Harris, C.; et al. Both decreased and increased SRPK1 levels promote cancer by interfering with PHLPP-mediated dephosphorylation of Akt. Mol. Cell 2014, 54, 378–391. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.F.; Ma, X.D.; Meng, L.Z.; Jing, R.N.; Wang, F.; Wang, S.; Cheng, Z.J.; Zhang, X.; Jiang, L.; et al. Disruption of gene SPL35, encoding a novel CUE domain-containing protein, leads to cell death and enhanced disease response in rice. Plant Biotechnol. J. 2019, 17, 1679–1693. [Google Scholar] [CrossRef]
| Population | No. of | Grain Length (mm) | Grain Width (mm) | 1000-Grain Weight (g) | Ratio of Grain Length to Width | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plants | Mean ± SD | D1 a | D2 b | Mean ± SD | D1 | D2 | Mean ± SD | D1 | D2 | Mean ± SD | D1 | D2 | |
| NILZS97 | 10 | 8.387 ± 0.074 | 3.074 ± 0.036 | 27.33 ± 0.82 | 2.738 ± 0.022 | ||||||||
| CKZS97 | 10 | 8.222 ± 0.068 | 3.063 ± 0.040 | 26.45 ± 0.37 | 2.696 ± 0.040 | ||||||||
| KO-ZS-1 | 10 | 8.121 ± 0.060 | −0.266 **** c | −0.102 ** | 2.983 ± 0.068 | −0.091 *** | −0.080 ** | 24.31 ± 1.18 | −3.02 **** | −2.14 **** | 2.737 ± 0.053 | −0.001 | 0.040 * |
| KO-ZS-2 | 10 | 8.018 ± 0.067 | −0.369 **** | −0.204 **** | 2.941 ± 0.032 | −0.133 **** | −0.122 **** | 23.78 ± 0.34 | −3.55 **** | −2.67 **** | 2.738 ± 0.020 | 0.000 | 0.042 **** |
| NILMY46 | 12 | 8.417 ± 0.018 | 3.109 ± 0.008 | 27.97 ± 0.16 | 2.715 ± 0.007 | ||||||||
| KO-MY-1 | 36 | 8.407 ± 0.012 | −0.010 | 2.934 ± 0.007 | −0.175 **** | 24.57 ± 0.16 | −3.40 **** | 2.884 ± 0.009 | 0.169 **** | ||||
| KO-MY-2 | 34 | 8.367 ± 0.015 | −0.050 * | 3.014 ± 0.007 | −0.095 **** | 26.25 ± 0.09 | −1.72 **** | 2.790 ± 0.008 | 0.075 **** | ||||
| KO-MY-3 | 35 | 8.389 ± 0.015 | −0.028 | 2.891 ± 0.015 | −0.218 **** | 24.03 ± 0.24 | −3.94 **** | 2.916 ± 0.013 | 0.201 **** | ||||
| KO-MY-4 | 36 | 8.314 ± 0.015 | −0.103 **** | 2.891 ± 0.015 | −0.218 **** | 23.74 ± 0.22 | −4.23 **** | 2.895 ± 0.013 | 0.180 **** | ||||
| KO-MY-5 | 34 | 8.166 ± 0.017 | −0.251 **** | 3.044 ± 0.007 | −0.065 **** | 25.77 ± 0.08 | −2.20 **** | 2.695 ± 0.008 | −0.020 | ||||
| KO-MY-6 | 29 | 8.260 ± 0.018 | −0.157 **** | 3.004 ± 0.009 | −0.105 **** | 25.69 ± 0.14 | −2.28 **** | 2.763 ± 0.009 | 0.048 **** | ||||
| KO-MY-7 | 35 | 8.147 ± 0.014 | −0.270 **** | 2.984 ± 0.006 | −0.125 **** | 24.59 ± 0.10 | −3.38 **** | 2.746 ± 0.006 | 0.031 ** | ||||
| ZH161 | 30 | 7.927 ± 0.091 | 2.578 ± 0.041 | 20.24 ± 0.53 | 3.092 ± 0.040 | ||||||||
| CKZH161 | 30 | 8.029 ± 0.097 | 2.552 ± 0.042 | 20.44 ± 0.35 | 3.164 ± 0.072 | ||||||||
| KO-ZH-1a | 30 | 8.116 ± 0.129 | 0.189 **** | 0.087 ** | 2.498 ± 0.034 | −0.080 **** | −0.054 **** | 19.94 ± 0.54 | −0.30 * | −0.50 **** | 3.271 ± 0.085 | 0.179 **** | 0.106 **** |
| KO-ZH-1b | 30 | 8.209 ± 0.103 | 0.282 **** | 0.180 **** | 2.510 ± 0.041 | −0.068 **** | −0.043 **** | 20.53 ± 0.61 | 0.29 * | 0.09 | 3.292 ± 0.056 | 0.201 **** | 0.128 **** |
| KO-ZH-2a | 30 | 8.049 ± 0.099 | 0.122 ** | 0.020 | 2.495 ± 0.031 | −0.082 **** | −0.057 **** | 19.90 ± 0.48 | −0.34 ** | −0.53 **** | 3.246 ± 0.055 | 0.154 **** | 0.081 **** |
| KO-ZH-2b | 30 | 7.904 ± 0.103 | −0.023 | −0.125 **** | 2.475 ± 0.042 | −0.103 **** | −0.077 **** | 19.32 ± 0.42 | −0.92 **** | −1.11 **** | 3.213 ± 0.071 | 0.122 **** | 0.049 ** |
| KO-ZH-3a | 30 | 8.089 ± 0.081 | 0.162 **** | 0.060 ** | 2.457 ± 0.051 | −0.120 **** | −0.095 **** | 19.58 ± 0.46 | −0.66 **** | −0.86 **** | 3.314 ± 0.065 | 0.222 **** | 0.149 **** |
| KO-ZH-3b | 30 | 8.156 ± 0.085 | 0.229 **** | 0.127 **** | 2.472 ± 0.037 | −0.106 **** | −0.080 **** | 19.85 ± 0.36 | −0.39 *** | −0.58 **** | 3.320 ± 0.049 | 0.229 **** | 0.156 **** |
| Population | Genotype a | No. of | Grain Length (mm) | Grain Width (mm) | 1000-Grain Weight (g) | Ratio of Grain Length to Width | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plants | Mean ± SD | D b | Mean ± SD | D | Mean ± SD | D | Mean ± SD | D | ||
| OE-1 | − | 11 | 8.355 ± 0.060 | 3.124 ± 0.027 | 27.71 ± 0.49 | 2.683 ± 0.018 | ||||
| + | 19 | 8.404 ± 0.075 | 0.049 *c | 3.085 ± 0.046 | −0.039 ** | 27.65 ± 0.59 | −0.07 | 2.733 ± 0.039 | 0.051 *** | |
| OE-2 | − | 7 | 8.271 ± 0.106 | 3.120 ± 0.045 | 26.25 ± 0.77 | 2.661 ± 0.027 | ||||
| + | 23 | 8.344 ± 0.076 | 0.073 * | 3.108 ± 0.058 | −0.012 | 27.09 ± 0.63 | 0.84 ** | 2.695 ± 0.054 | 0.033 | |
| OE-3 | − | 5 | 8.352 ± 0.058 | 3.081 ± 0.066 | 26.85 ± 0.91 | 2.720 ± 0.041 | ||||
| + | 25 | 8.435 ± 0.069 | 0.083 ** | 3.049 ± 0.057 | −0.032 | 26.99 ± 0.71 | 0.15 | 2.777 ± 0.041 | 0.057 ** | |
| OE-4 | − | 7 | 8.305 ± 0.055 | 3.089 ± 0.030 | 27.03 ± 0.40 | 2.696 ± 0.037 | ||||
| + | 22 | 8.409 ± 0.081 | 0.104 ** | 3.047 ± 0.024 | −0.042 *** | 27.49 ± 0.42 | 0.46 ** | 2.770 ± 0.039 | 0.074 **** | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.-H.; Wang, S.-L.; Zhu, Y.-J.; Fan, Y.-Y.; Huang, D.-R.; Zhu, A.-K.; Zhuang, J.-Y.; Liang, Y.; Zhang, Z.-H. Control of Grain Shape and Size in Rice by Two Functional Alleles of OsPUB3 in Varied Genetic Background. Plants 2022, 11, 2530. https://doi.org/10.3390/plants11192530
Li Z-H, Wang S-L, Zhu Y-J, Fan Y-Y, Huang D-R, Zhu A-K, Zhuang J-Y, Liang Y, Zhang Z-H. Control of Grain Shape and Size in Rice by Two Functional Alleles of OsPUB3 in Varied Genetic Background. Plants. 2022; 11(19):2530. https://doi.org/10.3390/plants11192530
Chicago/Turabian StyleLi, Zhu-Hao, Shi-Lin Wang, Yu-Jun Zhu, Ye-Yang Fan, De-Run Huang, Ai-Ke Zhu, Jie-Yun Zhuang, Yan Liang, and Zhen-Hua Zhang. 2022. "Control of Grain Shape and Size in Rice by Two Functional Alleles of OsPUB3 in Varied Genetic Background" Plants 11, no. 19: 2530. https://doi.org/10.3390/plants11192530
APA StyleLi, Z.-H., Wang, S.-L., Zhu, Y.-J., Fan, Y.-Y., Huang, D.-R., Zhu, A.-K., Zhuang, J.-Y., Liang, Y., & Zhang, Z.-H. (2022). Control of Grain Shape and Size in Rice by Two Functional Alleles of OsPUB3 in Varied Genetic Background. Plants, 11(19), 2530. https://doi.org/10.3390/plants11192530









