Using FIBexDB for In-Depth Analysis of Flax Lectin Gene Expression in Response to Fusarium oxysporum Infection
Abstract
:1. Introduction
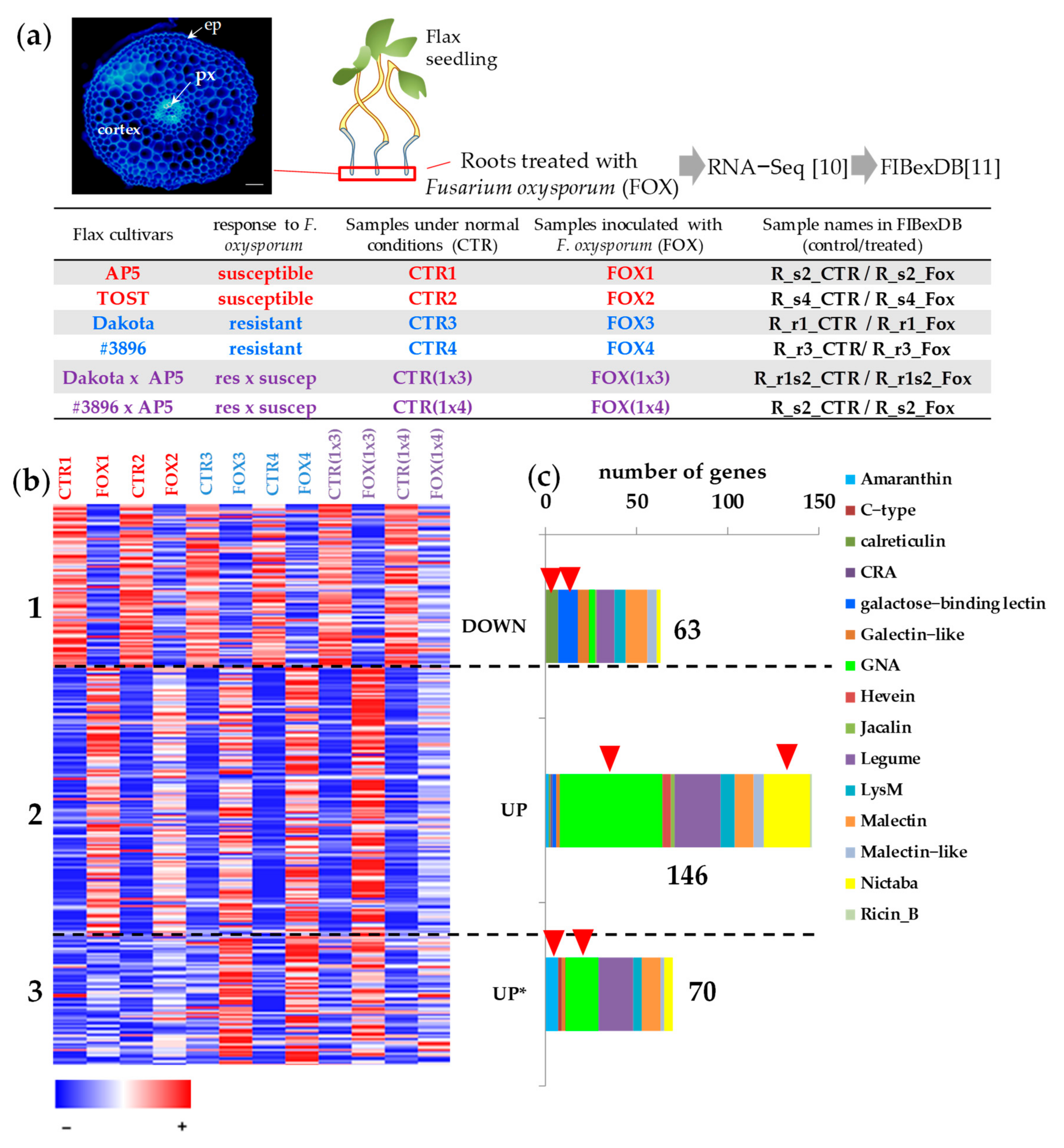
2. Results
2.1. Expression of Flax Genes for Lectins during Fungal Infection in Different Cultivars
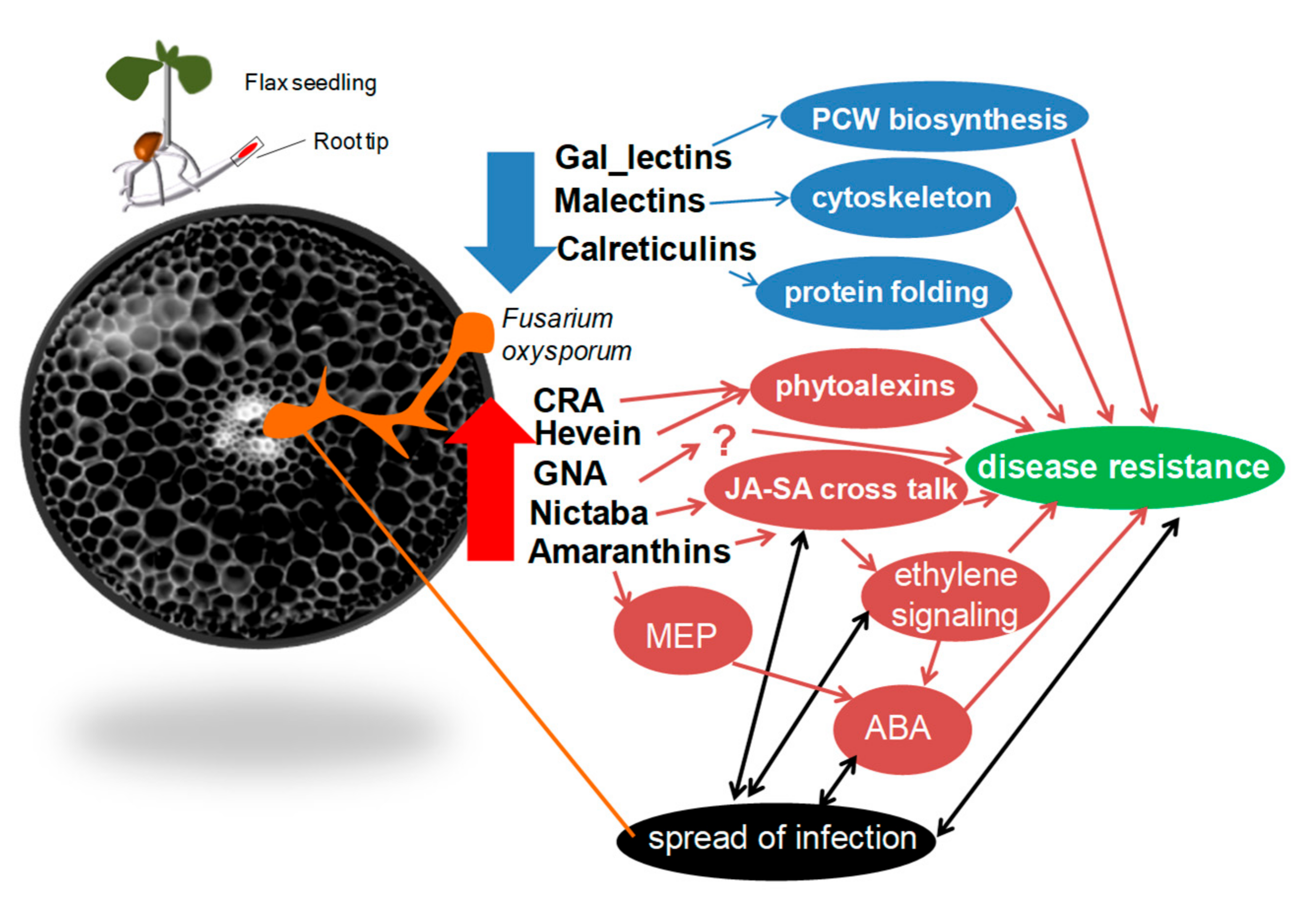
2.2. Cluster 1: Lectin Genes Downregulated in Flax Root Tips in Response to Fusarium oxysporum
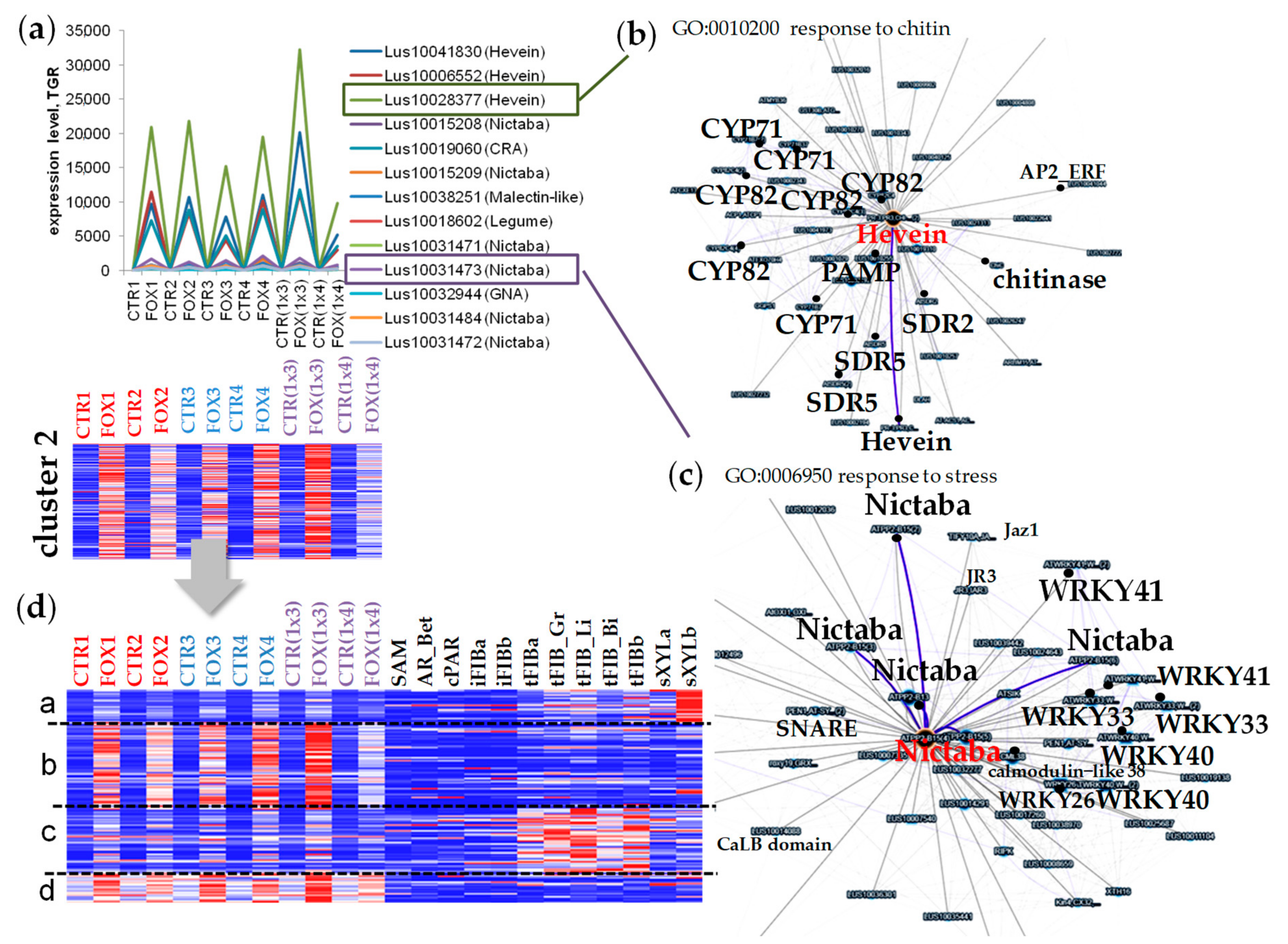
2.3. Cluster 2: Lectin Genes Upregulated in Flax Root Tips in Response to F. oxysporum
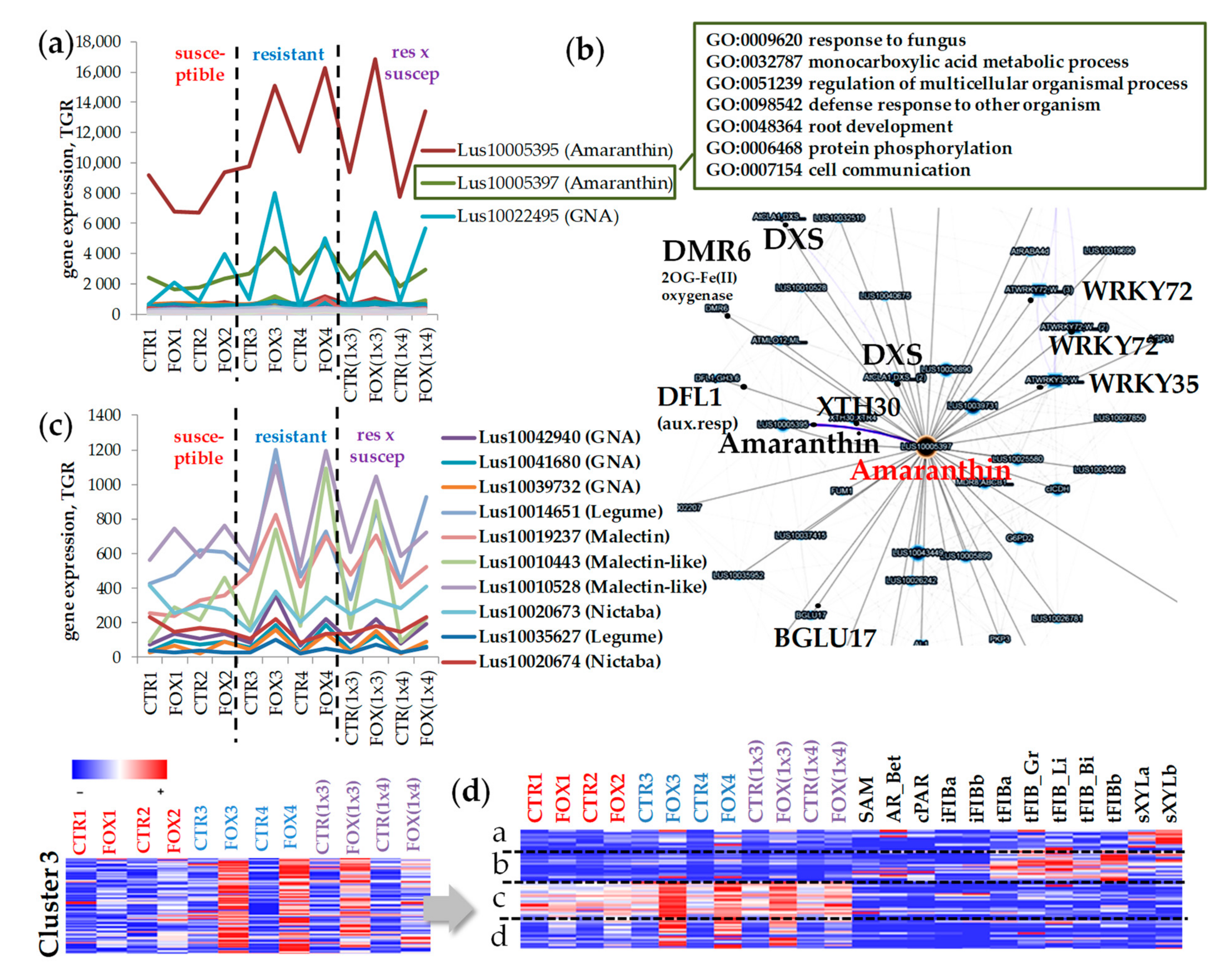
2.4. Cluster 3: Genes for Lectins Essentially Upregulated in Cultivars Resistant to Fusarium oxysporum
3. Discussion
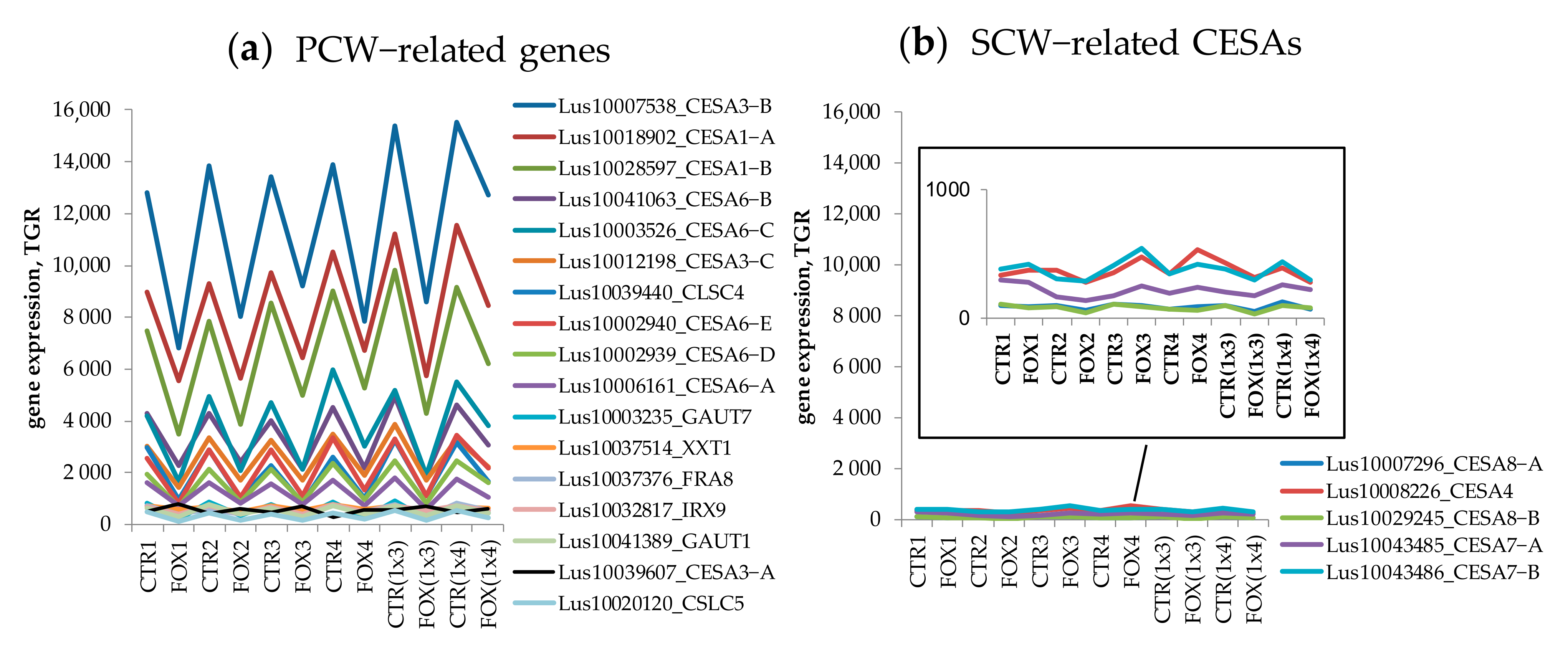
3.1. Genes Encoding Lectins Potentially Involved in the Biosynthesis of Cell Compounds
3.2. The Avant-Garde of Defense Flax Lectins in Response to Fusarium oxysporum
3.3. Lectins That Might Determine the Resistance of Flax to F. oxysporum
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaber, K.; Haubruge, É.; Francis, F. Development of entomotoxic molecules as control agents: Illustration of some protein potential uses and limits of lectins. Biotechnol. Agron. Soc. Environ. 2010, 14, 225–241. [Google Scholar]
- Newman, M.A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 2013, 4, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.; Zimmerli, L. Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 2013, 4, 124. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.F.S.; Da Silva, M.D.C.; Napoleão, T.H.; Paiva, P.M.G.; Correia, M.T.S.; Coelho, L.C.B.B. Lectins: Function, structure, biological properties andpotential applications. Curr. Top. Pept. Protein Res. 2014, 15, 41–62. [Google Scholar]
- Tsaneva, M.; Van Damme, E.J.M. 130 years of Plant Lectin Research. Glycoconj. J. 2020, 37, 533–551. [Google Scholar] [CrossRef] [PubMed]
- Petrova, N.; Nazipova, A.; Gorshkov, O.; Mokshina, N.; Patova, O.; Gorshkova, T. Gene expression patterns for proteins with lectin domains in flax stem tissues are related to deposition of distinct cell wall types. Front. Plant Sci. 2021, 12, 634594. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-Y.; Ma, Z.; Ramachandran, S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 2010, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- Lannoo, N.; Van Damme, E.J.M. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5, 397. [Google Scholar] [CrossRef] [Green Version]
- Schulze, B.; Mentzel, T.; Jehle, A.K.; Mueller, K.; Beeler, S.; Boller, T.; Felix, G.; Chinchilla, D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 2010, 285, 9444–9451. [Google Scholar] [CrossRef] [Green Version]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Novakovskiy, R.O.; Snezhkina, A.V.; Fedorova, M.S.; Yurkevich, O.Y.; Muravenko, O.V.; Bolsheva, N.; Kudryavtseva, A. Differential gene expression in response to Fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L.). BMC Plant Biol. 2017, 17, 253. [Google Scholar] [CrossRef]
- FIBexDB. Available online: https://ssl.cres-t.org/fibex/ (accessed on 8 December 2021).
- Mokshina, N.; Gorshkov, O.; Takasaki, H.; Onodera, H.; Sakamoto, S.; Gorshkova, T.; Mitsuda, N. FIBexDB: A new online transcriptome platform to analyze development of plant cellulosic fibers. New Phytol. 2021, 231, 512–515. [Google Scholar] [CrossRef]
- Galindo-González, L.; Deyholos, M.K. RNA-seq Transcriptome Response of Flax (Linum usitatissimum L.) to the Pathogenic Fungus Fusarium oxysporum f. sp. lini. Front. Plant Sci. 2016, 7, 1766. [Google Scholar] [CrossRef] [Green Version]
- Boba, A.; Kostyn, K.; Kozak, B.; Wojtasik, W.; Preisner, M.; Prescha, A.; Gola, E.; Lysh, D.; Dudek, B.; Szopa, J.; et al. Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 2020, 251, 50. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Deyholos, M.K. RNA-Seq analysis of the shoot apex of flax (Linum usitatissimum) to identify phloem fiber specification genes. Front. Plant Sci. 2016, 7, 950. [Google Scholar]
- Gorshkova, T.; Chernova, T.; Mokshina, N.; Gorshkov, V.; Kozlova, L.; Gorshkov, O. Transcriptome analysis of intrusively growing flax fibers isolated by laser microdissection. Sci. Rep. 2018, 8, 14570. [Google Scholar] [CrossRef]
- Gorshkov, O.; Chernova, T.; Mokshina, N.; Gogoleva, N.; Suslov, D.; Tkachenko, A.; Gorshkova, T. Intrusive growth of phloem fibers in flax stem: Integrated analysis of miRNA and mRNA expression profiles. Plants 2019, 8, 47. [Google Scholar] [CrossRef] [Green Version]
- Mokshina, N.; Gorshkov, O.; Galinousky, D.; Gorshkova, T. Genes with bast fiber-specific expression in flax plants—Molecular keys for targeted fiber crop improvement. Ind. Crop. Prod. 2020, 152, 112549. [Google Scholar] [CrossRef]
- Galinousky, D.; Mokshina, N.; Padvitski, T.; Ageeva, M.; Bogdan, V.; Kilchevsky, A.; Gorshkova, T. The toolbox for fiber flax breeding: A pipeline from gene expression to fiber quality. Front. Genet. 2020, 11, 1431. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Kishlyan, N.V.; Zyablitsin, A.V.; Sadritdinova, A.F.; Snezhkina, A.; Fedorova, M.; Yurkevich, O.; Muravenko, O.; et al. Glutathione S-transferases and UDP-glycosyltransferases are involved in response to aluminum stress in Flax. Front. Plant Sci. 2016, 7, 1920. [Google Scholar] [CrossRef] [Green Version]
- Métraux, J.-P.; Burkhart, W.; Moyer, M.; Dincher, S.; Middlesteadt, W.; Williams, S.; Payne, G.; Carnes, M.; Ryals, J. Isolation of a complementary DNA encoding a chitinase with structural homology to a bifunctional lysozyme/chitinase. Proc. Natl. Acad. Sci. USA 1989, 86, 896–900. [Google Scholar] [CrossRef] [Green Version]
- Melchers, L.S.; Apotheker-de Groot, M.; van der Knaap, J.A.; Ponstein, A.S.; Sela-Buurlage, M.B.; Bol, J.F.; Cornelissen, B.; van den Elzen, P.; Linthorst, H. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 1994, 5, 469–480. [Google Scholar] [CrossRef]
- García-Olmedo, F.; Molina, A.; Alamillo, J.M.; Rodríguez-Palenzuéla, P. Plant defense peptides. Biopolymers 1998, 47, 479–491. [Google Scholar] [CrossRef]
- Van Damme, E.J.; Culerrier, R.; Barre, A.; Alvarez, R.; Rougé, P.; Peumans, W.J. A novel family of lectins evolutionarily related to class V chitinases: An example of neofunctionalization in legumes. Plant Physiol. 2007, 144, 662–672. [Google Scholar] [CrossRef] [Green Version]
- Faruque, K.; Begam, R.; Deyholos, M.K. The amaranthin-like lectin (LuALL) genes of flax: A unique gene family with members inducible by defence hormones. Plant Mol. Biol. Rep. 2015, 33, 731–741. [Google Scholar] [CrossRef]
- Leach, M.R.; Williams, D.B. Calnexin and Calreticulin, Molecular Chaperones of the Endoplasmic Reticulum. In Calreticulin. Molecular Biology Intelligence; Eggleton, P., Michalak, M., Eds.; Springer: Boston, MA, USA, 2003; pp. 49–62. [Google Scholar]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [Green Version]
- Pröbsting, M.; Schenke, D.; Hossain, R.; Häder, C.; Thurau, T.; Wighardt, L.; Schuster, A.; Zhou, Z.; Ye, W.; Rietz, S.; et al. Loss of function of CRT1a (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotech. J. 2020, 18, 2328–2344. [Google Scholar] [CrossRef]
- Nouri, M.Z.; Hiraga, S.; Komatsu, S. Characterization of calnexin in soybean roots and hypocotyls under osmotic stress. Phytochemistry 2012, 74, 20–29. [Google Scholar] [CrossRef]
- Heilmann, I.; Shin, J.; Huang, J.; Perera, I.Y.; Davies, E. Transient dissociation of polyribosomes and concurrent recruitment of calreticulin and calmodulin transcripts in gravistimulated maize pulvini. Plant Physiol. 2001, 127, 1193–1203. [Google Scholar]
- Wojtasik, W.; Kulma, A.; Kostyn, K.; Szopa, J. The changes in pectin metabolism in flax infected with Fusarium. Plant Physiol. Biochem. 2011, 49, 862–872. [Google Scholar] [CrossRef]
- Sampedro, J.; Gianzo, C.; Iglesias, N.; Guitián, E.; Revilla, G.; Zarra, I. AtBGAL10 Is the Main Xyloglucan β-Galactosidase in Arabidopsis, and Its Absence Results in Unusual Xyloglucan Subunits and Growth Defects. Plant Physiol. 2012, 158, 1146–1157. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, A.A.; McCarthy, J.G. The structure of two N-methyltransferases from the caffeine biosynthetic pathway. Plant Physiol. 2007, 144, 879–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.; Li, H.Y.; Shen, J.; Wang, J.; Jiang, L. QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J. Exp. Bot. 2011, 62, 5063–5078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willats, W.G.; Orfila, C.; Limberg, G.; Buchholt, H.C.; van Alebeek, G.J.; Voragen, A.G.; Marcus, S.; Christensen, T.; Mikkelsen, J.; Murray, B.; et al. Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J. Biol. Chem. 2001, 276, 19404–19413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galindo-Trigo, S.; Grand, T.M.; Voigt, C.A.; Smith, L.M. A malectin domain kinesin functions in pollen and seed development in Arabidopsis. J. Exp. Bot. 2020, 71, 1828–1841. [Google Scholar] [CrossRef]
- Gu, F.; Bringmann, M.; Combs, J.R.; Yang, J.; Bergmann, D.C.; Nielsen, E. Arabidopsis CSLD5 Functions in Cell Plate Formation in a Cell Cycle-Dependent Manner. Plant Cell 2016, 28, 1722–1737. [Google Scholar] [CrossRef] [Green Version]
- Eggermont, L.; Verstraeten, B.; Van Damme, E.J.M. Genome-wide screening for lectin motifs in Arabidopsis thaliana. Plant Genome 2017, 10, 1–17. [Google Scholar] [CrossRef]
- Slavokhotova, A.A.; Shelenkov, A.A.; Andreev, Y.A.; Odintsova, T.I. Hevein-like antimicrobial peptides of plants. Biochem. (Mosc.) 2017, 82, 1659–1674. [Google Scholar] [CrossRef]
- Barad, S.; Sela, N.; Dubey, A.K.; Kumar, D.; Luria, N.; Ment, D.; Cohen, S.; Schaffer, A.; Prusky, D. Differential gene expression in tomato fruit and Colletotrichum gloeosporioides during colonization of the RNAi–SlPH tomato line with reduced fruit acidity and higher pH. BMC Genom. 2017, 18, 579. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Wang, X.Y.; Guo, W.Z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integrat. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef] [Green Version]
- Bathe, U.; Tissier, A. Cytochrome P450 enzymes: A driving force of plant diterpene diversity. Phytochemistry 2019, 161, 149–162. [Google Scholar] [CrossRef]
- Chen, Y.; Peumans, W.J.; Hause, B.; Bras, J.; Kumar, M.; Proost, P.; Barre, A.; Rouge, P.; Van Damme, E. Jasmonate methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB J. 2002, 16, 905–907. [Google Scholar] [CrossRef]
- Delporte, A.; De Zaeytijd, J.; De Storme, N.; Azmi, A.; Geelen, D.; Smagghe, G.; Guisez, Y.; Van Damme, E. Cell cycle-dependent O-GlcNAc modification of tobacco histones and their interaction with the tobacco lectin. Plant Physiol. Biochem. 2014, 83, 151–158. [Google Scholar] [CrossRef]
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Wang, H.; Xing, H.; Zhao, J.; Guo, N. GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol. 2019, 19, 598. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Yu, D. The WRKY57 Transcription Factor Affects the Expression of Jasmonate ZIM-Domain Genes Transcriptionally to Compromise Botrytis cinerea Resistance. Plant Physiol. 2016, 171, 2771–2782. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2021, 72, 1473–1489. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef] [Green Version]
- Stefanowicz, K.; Lannoo, N.; Zhao, Y.; Eggermont, L.; Hove, J.V.; Atalah, B.A.; Van Damme, E.J.M. Glycan-binding F-box protein from Arabidopsis thaliana protects plants from Pseudomonas syringae infection. BMC Plant Biol. 2016, 16, 213. [Google Scholar] [CrossRef] [Green Version]
- Zaltsman, A.; Krichevsky, A.; Loyter, A.; Citovsky, V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 2010, 7, 197–209. [Google Scholar] [CrossRef] [Green Version]
- Bishop, C.D.; Cooper, R.M. An ultrastructural study of vascular colonization in 3 vascular wilt diseases. 1. Colonization of susceptible cultivars. Physiol. Plant Pathol. 1983, 23, 323–343. [Google Scholar] [CrossRef]
- Ndimba, B.K.; Chivasa, S.; Hamilton, J.M.; Simon, W.J.; Slabas, A.R. Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 2003, 3, 1047–1059. [Google Scholar] [CrossRef]
- Cole, S.J.; Yoon, A.J.; Faull, K.F.; Diener, A.C. Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Mol. Plant Pathol. 2014, 15, 589–600. [Google Scholar] [CrossRef]
- Yamada, Y.; Nishida, S.; Shitan, N.; Sato, F. Genome-wide identification of AP2/ERF transcription factor-encoding genes in California poppy (Eschscholzia californica) and their expression profiles in response to methyl jasmonate. Sci. Rep. 2020, 10, 18066. [Google Scholar] [CrossRef]
- Dang, L.; Rougé, P.; Van Damme, E.J.M. Amaranthin-like proteins with aerolysin domains in plants. Front Plant Sci. 2017, 8, 1368. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.A.; Walter, M.H.; Ralph, S.G.; Dabrowska, P.; Luck, K.; Uros, E.M.; Boland, W.; Strack, D.; Rodriguez-Concepcion, M.; Bohlmann, J.; et al. Functional identification and differential expression of 1-deoxy-D-xylulose 5-phosphate synthase in induced terpenoid resin formation of Norway spruce (Picea abies). Plant Mol. Biol. 2007, 65, 243–257. [Google Scholar] [CrossRef]
- Zulak, K.; Bohlmann, J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J. Integr. Plant Biol. 2010, 52, 86–97. [Google Scholar] [CrossRef]
- Love, M.I.; Anders, S.; Kim, V.; Huber, W. RNA-Seq workflow: Gene-level exploratory analysis and differential expression. F1000Research 2015, 4, 1070. [Google Scholar] [CrossRef]
- SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat. Biotechnol. 2014, 32, 903–914. [Google Scholar] [CrossRef]
- TAIR. Available online: https://www.arabidopsis.org/ (accessed on 8 December 2021).
- Clemente, H.S.; Pont-Lezica, R.; Jamet, E. Bioinformatics as a tool for assessing the quality of sub-cellular proteomic strategies and inferring functions of proteins: Plant cell wall proteomics as a test case. Bioinform. Biol. Insights 2009, 3, 15–28. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrova, N.; Mokshina, N. Using FIBexDB for In-Depth Analysis of Flax Lectin Gene Expression in Response to Fusarium oxysporum Infection. Plants 2022, 11, 163. https://doi.org/10.3390/plants11020163
Petrova N, Mokshina N. Using FIBexDB for In-Depth Analysis of Flax Lectin Gene Expression in Response to Fusarium oxysporum Infection. Plants. 2022; 11(2):163. https://doi.org/10.3390/plants11020163
Chicago/Turabian StylePetrova, Natalia, and Natalia Mokshina. 2022. "Using FIBexDB for In-Depth Analysis of Flax Lectin Gene Expression in Response to Fusarium oxysporum Infection" Plants 11, no. 2: 163. https://doi.org/10.3390/plants11020163




