Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides
Abstract
:1. Introduction
2. Intracellular Transport of Proteins to the ER and Its Derivatives
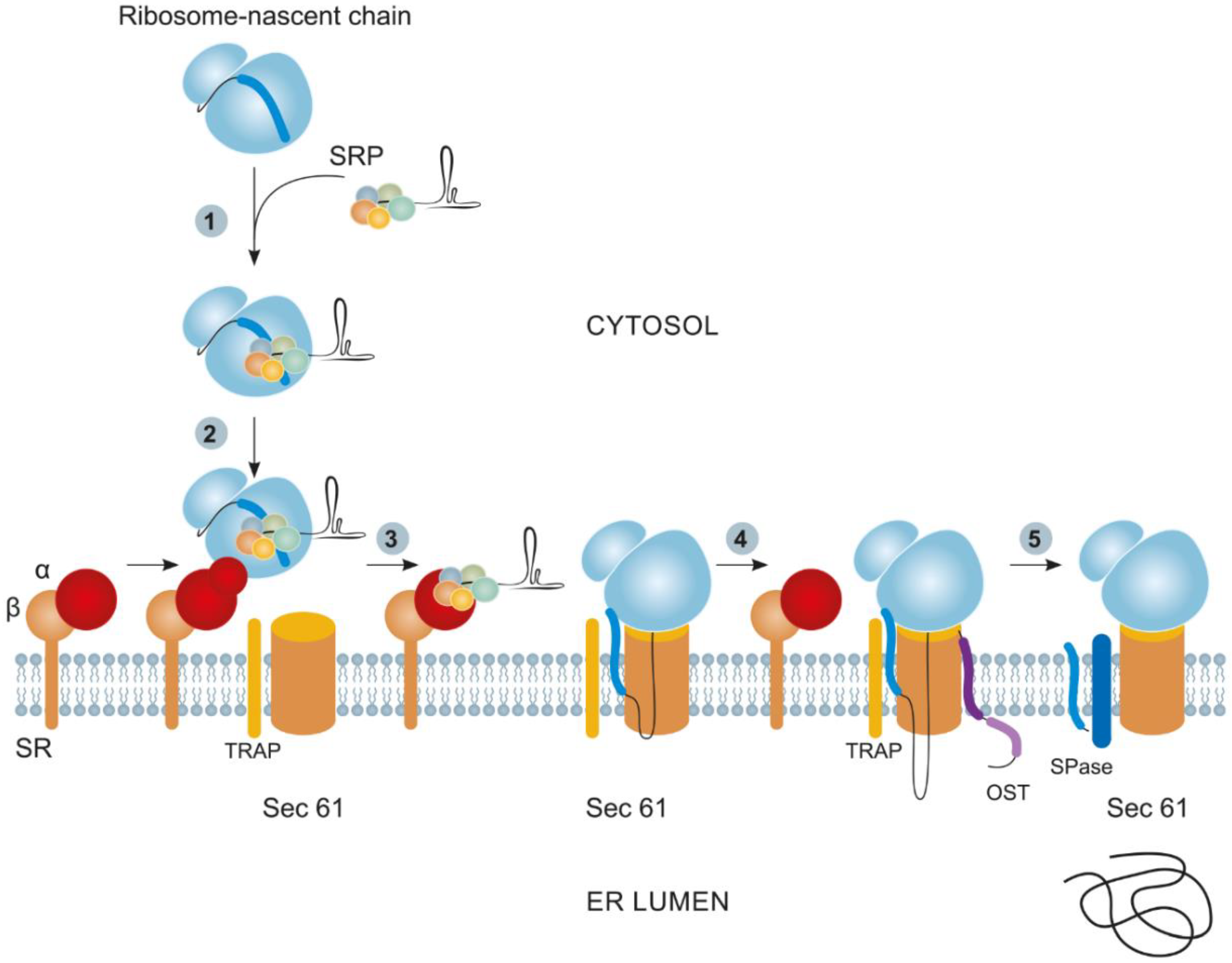
2.1. Cotranslational Protein Targeting to the ER and Its Derivatives
2.2. Post-Translational Targeting to the ER
2.3. Structure of SPs Targeting Proteins to the ER and Its Derivatives
2.4. Using SPs Targeting to the ER and Its Derivatives to Raise the Accumulation Efficiency of Recombinant Proteins
2.4.1. Retention of Recombinant Proteins in the ER
2.4.2. Targeting of Recombinant Proteins to PB-like Structures
2.4.3. Targeting of Recombinant Proteins to Vacuoles
2.4.4. Targeting of Recombinant Proteins to the Apoplast
3. Protein Translocation to Endosymbiotic Organelles
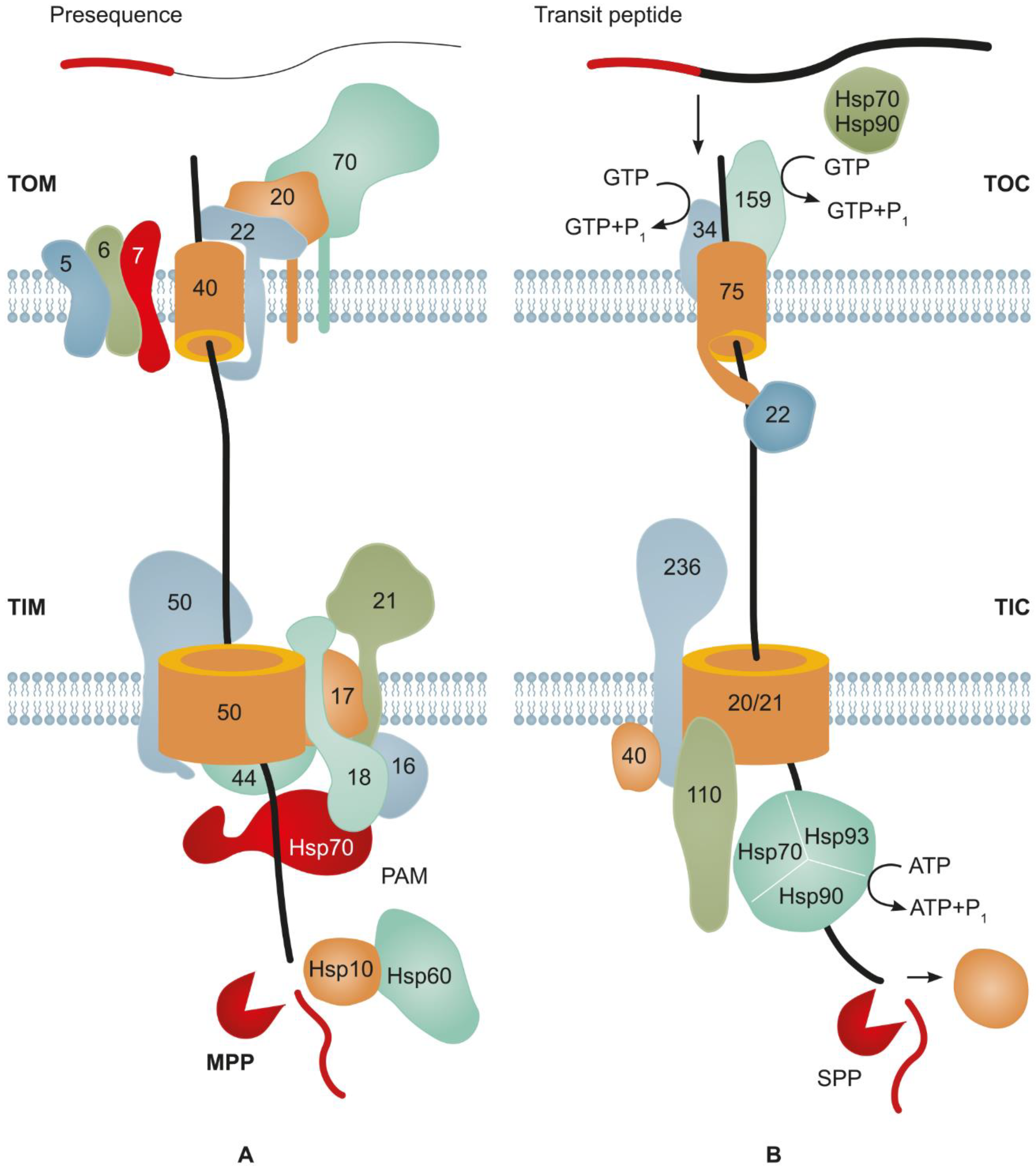
3.1. Major Mechanisms Underlying Protein Import into Mitochondria
3.2. Main Mechanisms of Protein Import into Plastids
3.3. Structure of SPs Targeting Proteins to Endosymbiotic Organelles
3.4. Targeting of Recombinant Proteins to Endosymbiotic Organelles
4. Prediction of Signal Sequences That Determine Protein Localization in the Cell
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Burnett, M.J.B.; Burnett, A.C. Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants People Planet 2020, 2, 121–132. [Google Scholar] [CrossRef]
- Huebbers, J.W.; Buyel, J.F. On the verge of the market—Plant factories for the automated and standardized production of biopharmaceuticals. Biotechnol. Adv. 2021, 46, 107681. [Google Scholar] [CrossRef] [PubMed]
- Schillberg, S.; Finnern, R. Plant molecular farming for the production of valuable proteins—Critical evaluation of achievements and future challenges. J. Plant Physiol. 2021, 258–259, 153359. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G.; Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. [Google Scholar] [CrossRef]
- Zhu, J. Mammalian cell protein expression for biopharmaceutical production. Biotechnol. Adv. 2012, 30, 1158–1170. [Google Scholar] [CrossRef]
- Fischer, R.; Buyel, J.F. Molecular farming—The slope of enlightenment. Biotechnol. Adv. 2020, 40, 107519. [Google Scholar] [CrossRef]
- Egelkrout, E.; Rajan, V.; Howard, J.A. Overproduction of recombinant proteins in plants. Plant Sci. 2012, 184, 83–101. [Google Scholar] [CrossRef]
- Mirzaee, M.; Osmani, Z.; Frébortová, J.; Frébort, I. Recent advances in molecular farming using monocot plants. Biotechnol. Adv. 2022, 58, 107913. [Google Scholar] [CrossRef]
- Karki, U.; Fang, H.; Guo, W.; Unnold-Cofre, C.; Xu, J. Cellular engineering of plant cells for improved therapeutic protein production. Plant Cell Rep. 2021, 40, 1087–1099. [Google Scholar] [CrossRef]
- Rozov, S.M.; Deineko, E.V. Strategies for optimizing recombinant protein synthesis in plant cells: Classical approaches and new directions. Mol. Biol. 2019, 53, 157–175. [Google Scholar] [CrossRef]
- Buyel, J.F.; Stöger, E.; Bortesi, L. Targeted genome editing of plants and plant cells for biomanufacturing. Transgenic Res. 2021, 30, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Rozov, S.M.; Permyakova, N.V.; Sidorchuk, Y.V.; Deineko, E.V. Optimization of genome knock-in method: Search for the most efficient genome regions for transgene expression in plants. Int. J. Mol. Sci. 2022, 23, 4416. [Google Scholar] [CrossRef] [PubMed]
- Faye, L.; Boulaflous, A.; Benchabane, M.; Gomord, V.; Michaud, D. Protein modifications in the plant secretory pathway: Current status and practical implications in molecular pharming. Vaccine 2005, 23, 1770–1778. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Conrad, U.; Fiedler, U. Compartment-specific accumulation of recombinant immunoglobulins in plant cells: An essential tool for antibody production and immunomodulation of physiological functions and pathogen activity. Plant Mol. Biol. 1998, 38, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Gils, M.; Kandzia, R.; Marillonnet, S.; Klimyuk, V.; Gleba, Y. High-yield production of authentic human growth hormone using a plant virus-based expression system. Plant Biotechnol. J. 2005, 3, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.C.; Asgian, J.L.; Ekici, O.D.; James, K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002, 102, 4639–4750. [Google Scholar] [CrossRef]
- Pillay, P.; Schlüter, U.; Van Wyk, S.; Kunert, K.J.; Vorster, B.J. Proteolysis of recombinant proteins in bioengineered plant cells. Bioengineered 2014, 5, 15–20. [Google Scholar] [CrossRef]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef]
- Jarvis, P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008, 179, 257–285. [Google Scholar] [CrossRef]
- Lee, D.W.; Jung, C.; Hwang, I. Cytosolic events involved in chloroplast protein targeting. Biochim. Biophys. Acta 2013, 1833, 245–252. [Google Scholar] [CrossRef] [PubMed]
- da Cunha, N.B.; Leite, M.L.; Dias, S.C.; Vianna, G.R.; Rech Filho, E.L. Plant genetic engineering: Basic concepts and strategies for boosting the accumulation of recombinant proteins in crops. Int. J. Eng. Sci. 2019, 8, 7–13. [Google Scholar]
- Muthamilselvan, T.; Kim, J.S.; Cheong, G.; Hwang, I. Production of recombinant proteins through sequestration in chloroplasts: A strategy based on nuclear transformation and post-translational protein import. Plant Cell Rep. 2019, 38, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhu, J. Recent development and optimization of pseudomonas aeruginosa exotoxin immunotoxins in cancer therapeutic applications. Int. Immunopharmacol. 2021, 96, 107759. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Joyia, F.A.; Mustafa, G. Seeds as economical production platform for recombinant proteins. Protein Pept. Lett. 2020, 27, 89–104. [Google Scholar] [CrossRef]
- Lee, D.W.; Kim, J.K.; Lee, S.; Choi, S.; Kim, S.; Hwang, I. Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 2008, 20, 1603–1622. [Google Scholar] [CrossRef]
- Lee, D.W.; Yoo, Y.-J.; Razzak, M.A.; Hwang, I. Prolines in transit peptides are crucial for efficient preprotein translocation into chloroplasts. Plant Physiol. 2018, 176, 663–677. [Google Scholar] [CrossRef]
- Adams, K.L.; Palmer, J.D. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef]
- Shi, L.X.; Theg, S.M. The chloroplast protein import system: From algae to trees. Biochim. Biophys. Acta 2013, 1833, 314–331. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef]
- Guerriero, C.J.; Brodsky, J.L. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 2012, 92, 537–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Nagata, K. Protein folding and quality control in the ER. Cold Spring Harbor Perspect. Biol. 2011, 3, a007526. [Google Scholar] [CrossRef] [PubMed]
- Luirink, J.; Dobberstein, B. Mammalian and Escherichia coli signal recognition particles. Mol. Microbiol. 1994, 11, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Egea, P.F.; Stroud, R.M.; Walter, P. Targeting proteins to membranes: Structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005, 15, 213–220. [Google Scholar] [CrossRef]
- Alberts, B.; Hopkin, K.; Johnson, A.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Essential Cell Biology, 5th ed.; W.W. Norton & Company: New York, NY, USA, 2018. [Google Scholar]
- Voorhees, R.M.; Fernández, I.S.; Scheres, S.H.; Hegde, R.S. Structure of the mammalian ribosome–Sec61 complex to 3.4 Å resolution. Cell 2014, 157, 1632–1643. [Google Scholar] [CrossRef]
- Nyathi, Y.; Wilkinson, B.M.; Pool, M.R. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 2392–2402. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Zimmermann, R.; Förster, F. Organization of the native ribosome–translocon complex at the mammalian endoplasmic reticulum membrane. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 2122–2129. [Google Scholar] [CrossRef]
- Sommer, N.; Junne, T.; Kalies, K.U.; Spiess, M.; Hartmann, E. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 3104–3111. [Google Scholar] [CrossRef]
- Mohorko, E.; Glockshuber, R.; Aebi, M. Oligosaccharyltransferase: The central enzyme of N-linked protein glycosylation. J. Inherit. Metab. Dis. 2011, 34, 869–878. [Google Scholar] [CrossRef]
- Johnson, N.; Powis, K.; High, S. Post-translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 2403–2409. [Google Scholar] [CrossRef] [PubMed]
- Haßdenteufel, S.; Nguyen, D.; Helms, V.; Lang, S.; Zimmermann, R. ER import of small human presecretory proteins: Components and mechanisms. FEBS Lett. 2019, 593, 2506–2524. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 2008, 134, 634–645. [Google Scholar] [CrossRef]
- Johnson, N.; Vilardi, F.; Lang, S.; Leznicki, P.; Zimmermann, R.; High, S. TRC40 can deliver short secretory proteins to the Sec61 translocon. J. Cell Sci. 2012, 125, 3612–3620. [Google Scholar] [CrossRef] [PubMed]
- Chartron, J.W.; VanderVelde, D.G.; Clemons Jr, W.M. Structures of the Sgt2/SGTA dimerization domain with the Get5/UBL4A UBL domain reveal an interaction that forms a conserved dynamic interface. Cell Rep. 2012, 2, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Mock, J.Y.; Chartron, J.W.; Zaslaver, M.A.; Xu, Y.; Ye, Y.; Clemons, W.M. Bag6 complex contains a minimal tail-anchor–targeting module and a mock BAG domain. Proc. Natl. Acad. Sci. USA 2015, 112, 106–111. [Google Scholar] [CrossRef]
- Shao, S.; Rodrigo-Brenni, M.C.; Kivlen, M.H.; Hegde, R.S. Mechanistic basis for a molecular triage reaction. Science 2017, 355, 298–302. [Google Scholar] [CrossRef]
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.C.; Chuartzman, S.G.; Jan, C.H.; Haßdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138. [Google Scholar] [CrossRef]
- Casson, J.; McKenna, M.; Haßdenteufel, S.; Aviram, N.; Zimmerman, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861. [Google Scholar] [CrossRef]
- Shao, S.; Hegde, R.S. A calmodulin-dependent translocation pathway for small secretory proteins. Cell 2011, 147, 1576–1588. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Schäuble, N.; Cassella, P.; Leznicki, P.; Müller, A.; High, S.; Jung, M.; Zimmermann, R. Ca2+-calmodulin inhibits tail-anchored protein insertion into the mammalian endoplasmic reticulum membrane. FEBS Lett. 2011, 585, 3485–3490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Heijne, G. The signal peptide. J. Membr. Biol. 1990, 115, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Hiss, J.A.; Schneider, G. Architecture, function and prediction of long signal peptides. Brief. Bioinform. 2009, 10, 569–578. [Google Scholar] [CrossRef]
- Van Voorst, F.; De Kruijff, B. Role of lipids in the translocation of proteins across membranes. Biochem. J. 2000, 347, 601–612. [Google Scholar] [CrossRef]
- Batey, R.T.; Rambo, R.P.; Lucast, L.; Rha, B.; Doudna, J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 2000, 287, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, I.; Lara, P.; Hessa, T.; Johnson, A.E.; von Heijne, G.; Karamyshev, A.L. The code for directing proteins for translocation across ER membrane: SRP cotranslationally recognizes specific features of a signal sequence. J. Mol. Biol. 2015, 427, 1191–1201. [Google Scholar] [CrossRef]
- Hegde, R.S.; Bernstein, H.D. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006, 31, 563–571. [Google Scholar] [CrossRef]
- Mordkovich, N.; Okorokova, N.; Veiko, V. Structural and functional organization of the signal peptide of pro-enterotoxin B from Staphylococcus aureus. Appl. Biochem. Microbiol. 2015, 51, 641–648. [Google Scholar] [CrossRef]
- Driessen, A.J.; Van der Does, C. Protein export in bacteria. In Protein Targeting, Transport and Translocation; Dalbey, R., von Heijne, G., Eds.; Academic Press: San Diego, CA, USA, 2002; pp. 47–73. [Google Scholar] [CrossRef]
- Low, K.O.; Mahadi, N.M.; Illias, R.M. Optimisation of signal peptide for recombinant protein secretion in bacterial hosts. Appl. Microbiol. Biotechnol. 2013, 97, 3811–3826. [Google Scholar] [CrossRef]
- Jonet, M.A.; Mahadi, N.M.; Murad, A.M.A.; Rabu, A.; Bakar, F.D.A.; Rahim, R.A.; Low, K.O.; Illias, R.M. Optimization of a heterologous signal peptide by site-directed mutagenesis for improved secretion of recombinant proteins in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2012, 22, 48–58. [Google Scholar] [CrossRef]
- Paetzel, M.; Karla, A.; Strynadka, N.C.; Dalbey, R.E. Signal peptidases. Chem. Rev. 2002, 102, 4549–4580. [Google Scholar] [CrossRef] [PubMed]
- Karamyshev, A.L.; Karamysheva, Z.N.; Kajava, A.V.; Ksenzenko, V.N.; Nesmeyanova, M.A. Processing of Escherichia coli alkaline phosphatase: Role of the primary structure of the signal peptide cleavage region. J. Mol. Biol. 1998, 277, 859–870. [Google Scholar] [CrossRef]
- Choo, K.H.; Tan, T.W.; Ranganathan, S. SPdb–a signal peptide database. BMC Bioinform. 2005, 6, 249. [Google Scholar] [CrossRef] [PubMed]
- Ting, Y.T.; Harris, P.W.; Batot, G.; Brimble, M.A.; Baker, E.N.; Young, P.G. Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization. IUCrJ 2016, 3, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Voss, M.; Schröder, B.; Fluhrer, R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim. Biophys. Acta (BBA)-Biomembr. 2013, 1828, 2828–2839. [Google Scholar] [CrossRef]
- Adams, B.M.; Oster, M.E.; Hebert, D.N. Protein quality control in the endoplasmic reticulum. Protein J. 2019, 38, 317–329. [Google Scholar] [CrossRef]
- Chrispeels, M.J.; Herman, E.M. Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol. 2000, 123, 1227–1234. [Google Scholar] [CrossRef]
- Cheung, A.Y.; Cosgrove, D.J.; Hara-Nishimura, I.; Jurgens, G.; Lloyd, C.; Robinson, D.G.; Staehelin, L.A.; Weijers, D. A rich and bountiful harvest: Key discoveries in plant cell biology. Plant Cell 2022, 34, 53–71. [Google Scholar] [CrossRef]
- Hara-Nishimura, I.; Matsushima, R.; Shimada, T.; Nishimura, M. Diversity and formation of endoplasmic reticulum-derived compartments in plants. Are these compartments specific to plant cells? Plant Physiol. 2004, 136, 3435–3439. [Google Scholar] [CrossRef]
- Rozov, S.M.; Permyakova, N.V.; Deineko, E.V. Main strategies of plant expression system glycoengineering for producing humanized recombinant pharmaceutical proteins. Biochemistry 2018, 83, 215–232. [Google Scholar] [CrossRef]
- Sabalza, M.; Christou, P.; Capell, T. Recombinant plant-derived pharmaceutical proteins: Current technical and economic bottlenecks. Biotechnol. Lett. 2014, 36, 2367–2379. [Google Scholar] [CrossRef]
- Newstead, S.; Barr, F. Molecular basis for KDEL-mediated retrieval of escaped ER-resident proteins–SWEET talking the COPs. J. Cell Sci. 2020, 133, jcs250100. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Huang, Y.; Davis, A.; Yin, Z.; Mi, Q.; Menassa, R.; Brandle, J.E.; Jevnikar, A.M. Production of biologically active human interleukin-4 in transgenic tobacco and potato. Plant Biotechnol. J. 2005, 3, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.L.; Goldstein, C.; Su, W.; Moore, P.H.; Albert, H.H. Production of biologically active GM-CSF in sugarcane: A secure biofactory. Transgenic Res. 2005, 14, 167–178. [Google Scholar] [CrossRef]
- Sharma, M.K.; Singh, N.K.; Jani, D.; Sisodia, R.; Thungapathra, M.; Gautam, J.K.; Meena, L.S.; Singh, Y.; Ghosh, A.; Tyagi, A.K.; et al. Expression of toxin co-regulated pilus subunit A (TCPA) of Vibrio cholerae and its immunogenic epitopes fused to cholera toxin B subunit in transgenic tomato (Solanum lycopersicum). Plant Cell Rep. 2008, 27, 307–318. [Google Scholar] [CrossRef]
- Virdi, V.; Coddens, A.; De Buck, S.; Millet, S.; Goddeeris, B.M.; Cox, E.; De Greve, H.; Depicker, A. Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc. Natl. Acad. Sci. USA 2013, 110, 11809–11814. [Google Scholar] [CrossRef]
- Choi, J.; Diao, H.; Feng, Z.C.; Lau, A.; Wang, R.; Jevnikar, A.M.; Ma, S. A fusion protein derived from plants holds promising potential as a new oral therapy for type 2 diabetes. Plant Biotechnol. J. 2014, 12, 425–435. [Google Scholar] [CrossRef]
- Lee, G.; Na, Y.J.; Yang, B.G.; Choi, J.P.; Seo, Y.B.; Hong, C.P.; Yun, C.H.; Kim, D.H.; Sohn, E.J.; Kim, J.H.; et al. Oral immunization of haemaggulutinin H5 expressed in plant endoplasmic reticulum with adjuvant saponin protects mice against highly pathogenic avian influenza A virus infection. Plant Biotechnol. J. 2015, 13, 62–72. [Google Scholar] [CrossRef]
- Petruccelli, S.; Otegui, M.S.; Lareu, F.; Tran Dinh, O.; Fitchette, A.C.; Circosta, A.; Rumbo, M.; Bardor, M.; Carcamo, R.; Gomord, V.; et al. A KDEL-tagged monoclonal antibody is efficiently retained in the endoplasmic reticulum in leaves, but is both partially secreted and sorted to protein storage vacuoles in seeds. Plant Biotechnol. J. 2006, 4, 511–527. [Google Scholar] [CrossRef]
- Van Droogenbroeck, B.; Cao, J.; Stadlmann, J.; Altmann, F.; Colanesi, S.; Hillmer, S.; Robinson, D.G.; Van Lerberge, E.; Terryn, N.; Van Montagu, M.; et al. Aberrant localization and underglycosylation of highly accumulating single-chain Fv-Fc antibodies in transgenic Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 2007, 104, 1430–1435. [Google Scholar] [CrossRef] [PubMed]
- Loos, A.; Van Droogenbroeck, B.; Hillmer, S.; Grass, J.; Kunert, R.; Cao, J.; Robinson, D.G.; Depicker, A.; Steinkellner, H. Production of monoclonal antibodies with a controlled N-glycosylation pattern in seeds of Arabidopsis thaliana. Plant Biotechnol. J. 2011, 9, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Sukenik, S.C.; Karuppanan, K.; Li, Q.; Lebrilla, C.B.; Nandi, S.; McDonald, K.A. Transient recombinant protein production in glycoengineered Nicotiana benthamiana cell suspension culture. Int. J. Mol. Sci. 2018, 19, 1205. [Google Scholar] [CrossRef]
- Robinson, D.G.; Oliviusson, P.; Hinz, G. Protein sorting to the storage vacuoles of plants: A critical appraisal. Traffic 2005, 6, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Wakasa, Y.; Takaiwa, F. The use of rice seeds to produce human pharmaceuticals for oral therapy. Biotechnol. J. 2013, 8, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Larkins, B.A.; Hurkman, W.J. Synthesis and deposition of zein in protein bodies of maize endosperm. Plant Physiol. 1978, 62, 256–263. [Google Scholar] [CrossRef]
- Lending, C.R.; Larkins, B. Changes in the zein composition of protein bodies during endosperm development. Plant Cell 1989, 1, 1011–1023. [Google Scholar] [CrossRef]
- Coleman, C.E.; Herman, E.M.; Takasaki, K.; Larkins, B.A. The maize γ-zein sequesters α zeins and stabilizes its accumulation in protein bodies of transgenic tobacco endosperm. Plant Cell 1996, 8, 2335–2345. [Google Scholar] [CrossRef]
- Kogan, M.J.; Dalcol, I.; Gorostiza, P.; Lopez-Iglesias, C.; Pons, R.; Pons, M.; Sanz, F.; Giralt, E. Supramolecular properties of the proline-rich gamma-zein N-terminal domain. Biophys. J. 2002, 83, 1194–1204. [Google Scholar] [CrossRef]
- Torrent, M.; Llop-Tous, I.; Ludevid, M.D. Protein body induction: A new tool to produce and recover recombinant proteins in plants. In Recombinant Proteins from Plants. Methods in Molecular Biology; Gomord, V., Faye, L., Eds.; Springer: Heidelberg/Berlin, Germany; Humana Press: Totowa, NJ, USA, 2009; Volume 483, pp. 193–208. [Google Scholar] [CrossRef]
- Torrent, M.; Llompart, B.; Lasserre-Ramassamy, S.; Llop-Tous, I.; Bastida, M.; Marzabal, P.; Westernholm-Parvinen, A.; Saloheimo, M.; Htietz, P.B.; Ludevid, M.D. Eukaryotic protein production in designed storage organelles. BMC Biol. 2009, 7, 5. [Google Scholar] [CrossRef]
- De Virgilio, M.; Marchis, D.; Bellucci, F.; Mainieri, M.; Rossi, D.; Benvenuto, M.; Arcioni, S.; Vitale, A. The human immunodeficiency virus antigen Nef forms protein bodies in leaves of transgenic tobacco when fused to zeolin. J. Exp. Bot. 2008, 59, 2815–2829. [Google Scholar] [CrossRef] [PubMed]
- Llop-Tous, I.; Madurga, S.; Giralt, E.; Marzabal, P.; Torrent, M.; Ludevid, M.D. Relevant elements of a maize gamma-zein domain involved in protein body biogenesis. J. Biol. Chem. 2010, 285, 35633–35644. [Google Scholar] [CrossRef] [PubMed]
- Llop-Tous, I.; Ortiz, M.; Torrent, M.; Ludevid, M.D. The expression of a xylanase targeted to ER-protein bodies provides a simple strategy to produce active insoluble enzyme polymers in tobacco plants. PLoS ONE 2011, 6, e19474. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, M.; Ohlschlager, P.; Almajhdi, F.N.; Alloza, L.; Marzabal, P.; Meyers, A.E.; Hitzeroth, I.I.; Rybicki, E.P. Human papillomavirus (HPV) type 16 E7 protein bodies cause tumour regression in mice. BMC Cancer 2014, 14, 367. [Google Scholar] [CrossRef]
- Conley, A.J.; Joensuu, J.J.; Jevnikar, A.M.; Menassa, R.; Brandle, J.E. Optimization of elastin-like polypeptide fusions for expression and purification of recombinant proteins in plants. Biotechnol. Bioeng. 2009, 103, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Dzuricky, M.; Chilkoti, A. Elastin-like polypeptides as models of intrinsically disordered proteins. FEBS Lett. 2015, 589, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Zhu, H.; Menassa, R.; Gyenis, L.; Richman, A.; Brandle, J. Elastin-like polypeptide fusions enhance the accumulation of recombinant proteins in tobacco leaves. Transgenic Res. 2007, 16, 239–249. [Google Scholar] [CrossRef]
- Kaldis, A.; Ahmad, A.; Reid, A.; McGarvey, B.; Brandle, J.; Ma, S.; Jevnikar, A.; Kohalmi, S.E.; Menassa, R. High-level production of human interleukin-10 fusions in tobacco cell suspension cultures. Plant Biotechnol. J. 2013, 11, 535–545. [Google Scholar] [CrossRef]
- Floss, D.M.; Sack, M.; Stadlmann, J.; Rademacher, T.; Scheller, J.; Stoger, E.; Fischer, R.; Conrad, U. Biochemical and functional characterization of anti-HIV antibody-ELP fusion proteins from transgenic plants. Plant Biotechnol. J. 2008, 6, 379–391. [Google Scholar] [CrossRef]
- Saberianfar, R.; Joensuu, J.J.; Conley, A.J.; Menassa, R. Protein body formation in leaves of Nicotiana benthamiana: A concentration-dependent mechanism influenced by the presence of fusion tags. Plant Biotechnol. J. 2015, 13, 927–937. [Google Scholar] [CrossRef]
- Linder, M.B.; Szilvay, G.R.; Nakari-Setala, T.; Penttila, M.E. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 877–896. [Google Scholar] [CrossRef] [PubMed]
- Linder, M.B.; Qiao, M.; Laumen, F.; Selber, K.; Hyytia, T.; Nakari-Setala, T.; Penttila, M.E. Efficient purification of recombinant proteins using hydrophobins as tags in surfactant-based two-phase systems. Biochemistry 2004, 43, 11873–11882. [Google Scholar] [CrossRef] [PubMed]
- Joensuu, J.J.; Conley, A.J.; Lienemann, M.; Brandle, J.E.; Linder, M.B.; Menassa, R. Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol. 2010, 152, 622–633. [Google Scholar] [CrossRef]
- Takaiwa, F.; Wakasa, Y.; Hayashi, S.; Kawakatsu, T. An overview on the strategies to exploit rice endosperm as production platform for biopharmaceuticals. Plant Sci. 2017, 263, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Viegas, V.S.M.; Ocampo, C.G.; Petruccelli, S. Vacuolar deposition of recombinant proteins in plant vegetative organs as a strategy to increase yields. Bioengineered 2017, 8, 203–211. [Google Scholar] [CrossRef]
- Ocampo, C.G.; Lareu, J.F.; Viegas, V.S.M.; Mangano, S.; Loos, A.; Steinkellner, H.; Petruccelli, S. Vacuolar targeting of recombinant antibodies in Nicotiana benthamiana. Plant Biotechnol. J. 2016, 14, 2265–2275. [Google Scholar] [CrossRef] [PubMed]
- Shaaltiel, Y.; Bartfeld, D.; Hashmueli, S.; Baum, G.; Brill-Almon, E.; Galili, G.; Dym, O.; Boldin-Adamsky, S.A.; Silman, I.; Sussman, J.L.; et al. Production of glucocerebrosidase with terminal mannose glycans for enzyme replacement therapy of Gaucher’s disease using a plant cell system. Plant Biotechnol. J. 2007, 5, 579–590. [Google Scholar] [CrossRef]
- Pereira, E.O.; Kolotilin, I.; Conley, A.J.; Menassa, R. Production and characterization of in planta transiently produced polygalacturonase from Aspergillus niger and its fusions with hydrophobin or ELP tags. BMC Biotechnol. 2014, 14, 59. [Google Scholar] [CrossRef]
- Palaniswamy, H.; Syamaladevi, D.P.; Mohan, C.; Philip, A.; Petchiyappan, A.; Narayanan, S. Vacuolar targeting of r-proteins in sugarcane leads to higher levels of purifiable commercially equivalent recombinant proteins in cane juice. Plant Biotechnol. J. 2016, 14, 791–807. [Google Scholar] [CrossRef]
- Neumann, U.; Brandizzi, F.; Hawes, C. Protein transport in plant cells: In and out of the Golgi. Ann. Bot. 2003, 92, 167–180. [Google Scholar] [CrossRef]
- Drakakaki, G.; Dandekar, A. Protein secretion: How many secretory routes does a plant cell have? Plant Sci. 2013, 203, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Rapoport, T.A. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 2012, 41, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Alexandersson, E.; Ali, A.; Resjö, S.; Andreasson, E. Plant secretome proteomics. Front. Plant Sci. 2013, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Robinson, D.G. Transport vesicle formation in plant cells. Curr. Opin. Plant Biol. 2009, 12, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.B.; Abranches, R.; Fischer, R.; Sack, M.; Holland, T. Putting the spotlight back on plant suspension cultures. Front. Plant Sci. 2016, 7, 297. [Google Scholar] [CrossRef] [PubMed]
- Klimyuk, V.; Pogue, G.; Herz, S.; Butler, J.; Haydon, H. Production of recombinant antigens and antibodies in Nicotiana benthamiana using ‘magnifection’technology: GMP-compliant facilities for small-and large-scale manufacturing. In Plant viral vectors; Palmer, K., Gleba, Y., Eds.; Current Topics in Microbiology and Immunology; Springer: Heidelberg/Berlin, Germany, 2012; volume 375, pp. 127–154. [Google Scholar] [CrossRef]
- Sariyatun, R.; Florence; Kajiura, H.; Ohashi, T.; Misaki, R.; Fujiyama, K. Production of human acid-alpha glucosidase with a paucimannose structure by glycoengineered arabidopsis cell culture. Front. Plant Sci. 2021, 12, 703020. [Google Scholar] [CrossRef]
- Huang, L.F.; Tan, C.C.; Yeh, J.F.; Liu, H.Y.; Liu, Y.K.; Ho, S.L.; Lu, C.A. Efficient secretion of recombinant proteins from rice suspension-cultured cells modulated by the choice of signal peptide. PLoS ONE 2015, 10, e0140812. [Google Scholar] [CrossRef]
- Margulis, L. Symbiosis in Cell Evolution: Life and Its Environment on the Early Earth; Freeman Press: San Francisco, CA, USA, 1981. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Zoschke, R.; Bock, R. Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell 2018, 30, 745–770. [Google Scholar] [CrossRef]
- Schleiff, E.; Becker, T. Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 2011, 12, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Spenning, E.; Glaser, E.; Wieslander, E. Import determinants of organelle-specific and dual targeting peptides of mitochondria and chloroplasts in Arabidopsis thaliana. Mol. Plant. 2014, 7, 121–136. [Google Scholar] [CrossRef]
- Yogev, O.; Karniely, S.; Pines, O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 2007, 282, 29222–29229. [Google Scholar] [CrossRef] [PubMed]
- Eliyahu, E.; Pnueli, L.; Melamed, D.; Scherrer, T.; Gerber, A.P.; Pines, O.; Rapaport, D.; Arava, Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol. Cell. Biol. 2010, 30, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Weis, B.L.; Schleiff, E.; Zerges, W. Protein targeting to subcellular organelles via MRNA localization. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 260–273. [Google Scholar] [CrossRef]
- Lashkevich, K.A.; Dmitriev, S.E. mRNA targeting, transport and local translation in eukaryotic cells: From the classical view to a diversity of new concepts. Mol. Biol. 2021, 55, 507–537. [Google Scholar] [CrossRef]
- Maity, S.; Chakrabarti, O. Mitochondrial protein import as a quality control sensor. Biol. Cell 2021, 113, 375–400. [Google Scholar] [CrossRef]
- Tucker, K.; Park, E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 2019, 26, 1158–1166. [Google Scholar] [CrossRef]
- Edwards, R.; Gerlich, S.; Tokatlidis, K. The biogenesis of mitochondrial intermembrane space proteins. Biol. Chem. 2020, 401, 737–747. [Google Scholar] [CrossRef]
- Becker, T.; Song, J.; Pfanner, N. Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell. Biol. 2019, 29, 534–548. [Google Scholar] [CrossRef]
- Bykov, Y.S.; Rapaport, D.; Herrmann, J.M.; Schuldiner, M. Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem. Sci. 2020, 45, 650–667. [Google Scholar] [CrossRef] [PubMed]
- Backes, S.; Bykov, Y.S.; Flohr, T.; Räschle, M.; Zhou, J.; Lenhard, S.; Krämer, L.; Mühlhaus, T.; Bibi, C.; Jann, C.; et al. The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell Rep. 2021, 35, 108936. [Google Scholar] [CrossRef] [PubMed]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.A.; Marszalek, J. How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 2017, 42, 355–368. [Google Scholar] [CrossRef]
- Opalinski, L.; Song, J.; Priesnitz, C.; Wenz, L.S.; Oeljeklaus, S.; Warscheid, B.; Pfanner, N.; Becker, T. Recruitment of cytosolic J-proteins by TOM receptors promotes mitochondrial protein biogenesis. Cell Rep. 2018, 25, 2036–2043. [Google Scholar] [CrossRef]
- Hansen, K.G.; Aviram, N.; Laborenz, J.; Bibi, C.; Meyer, M.; Spang, A.; Schuldiner, M.; Herrmann, J.M. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 2018, 361, 1118–1122. [Google Scholar] [CrossRef]
- Gold, V.A.; Chroscicki, P.; Bragoszewski, P.; Chacinska, A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep. 2017, 18, 1786–1800. [Google Scholar] [CrossRef]
- Williams, C.C.; Jan, C.H.; Weissman, J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 2014, 346, 748–751. [Google Scholar] [CrossRef]
- Marc, P.; Margeot, A.; Devaux, F.; Blugeon, C.; Corral-Debrinski, M.; Jacq, C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002, 3, 159–164. [Google Scholar] [CrossRef]
- Lapointe, C.P.; Stefely, J.A.; Jochem, A.; Hutchins, P.D.; Wilson, G.M.; Kwiecien, N.W.; Coon, J.J.; Wickens, M.; Pagliarini, D.J. Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 2018, 6, 125–135. [Google Scholar] [CrossRef]
- Saint-Georges, Y.; Garcia, M.; Delaveau, T.; Jourdren, L.; Le Crom, S.; Lemoine, S.; Tanty, V.; Devaux, F.; Jacq, C. Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE 2008, 3, e2293. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M.; Rasmusson, A.G.; Van Aken, O. Plant mitochondria–past, present and future. Plant J. 2021, 108, 912–959. [Google Scholar] [CrossRef] [PubMed]
- Lesnik, C.; Cohen, Y.; Atir-Lande, A.; Schuldiner, M.; Arava, Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun. 2014, 5, 5711. [Google Scholar] [CrossRef]
- Ponce-Rojas, J.C.; Avendaño-Monsalve, M.C.; Yañez-Falcón, A.R.; Jaimes-Miranda, F.; Garay, E.; Torres-Quiroz, F.; DeLuna, A.; Funes, S. ab’-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 2017, 284, 814–830. [Google Scholar] [CrossRef]
- Lee, D.W.; Hwang, I. Evolution and design principles of the diverse chloroplast transit peptides. Mol. Cells 2018, 41, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Zhong, R.; Lamppa, G. Chloroplast stromal processing peptidase activity is modulated by transit peptide determinants that include inhibitory roles for its N-terminal domain and initial Met. Biochem. Biophys. Res. Commun. 2018, 503, 3149–3154. [Google Scholar] [CrossRef]
- Bölter, B. En route into chloroplasts: Preproteins’ way home. Photosyn. Res. 2018, 138, 263–275. [Google Scholar] [CrossRef]
- Day, P.M.; Theg, S.M. Evolution of protein transport to the chloroplast envelope membranes. Photosyn. Res. 2018, 138, 315–326. [Google Scholar] [CrossRef]
- Chotewutmontri, P.; Bruce, B.D. Non-native, N-terminal Hsp70 molecular motor recognition elements in transit peptides support plastid protein translocation. J. Biol. Chem. 2015, 290, 7602–7621. [Google Scholar] [CrossRef]
- Chotewutmontri, P.; Holbrook, K.; Bruce, B.D. Plastid protein targeting: Preprotein recognition and translocation. Int. Rev. Cell Mol. Biol. 2017, 330, 227–294. [Google Scholar] [CrossRef]
- Chang, W.L.; Soll, J.; Bölter, B. The gateway to chloroplast: Re-defining the function of chloroplast receptor proteins. Biol. Chem. 2012, 393, 1263–1277. [Google Scholar] [CrossRef]
- Wiesemann, K.; Simm, S.; Mirus, O.; Ladig, R.; Schleiff, E. Regulation of two GTPases Toc159 and Toc34 in the translocon of the outer envelope of chloroplasts. Biochim. Biophys. Acta 2019, 1867, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, I.; Theg, S.M. Structural considerations of folded protein import through the chloroplast TOC/TIC translocons. FEBS Lett. 2019, 593, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.G.L.; Small, E.L.; Inoue, H.; Schnell, D.J. Molecular topology of the transit peptide during chloroplast protein import. Plant Cell 2018, 30, 1789–1806. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Chen, L.J.; Chu, C.C.; Huang, P.K.; Wen, J.R.; Li, H.M. TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 2018, 564, 125–129. [Google Scholar] [CrossRef]
- Rudolf, M.; Machettira, A.B.; Gross, L.E.; Weber, R.L.; Bolte, K.; Bionda, T.; Sommer, M.S.; Maier, U.G.; Weber, A.P.M.; Shleiff, E.; et al. In vivo function of Tic22, a protein import component of the intermembrane space of chloroplasts. Mol. Plant 2013, 6, 817–829. [Google Scholar] [CrossRef]
- Kasmati, A.R.; Topel, M.; Patel, R.; Murtaza, G.; Jarvis, P. Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 2011, 66, 877–889. [Google Scholar] [CrossRef] [Green Version]
- Flores-Perez, U.; Jarvis, P. Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 2013, 1833, 332–340. [Google Scholar] [CrossRef]
- Uniacke, J.; Zerges, W. Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2009, 106, 1439–1444. [Google Scholar] [CrossRef]
- Sun, Y.; Bakhtiari, S.; Valente-Paterno, M.; Wu, Y.; Law, C.; Dai, D.; Dhaliwal, J.; Bui, K.H.; Zerges, W. Chloroplast-localized translation for protein targeting in Chlamydomonas reinhardtii. bioRxiv 2021. [Google Scholar] [CrossRef]
- Villarejo, A.; Buren, S.; Larsson, S.; Dejardin, A.; Monne, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005, 7, 1224–1231. [Google Scholar] [CrossRef]
- Buren, S.; Ortega-Villasante, C.; Blanco-Rivero, A.; Martinez-Bernardini, A.; Shutova, T.; Shevela, D.; Messinger, J.; Bako, L.; Villarejo, A.; Samuelsson, G. Importance of post-translational modifications for functionality of a chloroplast-localized carbonic anhydrase (CAH1) in Arabidopsis thaliana. PLoS ONE 2011, 6, e21021. [Google Scholar] [CrossRef]
- Bhushan, S.; Kuhn, C.; Berglund, A.K.; Roth, C.; Glaser, E. The role of the N-terminal domain of chloroplast targeting peptides in organellar protein import and miss-sorting. FEBS Lett. 2006, 580, 3966–3972. [Google Scholar] [CrossRef]
- Lee, D.W.; Lee, S.; Lee, J.; Woo, S.; Razzak, M.A.; Vitale, A.; Hwang, I. Molecular mechanism of the specificity of protein import into chloroplasts and mitochondria in plant cells. Mol. Plant 2019, 12, 951–966. [Google Scholar] [CrossRef]
- Lee, D.W.; Hwang, I. Understanding the evolution of endosymbiotic organelles based on the targeting sequences of organellar proteins. New Phytol. 2021, 230, 924–930. [Google Scholar] [CrossRef]
- Garg, S.G.; Gould, S.B. The role of charge in protein targeting evolution. Trends Cell Biol. 2016, 26, 894–905. [Google Scholar] [CrossRef]
- Sharma, M.; Bennewitz, B.; Klösgen, R.B. Rather rule than exception? How to evaluate the relevance of dual protein targeting to mitochondria and chloroplasts. Photosynth. Res. 2018, 138, 335–343. [Google Scholar] [CrossRef]
- McKinnon, L.; Theg, S.M. Determinants of the specificity of protein targeting to chloroplasts or mitochondria. Mol. Plant 2019, 12, 893–895. [Google Scholar] [CrossRef]
- Bruce, B.D. Chloroplast transit peptides: Structure, function and evolution. Trends Cell Biol. 2000, 10, 440–447. [Google Scholar] [CrossRef]
- Daras, G.; Rigas, S.; Tsitsekian, D.; Zur, H.; Tuller, T.; Hatzopoulos, P. Alternative transcription initiation and the AUG context configuration control dual-organellar targeting and functional competence of Arabidopsis Lon1 protease. Mol. Plant 2014, 7, 989–1005. [Google Scholar] [CrossRef]
- Ye, W.; Spanning, E.; Glaser, E.; Maler, L. Interaction of the dual targeting peptide of Thr-tRNA synthetase with the chloroplastic receptor Toc34 in Arabidopsis thaliana. FEBS Open Biol. 2015, 5, 405–412. [Google Scholar] [CrossRef]
- Lee, D.W.; Lee, S.; Min, C.K.; Park, C.; Kim, J.M.; Hwang, C.S.; Park, S.K.; Cho, N.-H.; Hwang, I. Cross-species functional conservation and possible origin of the N-terminal specificity domain of mitochondrial presequences. Front. Plant Sci. 2020, 11, 64. [Google Scholar] [CrossRef]
- Garrido, C.; Caspari, O.D.; Choquet, Y.; Wollman, F.A.; Lafontaine, I. Evidence supporting an antimicrobial origin of targeting peptides to endosymbiotic organelles. Cells 2020, 9, 1795. [Google Scholar] [CrossRef]
- Caspari, O.D.; Lafontaine, I. The role of antimicrobial peptides in the evolution of endosymbiotic protein import. PLoS Pathog. 2021, 17, e1009466. [Google Scholar] [CrossRef]
- Rascón-Cruz, Q.; González-Barriga, C.D.; Iglesias-Figueroa, B.F.; Trejo-Muñoz, J.C.; Siqueiros-Cendón, T.; Sinagawa-García, S.R.; Alevaro-Gallegos, S.; Espinoza-Sánchez, E.A. Plastid transformation: Advances and challenges for its implementation in agricultural crops. Electron. J. Biotechnol. 2021, 51, 95–109. [Google Scholar] [CrossRef]
- Rozov, S.M.; Sidorchuk, Y.V.; Deineko, E.V. Transplastomic plants: Problems of production and their solution. Russ. J. Plant Physiol. 2022, 69, 20. [Google Scholar] [CrossRef]
- Nishimura, K.; Kato, Y.; Sakamoto, W. Chloroplast proteases: Updates on proteolysis within and across suborganellar compartments. Plant Physiol. 2016, 171, 2280–2293. [Google Scholar] [CrossRef] [Green Version]
- Rybicki, E.P. Plant-made vaccines for humans and animals. Plant Biotechnol. J. 2010, 8, 620–637. [Google Scholar] [CrossRef]
- Sticklen, M. Plant genetic engineering to improve biomass characteristics for biofuels. Curr. opinion biotechnol. 2006, 17, 315–319. [Google Scholar] [CrossRef]
- Hyunjong, B.; Lee, D.-S.; Hwang, I. Dual targeting of xylanase to chloroplasts and peroxisomes as a means to increase protein accumulation in plant cells. J. Exp. Bot. 2005, 57, 161–169. [Google Scholar] [CrossRef]
- Kavanagh, T.A.; Jefferson, R.A.; Bevan, M.W. Targeting a foreign protein to chloroplasts using fusions to the transit peptide of a chlorophyll a/b protein. Mol. Gen. Genet. 1988, 215, 38–45. [Google Scholar] [CrossRef]
- Köhler, R.H.; Cao, J.; Zipfel, W.R.; Webb, W.W.; Hanson, M.R. Exchange of protein molecules through connections between higher plant plastids. Science 1997, 276, 2039–2042. [Google Scholar] [CrossRef]
- Panstruga, R.; Hippe-Sanwald, S.; Lee, Y.-K.; Lataster, M.; Lipka, V.; Fischer, R.; Liao, Y.C.; Häusler, R.E.; Kreuzaler, F.; Hirsch, H.-J. Expression and chloroplast-targeting of active phosphoenolpyruvate synthetase from Escherichia coli in Solanum tuberosum. Plant Sci. 1997, 127, 191–205. [Google Scholar] [CrossRef]
- Jang, I.-C.; Nahm, B.H.; Kim, J.-K. Subcellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol. Breed. 1999, 5, 453–461. [Google Scholar] [CrossRef]
- Kim, E.H.; Suh, S.C.; Park, B.S.; Shin, K.S.; Kweon, S.J.; Han, E.J.; Park, S.H.; Kim, Y.S.; Kim, J.K. Chloroplast-targeted expression of synthetic cry1Ac in transgenic rice as an alternative strategy for increased pest protection. Planta 2009, 230, 397–405. [Google Scholar] [CrossRef]
- Kim, S.; Lee, D.-S.; Choi, I.S.; Ahn, S.-J.; Kim, Y.-H.; Bae, H.-J. Arabidopsis thaliana Rubisco small subunit transit peptide increases the accumulation of Thermotoga maritima endoglucanase Cel5A in chloroplasts of transgenic tobacco plants. Transgenic Res. 2010, 19, 489–497. [Google Scholar] [CrossRef]
- Meyers, A.; Chakauya, E.; Shephard, E.; Tanzer, F.L.; Maclean, J.; Lynch, A.; Williamson, A.-L.; Rybicki, E.P. Expression of HIV-1 antigens in plants as potential subunit vaccines. BMC Biotechnol. 2008, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Maclean, J.; Koekemoer, M.; Olivier, A.J.; Stewart, D.; Hitzeroth, I.I.; Rademacher, T.; Fischer, R.; Williamson, A.L.; Rybicki, E.P. Optimization of human papillomavirus type 16 (HPV-16) L1 expression in plants: Comparison of the suitability of different HPV-16 L1 gene variants and different cell-compartment localization. J. Gen. Virol. 2007, 88, 1460–1469. [Google Scholar] [CrossRef]
- Zahin, M.; Joh, J.; Khanal, S.; Husk, A.; Mason, H.; Warzecha, H.; Ghim, S.J.; Miller, D.M.; Matoba, N.; Jenson, A.B. Scalable production of HPV16 L1 protein and VLPs from tobacco leaves. PLoS ONE 2016, 11, e0160995. [Google Scholar] [CrossRef]
- Yanez, R.J.R.; Lamprecht, R.; Granadillo, M.; Torrens, I.; Arcalis, E.; Stoger, E.; Rybicki, E.P.; Hitzeroth, I.I. LALF32-51-E7, a HPV-16 therapeutic vaccine candidate, forms protein body-like structures when expressed in Nicotiana benthamiana leaves. Plant Biotechnol. J. 2018, 16, 628–637. [Google Scholar] [CrossRef]
- Buchke, S.; Sharma, M.; Bora, A.; Relekar, M.; Bhanu, P.; Kumar, J. Mitochondria-targeted, nanoparticle-based drug-delivery systems: Therapeutics for mitochondrial disorders. Life 2022, 12, 657. [Google Scholar] [CrossRef]
- Palmer, C.S.; Anderson, A.J.; Stojanovski, D. Mitochondrial protein import dysfunction: Mitochondrial disease, neurodegenerative disease and cancer. FEBS Lett. 2021, 595, 1107–1131. [Google Scholar] [CrossRef]
- Atkin, O.K.; Macherel, D. The crucial role of plant mitochondria in orchestrating drought tolerance. Ann. Bot. 2009, 103, 581–597. [Google Scholar] [CrossRef]
- Lopez-Torrejon, G.; Jimenez-Vicente, E.; Buesa, J.M.; Hernandez, J.A.; Verma, H.K.; Rubio, L.M. Expression of a functional oxygen-labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast. Nat. Commun. 2016, 7, 11426. [Google Scholar] [CrossRef]
- Baysal, C.; Pérez-González, A.; Eseverri, Á.; Jiang, X.; Medina, V.; Caro, E.; Rubio, L.; Christou, P.; Zhu, C. Recognition motifs rather than phylogenetic origin influence the ability of targeting peptides to import nuclear-encoded recombinant proteins into rice mitochondria. Transgenic Res. 2020, 29, 37–52. [Google Scholar] [CrossRef]
- Jiang, X.; Coroian, D.; Barahona, E.; Echavarri-Erasun, C.; Castellanos-Rueda, R.; Eseverri, Á.; Aznar-Moreno, J.A.; Buren, S.; Rubio, L.M. Functional nitrogenase cofactor maturase NifB in mitochondria and chloroplasts of Nicotiana benthamiana. mBio 2022, 13, e00268-22. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their n-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Small, I.; Peeters, N.; Legeai, F.; Lurin, C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 2004, 4, 1581–1590. [Google Scholar] [CrossRef]
- Petsalaki, E.I.; Bagos, P.G.; Litou, Z.I.; Hamodrakas, S.J. PredSL: A tool for the N-terminal sequence-based prediction of protein subcellular localization. Genom. Proteom. Bioinf. 2006, 4, 48–55. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Tsuji, J.; Fu, S.-C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Casadio, R. TPpred3 detects and discriminates mitochondrial and chloroplastic targeting peptides in eukaryotic proteins. Bioinformatics 2015, 31, 3269–3275. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, D.; Wang, W.; Xu, D. Computational methods for protein localization prediction. Comput. Struct. Biotechnol. J. 2021, 19, 5834–5844. [Google Scholar] [CrossRef]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A brief history of protein sorting prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winter, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 achieves signal peptide prediction across all types using protein language models. bioRxiv 2021. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Thumuluri, V.; Armenteros, J.J.A.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022, W1, W228–W234. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozov, S.M.; Deineko, E.V. Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides. Plants 2022, 11, 2561. https://doi.org/10.3390/plants11192561
Rozov SM, Deineko EV. Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides. Plants. 2022; 11(19):2561. https://doi.org/10.3390/plants11192561
Chicago/Turabian StyleRozov, Sergey M., and Elena V. Deineko. 2022. "Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides" Plants 11, no. 19: 2561. https://doi.org/10.3390/plants11192561



