Few-Shot Learning for Plant-Disease Recognition in the Frequency Domain
Abstract
1. Introduction
- (1)
- This is the first work to introduce plant-disease recognition in the frequency domain.
- (2)
- We adopted DCT to transform data to the frequency domain. For the DCT module, we propose a learning-based frequency selection method to improve the adaptability to different datasets or data settings. In addition, we designed a GC module to align the skewed distributions of feature vectors to a Gaussian-like distribution. The two modules can be flexibly ported to other methods or networks.
- (3)
- We conduct extensive experiments to explore the DCT module and GC module. Compared with the related methods, the accuracy our method is state-of-the-art.
2. Experiments and Results
- RandomResizedCrop;
- RandomHorizontalFlip;
- Normalize.
- Resize(int(filter_size*image_size*1.15)) (before DCT);
- CenterCrop(filter_size*image_size) (before DCT);
- Normalize (after DCT).
2.1. Comparison and Ablation Study
2.2. Channel Selection
2.3. The Effect of Top-N
2.4. The Effect of High Frequency
2.5. The Effect of Filter-Size
2.6. Ablation Experiment of the GC Module
2.7. Setting of
2.8. Integrating Spatial and Frequency Representation
2.9. Efficiency Analysis
2.10. Comparison with Related Works
3. Discussion
3.1. Motivation, Works and Contributions
3.2. Findings
- 1.
- The performance was improved in the frequency domain compared to in the spatial domain, which indicates that better features can be obtained from frequency representation. In other words, CNNs can extract more critical features from a frequency representation. Another proof demonstrates this point, which is that the convergent speed was much faster for the frequency domain than for the spatial domain, and the loss was smaller as well. This important finding suggests that the frequency representation in the frequency domain can be attempted not only for classification tasks in FSL, but also for other classification tasks, and even for any other task that uses a CNN to extract features.
- 2.
- In this work, we used cosine similarity to calculate the distance, and the GC module has shown prominent improvements in both the frequency domain and the spatial domain. Our method illustrates the use of Gaussian-like calibration in a CNN pipeline. In fact, the GC module can be used in a broader range of machine learning. Several researches have shown that not only cosine similarity, but also other distance-based methods, such as KNN, K-means and the SVM classifier, are worth using on PT to improve performance.
- 3.
- We have demonstrated that channel selection in DCT is critical to determine the performance. Different frequency combinations lead to very different results. As the frequencies are considered as channels, the channel attention can be used to calculate the weights of different frequencies. We provided an idea for frequency selection. In this work, the SE module was used. Actually, it can be replaced with other channel attention modules, such as the channel attention module in the CABM, thereby providing great flexibly [42].
- 4.
- In principle, performance is always lifted when adding more information. However, the experiment of combining the spatial and frequency representations together did not show an improvement, but a detriment. According to our analysis, the spatial representation and the frequency representation are two ways of representing the same entity, but the substance of the content is the same. In the whole pipeline, the performance is also decided by the parsing ability of the backbone network to a certain representation of sample. The combination of the spatial representation and frequency representation does not enrich the features or generate extra useful information. Conversely, redundant representation become the interference factor.
- 5.
- In recent years, the architectures of networks have gone deeper and deeper. However, it does not mean that deeper networks always outperform shallower networks. First, FSL is a kind of learning task with limited data-scale. For a deeper network, it always has a large number of parameters that need to be updated. Under the data-limitation condition, too deep a network could result in insufficient updating of parameters in back-propagation due to the overly long back-propagation path. In parameter updating, shallower networks are more flexible, and the deeper networks are bulky. In addition, our specific task identified diseases while relying on the colors, shapes and textures of lesions. These simpler and more basic features are learnt in shallower layers. Hence, the very deep networks are not helpful. In short, the size of network should match the specific task and data resources.
3.3. Limitations and Future Works
- 1.
- Multi-disease cases are not involved in this work. In short, the samples of PV were taken under controlled conditions (laboratory settings). These settings make the samples relatively simple and differ significantly from those obtained under in-field conditions. That is the reason many researches have already achieved high accuracy by using deep-learning CNNs on PV [43]. Since the aim of this work was to explore methods in the frequency domain and we did not want the content of this paper to be scattered, we only used PV in our experiments. Therefore, the multi-disease cases were not taken into account in this work. In fact, once infected by the first disease, the plant is vulnerable to other diseases, as the immune system is attacked and becomes weak [44]. In real field conditions, it is relatively common for multiple diseases to occur in one plant. However, the combinations of different diseases are too many to collect sufficient samples for each category from classification perspective (e.g., three diseases of a species generate seven categories). Hence, if using classification to solve this problem, requirements of data are difficult to meet. We recommend using semantic segmentation to solve this problem.
- 2.
- As shown in Section 4.1, the images of PV have simple backgrounds. Complex background cases were not considered in this work. In the application, we could not predict the scenario in the test. Images may be taken from fields with complex backgrounds. In order to improve the accuracy, it is necessary to perform pre-processing to reduce the influence of the background, which involves the research direction of object detection (e.g., leaves, fruits or lesions detection). In future work, research on plant disease identification should shift to real field environments. The treatment of complex environments is important to advance the technology into practical applications.
- 3.
- In this work, we set up three data settings to mimic different application scenarios. The results of setting-1 and setting-2 are far superior to those of setting-3. setting-1 is the easiest setting because the features of different plants are very distinguishable. Setting-1 covers 10 plants, and setting-2 covers 3 plants. The results of setting-2 got close to those of the setting-1, which indicates that the model not only identified the plants but also the diseases. The performance of setting-3 with 10 test categories belonging to tomato dropped. This kind of setting is more meaningful to farmers but is a difficult task. To some extent, this is the purest form of disease identification, without any notion of species. It is sub-class classification (fine-grained vision classification), which is another research topic for computer vision. It needs more finely distinguishable features, which like humans, distinguish two similar objects. The lesion detection can help the networks focusing on the fine-grained features and improve the sub-class classification task.
- 4.
- Cross-domain research is a hot topic in FSL recently, which is defined as training with one dataset and generalizing to another dataset in the test. Our work does not refer to this topic because only one dataset was used. Further mining the benefits of feature distribution may solve the cross-domain problem. In PV, although there are differences between categories, the distributions of the samples are similar because these samples were collected under the same conditions. The data from different datasets would lead to more complex distributions. The feature distributions of different datasets and the distribution calibration of the different domains are worth studying.
- 5.
- Another focus is the fine research of the frequency channels. In this work, although we evaluated the weights of each channel, we did not do further research of these channels. In future work, some visualization works and fine studies of each channel could help to better understand the frequency representation.
- 6.
- Even though we demonstrated that CNNs can extract better features from frequency representations, networks that are more suitable for frequency representations are worth being investigated.
4. Materials and Methods
4.1. Materials
4.2. Method
4.2.1. Problem Definition
4.2.2. Framework
4.2.3. DCT Module
4.2.4. Gaussian-like Calibrator
4.2.5. Distance Measurement
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FSL | few-shot learning |
| PV | PlantVillage (dataset) |
| CNN | convolutional neural network |
| GC | Gaussian-like calibration |
| DCT | discrete cosine transform |
| PT | power transform |
| SE | Squeeze-and-Excitation (attention module) |
References
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef] [PubMed]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Q.; Kwok, J.T.; Ni, L.M. Generalizing from a few examples: A survey on few-shot learning. ACM Comput. Surv. (CSUR) 2020, 53, 1–34. [Google Scholar] [CrossRef]
- Koch, G.; Zemel, R.; Salakhutdinov, R. Siamese neural networks for one-shot image recognition. In Proceedings of the ICML Deep Learning Workshop, Lille, France, 6–11 July 2015; Volume 2. [Google Scholar]
- Vinyals, O.; Blundell, C.; Lillicrap, T.; Wierstra, D. Matching networks for one shot learning. Adv. Neural Inf. Process. Syst. 2016, 29, 3630–3638. [Google Scholar] [CrossRef]
- Snell, J.; Swersky, K.; Zemel, R.S. Prototypical networks for few-shot learning. arXiv 2017, arXiv:1703.05175. [Google Scholar]
- Sung, F.; Yang, Y.; Zhang, L.; Xiang, T.; Torr, P.H.; Hospedales, T.M. Learning to compare: Relation network for few-shot learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 1199–1208. [Google Scholar] [CrossRef]
- Li, W.; Xu, J.; Huo, J.; Wang, L.; Gao, Y.; Luo, J. Distribution consistency based covariance metric networks for few-shot learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January 27–1 February 2019; Volume 33, pp. 8642–8649. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X.; Liu, Z.; Xu, H.; Darrell, T. A new meta-baseline for few-shot learning. arXiv 2020, arXiv:2003.04390. [Google Scholar]
- Argüeso, D.; Picon, A.; Irusta, U.; Medela, A.; San-Emeterio, M.G.; Bereciartua, A.; Alvarez-Gila, A. Few-Shot Learning approach for plant disease classification using images taken in the field. Comput. Electron. Agric. 2020, 175, 105542. [Google Scholar] [CrossRef]
- Jadon, S. SSM-Net for Plants Disease Identification in Low Data Regime. In Proceedings of the 2020 IEEE/ITU International Conference on Artificial Intelligence for Good (AI4G), Geneva, Switzerland, 21–25 September 2020; pp. 158–163. [Google Scholar] [CrossRef]
- Zhong, F.; Chen, Z.; Zhang, Y.; Xia, F. Zero-and few-shot learning for diseases recognition of Citrus aurantium L. using conditional adversarial autoencoders. Comput. Electron. Agric. 2020, 179, 105828. [Google Scholar] [CrossRef]
- Li, Y.; Chao, X. Semi-supervised few-shot learning approach for plant diseases recognition. Plant Methods 2021, 17, 1–10. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J. Meta-learning baselines and database for few-shot classification in agriculture. Comput. Electron. Agric. 2021, 182, 106055. [Google Scholar] [CrossRef]
- Chen, W.Y.; Liu, Y.C.; Kira, Z.; Wang, Y.C.F.; Huang, J.B. A closer look at few-shot classification. arXiv 2019, arXiv:1904.04232. [Google Scholar]
- Duan, Y.; Zheng, W.; Lin, X.; Lu, J.; Zhou, J. Deep adversarial metric learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 2780–2789. [Google Scholar] [CrossRef]
- Afifi, A.; Alhumam, A.; Abdelwahab, A. Convolutional neural network for automatic identification of plant diseases with limited data. Plants 2021, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cui, X.; Li, W. Meta-Learning for Few-Shot Plant Disease Detection. Foods 2021, 10, 2441. [Google Scholar] [CrossRef] [PubMed]
- Nuthalapati, S.V.; Tunga, A. Multi-Domain Few-Shot Learning and Dataset for Agricultural Applications. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 1399–1408. [Google Scholar] [CrossRef]
- Lin, H.; Tse, R.; Tang, S.K.; Qiang, Z.P.; Pau, G. The Positive Effect of Attention Module in Few-Shot Learning for Plant Disease Recognition. In Proceedings of the 5th International Conference on Pattern Recognition and Artificial Intelligence (PRAI), IEEE, Chengdu, China, 19–21 August 2022; pp. 114–120. [Google Scholar] [CrossRef]
- Lin, H.; Tse, R.; Tang, S.K.; Qiang, Z.P.; Pau, G. Few-shot learning approach with multi-scale feature fusion and attention for plant disease recognition. Front. Plant Sci. 2022, 13, 907916. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Y.; Wang, X.; Wang, W.; Li, J. Development of convolutional neural network and its application in image classification: A survey. Opt. Eng. 2019, 58, 040901. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.Y.; Porikli, F.; Plaza, A.J.; Kehtarnavaz, N.; Terzopoulos, D. Image segmentation using deep learning: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar] [CrossRef]
- Liu, L.; Ouyang, W.; Wang, X.; Fieguth, P.; Chen, J.; Liu, X.; Pietikäinen, M. Deep learning for generic object detection: A survey. Int. J. Comput. Vis. 2020, 128, 261–318. [Google Scholar] [CrossRef]
- Vijayvargiya, G.; Silakari, S.; Pandey, R. A survey: Various techniques of image compression. arXiv 2013, arXiv:1311.6877. [Google Scholar]
- Wallace, G.K. The JPEG still picture compression standard. IEEE Trans. Consum. Electron. 1992, 38, xviii–xxxiv. [Google Scholar] [CrossRef]
- Kim, J.; Lee, S. Deep learning of human visual sensitivity in image quality assessment framework. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1676–1684. [Google Scholar] [CrossRef]
- Xu, K.; Qin, M.; Sun, F.; Wang, Y.; Chen, Y.K.; Ren, F. Learning in the frequency domain. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 1740–1749. [Google Scholar] [CrossRef]
- Chen, X.; Wang, G. Few-shot learning by integrating spatial and frequency representation. In Proceedings of the 2021 18th Conference on Robots and Vision (CRV), Burnaby, BC, Canada, 26–28 May 2021; pp. 49–56. [Google Scholar] [CrossRef]
- Ahmed, N.; Natarajan, T.; Rao, K.R. Discrete cosine transform. IEEE Trans. Comput. 1974, 100, 90–93. [Google Scholar] [CrossRef]
- Natarajan, B.K.; Vasudev, B. A fast approximate algorithm for scaling down digital images in the DCT domain. In Proceedings of the International Conference on Image Processing, Washington, DC, USA, 23–26 October 1995; Volume 2, pp. 241–243. [Google Scholar] [CrossRef]
- Hussain, A.J.; Al-Fayadh, A.; Radi, N. Image compression techniques: A survey in lossless and lossy algorithms. Neurocomputing 2018, 300, 44–69. [Google Scholar] [CrossRef]
- Gueguen, L.; Sergeev, A.; Kadlec, B.; Liu, R.; Yosinski, J. Faster neural networks straight from jpeg. Adv. Neural Inf. Process. Syst. 2018, 31, 3937–3948. Available online: https://proceedings.neurips.cc/paper/2018/file/7af6266cc52234b5aa339b16695f7fc4-Paper.pdf (accessed on 14 October 2022).
- Yang, S.; Liu, L.; Xu, M. Free lunch for few-shot learning: Distribution calibration. arXiv 2021, arXiv:2101.06395. [Google Scholar]
- Tukey, J.W. Exploratory Data Analysis; Addison-Wesley Pub. Co.: Reading, MA, USA, 1977; Volume 2. [Google Scholar] [CrossRef]
- Hu, Y.; Gripon, V.; Pateux, S. Leveraging the feature distribution in transfer-based few-shot learning. In Proceedings of the International Conference on Artificial Neural Networks, Bratislava, Slovakia, 14–17 September 2021; Springer: Cham, Switzerland, 2021; pp. 487–499. [Google Scholar] [CrossRef]
- Hu, Y.; Pateux, S.; Gripon, V. Squeezing backbone feature distributions to the max for efficient few-shot learning. Algorithms 2022, 15, 147. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar] [CrossRef]
- Stanković, R.S.; Astola, J.T. Reprints from the Early Days of Information Sciences (Reminiscences of the Early Work in DCT); Technical Report; Tampere International Center for Signal Processing: Tampere, Finland, 2012; Available online: https://ethw.org/w/images/1/19/Report-60.pdf (accessed on 14 October 2022).
- Wang, Y.; Wang, S. IMAL: An Improved Meta-learning Approach for Few-shot Classification of Plant Diseases. In Proceedings of the 2021 IEEE 21st International Conference on Bioinformatics and Bioengineering (BIBE), Kragujevac, Serbia, 25–27 October 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Finn, C.; Abbeel, P.; Levine, S. Model-agnostic meta-learning for fast adaptation of deep networks. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 1126–1135. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar] [CrossRef]
- Hasan, R.I.; Yusuf, S.M.; Alzubaidi, L. Review of the state of the art of deep learning for plant diseases: A broad analysis and discussion. Plants 2020, 9, 1302. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Kahu, S.Y.; Raut, R.B.; Bhurchandi, K.M. Review and evaluation of color spaces for image/video compression. Color Res. Appl. 2019, 44, 8–33. [Google Scholar] [CrossRef]
- Chan, G. Toward Better Chroma Subsampling: Recipient of the 2007 SMPTE Student Paper Award. SMPTE Motion Imaging J. 2008, 117, 39–45. [Google Scholar] [CrossRef]
- Florea, C.; Gordan, M.; Orza, B.; Vlaicu, A. Compressed domain computationally efficient processing scheme for JPEG image filtering. In Advanced Engineering Forum; Trans Tech Publications Ltd.: Stafa-Zurich, Switzerland, 2013; Volume 8, pp. 480–489. [Google Scholar] [CrossRef]
- Chen, S.; Ma, B.; Zhang, K. On the similarity metric and the distance metric. Theor. Comput. Sci. 2009, 410, 2365–2376. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M.; Pei, J. 2-Getting to Know Your Data. In Data Mining, 3rd ed.; Han, J., Kamber, M., Pei, J., Eds.; The Morgan Kaufmann Series in Data Management Systems; Morgan Kaufmann: Boston, MA, USA, 2012; pp. 39–82. [Google Scholar] [CrossRef]









| ID | Method | Channel | 1-Shot | 5-Shot | 10-Shot | 20-Shot | 30-Shot | 40-Shot | 50-Shot |
|---|---|---|---|---|---|---|---|---|---|
| setting-1 | |||||||||
| e1 | s | - | 79.32 | 91.19 | 92.53 | 93.55 | 93.84 | 93.97 | 93.86 |
| e2 | s + GC | - | 83.08 | 93.15 | 94.71 | 95.75 | 95.90 | 96.01 | 95.84 |
| e3 | f | top-24 | 84.99 | 94.92 | 96.71 | 95.91 | 97.15 | 97.31 | 97.62 |
| e4 | f + GC | top-24 | 86.34 | 95.30 | 96.93 | 97.48 | 97.62 | 97.83 | 98.01 |
| setting-2 | |||||||||
| e5 | s | - | 78.27 | 90.96 | 92.69 | 93.60 | 93.76 | 94.02 | 94.03 |
| e6 | s + GC | - | 79.81 | 92.11 | 93.58 | 94.26 | 94.51 | 94.50 | 94.74 |
| e7 | f | top-24 | 82.85 | 94.48 | 95.58 | 96.35 | 96.59 | 96.78 | 96.77 |
| e8 | f + GC | top-24 | 85.41 | 95.21 | 96.17 | 96.66 | 96.90 | 97.04 | 97.15 |
| setting-3 | |||||||||
| e9 | s | - | 57.46 | 75.12 | 79.32 | 81.41 | 82.48 | 83.32 | 83.50 |
| e10 | s + GC | - | 60.69 | 79.38 | 82.78 | 85.19 | 86.18 | 86.87 | 86.86 |
| e11 | f | top-24 | 62.32 | 80.37 | 83.57 | 85.75 | 86.51 | 87.11 | 87.22 |
| e12 | f + GC | top-24 | 64.54 | 80.89 | 84.06 | 85.91 | 86.67 | 87.01 | 87.33 |
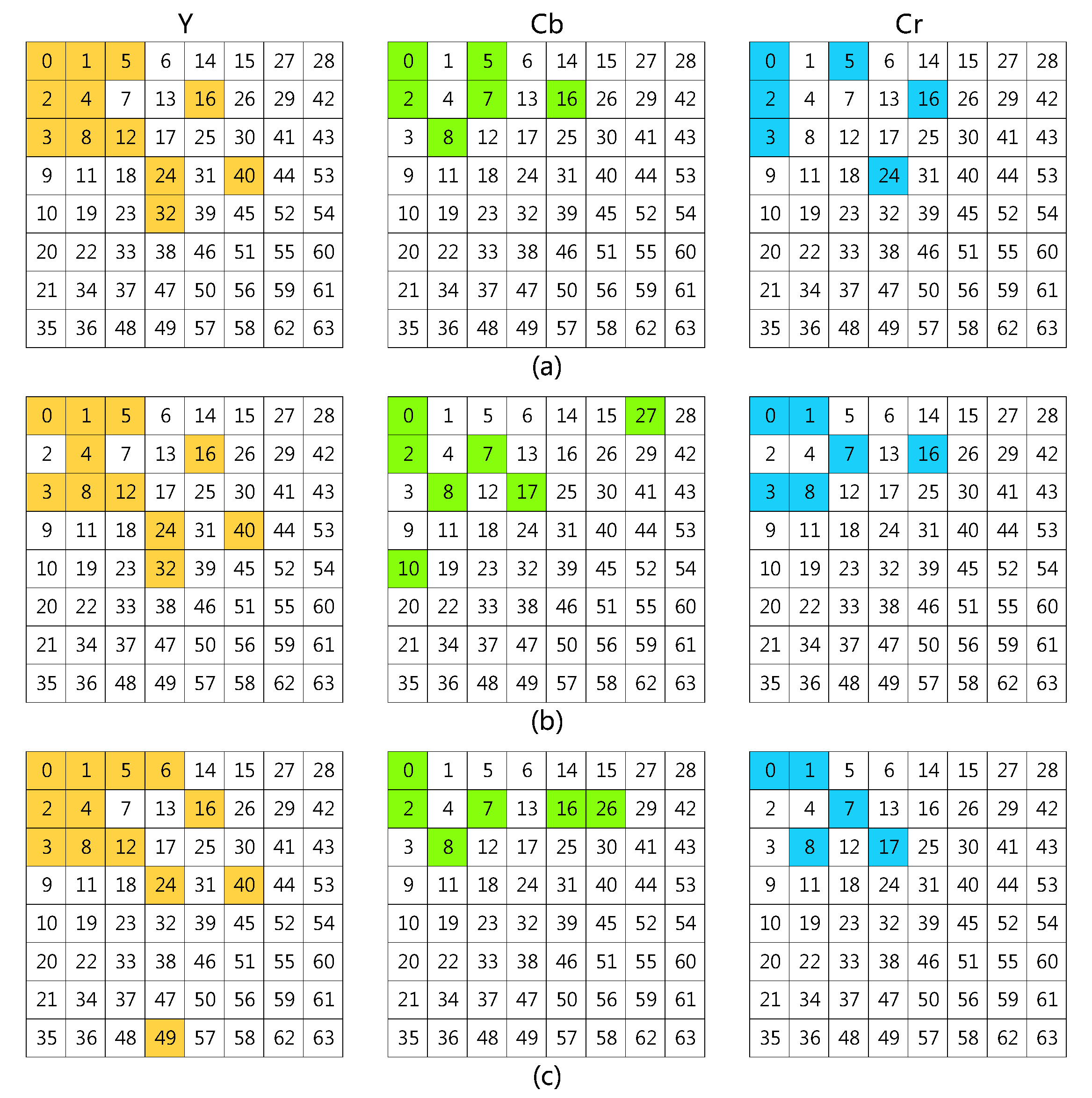
| ID | Channel | Y-Cb-Cr | Y | Cb | Cr | 1-Shot | 5-Shot | 10-Shot |
|---|---|---|---|---|---|---|---|---|
| e13 | top-24 | 14-5-5 | 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 14, 15, 20 | 0, 1, 2, 6, 9 | 0, 1, 2, 6, 9 | 79.66 | 93.56 | 95.24 |
| e7 | top-24 | 11-7-6 | 0, 1, 3, 4, 5, 8, 12, 16, 24, 32, 40 | 0, 2, 7, 8, 10, 17, 27 | 0, 1, 3, 7, 8, 16 | 82.85 | 94.48 | 95.58 |
| ID | Channel | Y-Cb-Cr | Y | Cb | Cr | 1-Shot | 5-Shot | 10-Shot |
|---|---|---|---|---|---|---|---|---|
| e14 | top-8 | 4-2-2 | 0, 8, 24, 40 | 0, 7 | 0, 16 | 80.53 | 93.08 | 94.39 |
| e15 | top-16 | 8-4-4 | 0, 1, 4, 5, 8, 24, 32, 40 | 0, 7, 8, 17 | 0, 7, 8, 16 | 81.34 | 93.73 | 95.05 |
| e7 | top-24 | 11-7-6 | 0, 1, 3, 4, 5, 8, 12, 16, 24, 32, 40 | 0, 2, 7, 8, 10, 17, 27 | 0, 1, 3, 7, 8, 16 | 82.85 | 94.48 | 95.58 |
| e16 | top-32 | 14-8-10 | 0, 1, 2, 3, 4, 5, 8, 12, 16, 24, 32, 34, 35, 40 | 0, 2, 7, 8, 10, 16, 17, 27 | 0, 1, 2, 3, 7, 8, 16, 17, 24, 32 | 82.25 | 94.10 | 95.36 |
| e17 | top-48 | 25-9-14 | 0, 1, 2, 3, 4, 5, 6, 8, 12, 16, 20, 24, 32, 34, 35, 36, 39, 40, 43, 49, 51, 56, 58, 59, 60 | 0, 2, 7, 8, 10, 16, 17, 18, 27 | 0, 1, 2, 3, 5, 7, 8, 10, 16, 17, 18, 19, 24, 32 | 81.84 | 93.71 | 94.94 |
| e18 | top-64 | 38-11-15 | 0, 1, 2, 3, 4, 5, 6, 7, 8, 12, 16, 20, 22, 24, 31, 32, 34, 35, 36, 38, 39, 40, 41, 43, 45, 46, 48, 49, 51, 53, 54, 55, 56, 58, 59, 60, 61, 62 | 0, 2, 7, 8, 10, 16, 17, 18, 19, 26, 27 | 0, 1, 2, 3, 4, 5, 7, 8, 10, 16, 17, 18, 19, 24, 32 | 82.84 | 94.49 | 95.77 |
| e19 | 64(Y) | 64-0-0 | 0-63 | - | - | 71.01 | 86.61 | 88.71 |
| ID | Channel | Y-Cb-Cr | Y | Cb | Cr | 1-Shot | 5-Shot | 10-Shot |
|---|---|---|---|---|---|---|---|---|
| e7 | top-24/192 | 11-7-6 | 0, 1, 3, 4, 5, 8, 12, 16, 24, 32, 40 | 0, 2, 7, 8, 10, 17, 27 | 0, 1, 3, 7, 8, 16 | 82.85 | 94.48 | 95.58 |
| e20 | top-24/108 | 11-6-7 | 0, 1, 3, 4, 5, 8, 12, 16, 20, 24, 32 | 0, 7, 8, 10, 16, 27 | 0, 1, 2, 7, 8, 16, 24 | 80.20 | 92.50 | 94.24 |
| e21 | top-24/84 | 11-6-7 | 0, 1, 2, 3, 4, 5, 8, 12, 16, 20, 24 | 0, 7, 8, 16, 19, 27 | 0, 1, 2, 7, 8, 16, 24 | 81.23 | 93.79 | 95.19 |
| ID | Channel | Y-Cb-Cr | Y | Cb | Cr | 1-Shot | 5-Shot | 10-Shot |
|---|---|---|---|---|---|---|---|---|
| filter-size: | ||||||||
| e22 | top-6 | 3-2-1 | 0, 3, 8 | 0, 8 | 0 | 81.15 | 93.42 | 94.87 |
| e23 | top-12 | 5-3-4 | 0, 1, 2, 3, 8 | 0, 2, 8 | 0, 3, 5, 8 | 80.65 | 92.67 | 94.26 |
| e24 | full-48 | 16-16-16 | 0-15 | 0-15 | 0-15 | 80.84 | 92.59 | 94.12 |
| filter-size: | ||||||||
| e7 | top-24 | 11-7-6 | 0, 1, 3, 4, 5, 8, 12, 16, 24, 32, 40 | 0, 2, 7, 8, 10, 17, 27 | 0, 1, 3, 7, 8, 16 | 82.85 | 94.48 | 95.58 |
| e25 | top-48 | 25-9-14 | 0, 1, 2, 3, 4, 5, 6, 8, 12, 16, 20, 24, 32, 34, 35, 36, 39, 40, 43, 49, 51, 56, 58, 59, 60 | 0, 2, 7, 8, 10, 16, 17, 18, 27 | 0, 1, 2, 3, 5, 7, 8, 10, 16, 17, 18, 19, 24, 32 | 81.84 | 93.71 | 94.94 |
| e26 | full-192 | 64-64-64 | 0-63 | 0-63 | 0-63 | 81.62 | 93.55 | 95.04 |
| filter-size: | ||||||||
| e27 | top-96 | 45-26-25 | 0, 1, 2, 3, 4, 17, 19, 32, 34, 37, 40, 43, 52, 54, 56, 58, 71, 73, 82, 85, 96, 103, 106, 107, 113, 117, 142, 144, 145, 150, 158, 170, 173, 187, 188, 190, 200, 201, 208, 219, 240, 242, 244, 253, 254 | 0, 1, 2, 17, 23, 46, 69, 75, 76, 90, 126, 129, 131, 140, 160, 164, 193, 196, 200, 207, 216, 217, 224, 229, 231, 252 | 0, 33, 46, 51, 54, 59, 71, 74, 85, 87, 100, 111, 112, 164, 166, 169, 172, 189, 199, 234, 237, 238, 249, 252, 254 | 80.93 | 92.84 | 94.42 |
| e28 | top-192 | 82-50-60 | 0, 1, 2, 3, 4, 13, 17, 19, 25, 27, 28, 29, 32, 34, 37, 38, 40, 41, 43, 46, 52, 53, 54, 56, 58, 60, 62, 71, 73, 82, 85, 94, 96, 99, 103, 106, 107, 113, 117, 122, 126, 141, 142, 144, 145, 150, 154, 158, 161, 168, 170, 173, 174, 179, 180, 184, 187, 188, 190, 192, 200, 201, 206, 208, 209, 211, 212, 218, 219, 223, 224, 240, 242, 244, 252, 253, 254, 255 | 0, 1, 2, 17, 23, 36, 45, 46, 58, 66, 69, 75, 76, 79, 90, 92, 97, 112, 115, 117, 119, 126, 129, 131, 132, 140, 143, 147, 157, 160, 161, 164, 165, 169, 180, 193, 196, 200, 205, 207, 210, 212, 216, 217, 219, 224, 229, 231, 235, 238, 240, 252 | 0, 7, 17, 18, 26, 32, 33, 34, 36, 39, 45, 46, 48, 51, 52, 53, 54, 59, 60, 71, 74, 82, 85, 87, 90, 98, 99, 100, 103, 111, 112, 135, 153, 154, 156, 158, 164, 166, 169, 172, 173, 177, 181, 183, 185, 189, 196, 198, 199, 210, 219, 229, 234, 237, 238, 245, 246, 249, 250, 252, 254 | 81.73 | 93.63 | 95.05 |
| e29 | full-768 | 256-256-256 | 0-255 | 0-255 | 0-255 | 81.47 | 93.41 | 94.87 |
| ID | Components of GC Module | 1-Shot | 5-Shot | 10-Shot |
|---|---|---|---|---|
| e30 | PT | 81.82 | 93.27 | 94.91 |
| e31 | PT + normalization | 82.60 | 94.55 | 95.95 |
| e32 | PT + centralization | 85.36 | 94.94 | 96.08 |
| e8 | PT + normalization + centralization | 85.41 | 95.21 | 96.17 |
| e33 | normalization + PT + centralization | 85.04 | 94.76 | 95.92 |
| ID | 1-Shot | 5-Shot | 10-Shot | |
|---|---|---|---|---|
| e34 | 1.5 | 83.13 | 93.57 | 94.77 |
| e35 | 1 | 84.74 | 94.82 | 95.88 |
| e8 | 0.5 | 85.41 | 95.21 | 96.17 |
| e36 | 0 | 84.75 | 95.08 | 96.37 |
| e37 | −0.5 | 69.44 | 78.84 | 79.73 |
| e38 | −1 | 68.10 | 77.19 | 78.22 |
| ID | Method | 1-Shot | 5-Shot | 10-Shot |
|---|---|---|---|---|
| e6 | s + GC | 79.81 | 92.11 | 93.58 |
| e8 | f + GC | 85.36 | 94.86 | 96.13 |
| e39 | s + f + GC | 84.08 | 94.35 | 95.92 |
| Data Setting and K-Shot | |||||||
|---|---|---|---|---|---|---|---|
| ID | Method | 1-Shot | 5-Shot | 10-Shot | Y | Cb | Cr |
| Data setting in [10] | |||||||
| Siamese Contrastive [10] | 50.2 | 64.2 | 70.2 |  | |||
| Siamese Triplet [10] | 65.2 | 72.3 | 76.8 | ||||
| Single SS [13] | 74.5 | 89.7 | 92.6 | ||||
| Iterative SS [13] | 75.1 | 90.0 | 92.7 | ||||
| MAML [40,41] | 58.8 | 79.3 | 87.1 | ||||
| IMAL [40] | 63.8 | 83.5 | 89.9 | ||||
| e40 | Ours s | 76.4 | 91.0 | 93.2 | |||
| e41 | Ours s + GC | 78.7 | 91.0 | 92.3 | |||
| e42 | Ours f | 79.8 | 92.5 | 94.3 | |||
| e43 | Ours f + GC | 81.6 | 93.1 | 94.3 | |||
| Split-1 of [13] | |||||||
| Single SS [13] | 33.7 | 50.9 | 66.7 |  | |||
| Iterative SS [13] | 34.0 | 53.1 | 68.8 | ||||
| [19] | 46.6 | 63.5 | - | ||||
| e9 | Ours s | 57.5 | 75.1 | 79.3 | |||
| e10 | Ours s + GC | 60.7 | 79.4 | 82.8 | |||
| e11 | Ours f | 62.3 | 80.4 | 83.6 | |||
| e12 | Ours f + GC | 64.5 | 80.9 | 84.1 | |||
| Split-2 of [13] | |||||||
| Single SS [13] | 44.7 | 74.7 | 85.7 |  | |||
| Iterative SS [13] | 46.4 | 76.9 | 89.2 | ||||
| [19] | 70.9 | 87.0 | - | ||||
| e5 | Ours s | 78.3 | 91.0 | 92.7 | |||
| e6 | Ours s + GC | 79.8 | 92.1 | 93.6 | |||
| e7 | Ours f | 82.9 | 94.5 | 95.6 | |||
| e8 | Ours f + GC | 85.4 | 94.9 | 96.1 | |||
| Split-3 of [13] | |||||||
| Single SS [13] | 52.3 | 67.6 | 79.9 |  | |||
| Iterative SS [13] | 55.2 | 69.3 | 80.8 | ||||
| [19] | 75.4 | 88.5 | - | ||||
| e44 | Ours s | 76.6 | 88.6 | 90.7 | |||
| e45 | Ours s + GC | 81.2 | 90.0 | 91.5 | |||
| e46 | Ours f | 79.9 | 90.8 | 92.7 | |||
| e47 | Ours f + GC | 82.7 | 92.1 | 93.7 | |||
| Data Setting | Source Set (28) | Target Set (10) | Target Species |
|---|---|---|---|
| setting-1 | 2, 3, 4, 5, 7, 9, 10, 11, 13, 14, 15, 18, 20, 22, 23, 24, 25, 26, 28, 30, 31, 32, 33, 34, 35, 36, 37, 38 | 1, 6, 8, 12, 16, 17, 19, 21, 27, 29 | apple, cherry, corn, grape, peach, pepper, potato, strawberry, tomato, orange |
| setting-2 | 5, 8, 9, 10, 11, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 | 1, 2, 3, 4, 6, 7, 12, 13, 14, 15 | apple, grape, cherry |
| setting-3 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 | tomato |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.; Tse, R.; Tang, S.-K.; Qiang, Z.; Pau, G. Few-Shot Learning for Plant-Disease Recognition in the Frequency Domain. Plants 2022, 11, 2814. https://doi.org/10.3390/plants11212814
Lin H, Tse R, Tang S-K, Qiang Z, Pau G. Few-Shot Learning for Plant-Disease Recognition in the Frequency Domain. Plants. 2022; 11(21):2814. https://doi.org/10.3390/plants11212814
Chicago/Turabian StyleLin, Hong, Rita Tse, Su-Kit Tang, Zhenping Qiang, and Giovanni Pau. 2022. "Few-Shot Learning for Plant-Disease Recognition in the Frequency Domain" Plants 11, no. 21: 2814. https://doi.org/10.3390/plants11212814
APA StyleLin, H., Tse, R., Tang, S.-K., Qiang, Z., & Pau, G. (2022). Few-Shot Learning for Plant-Disease Recognition in the Frequency Domain. Plants, 11(21), 2814. https://doi.org/10.3390/plants11212814










