From a Lose–Lose to a Win–Win Situation: User-Friendly Biomass Models for Acacia longifolia to Aid Research, Management and Valorisation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Database Search for Allometric Biomass Equations
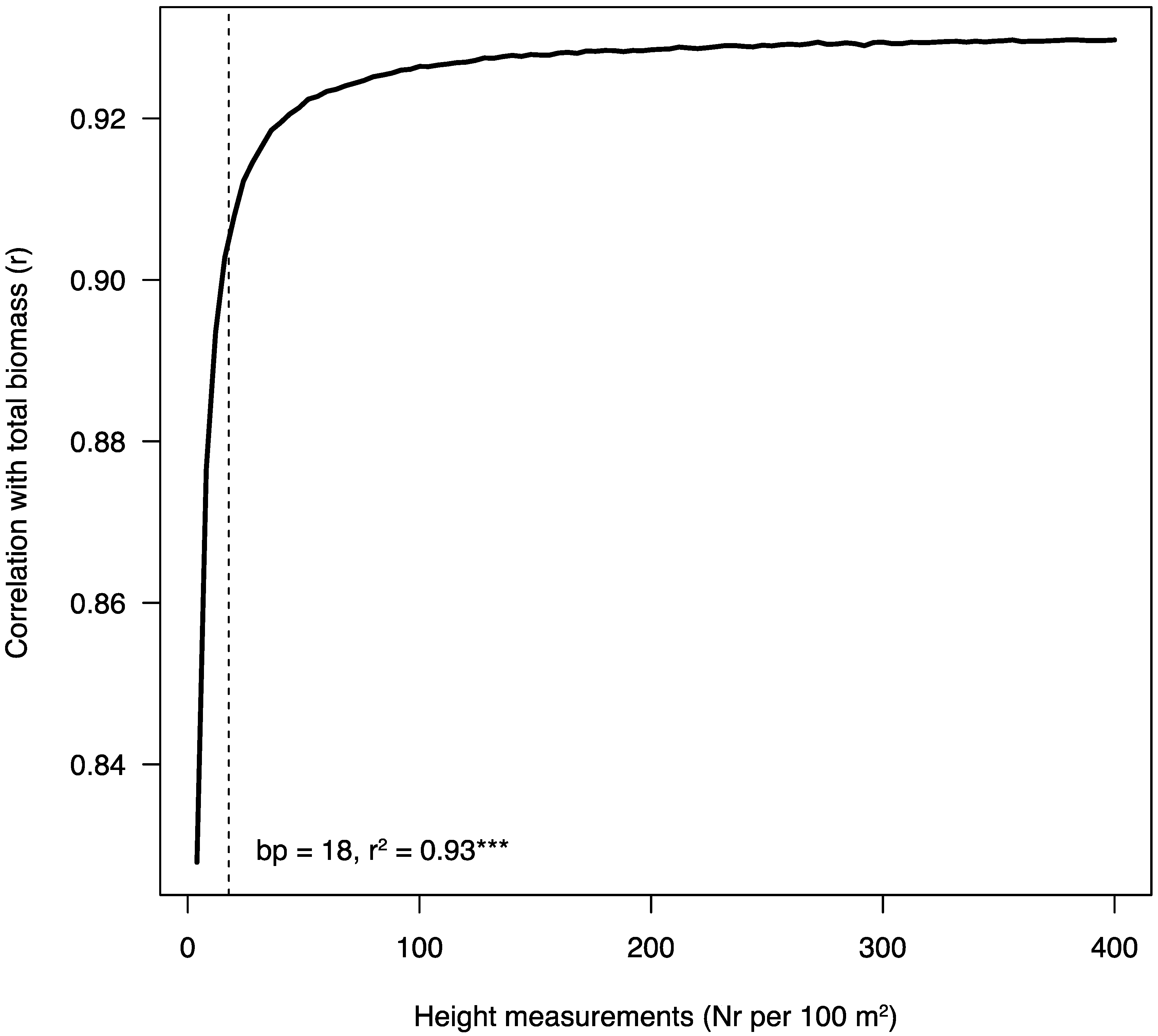
2.2. Tree and Stand Measurements
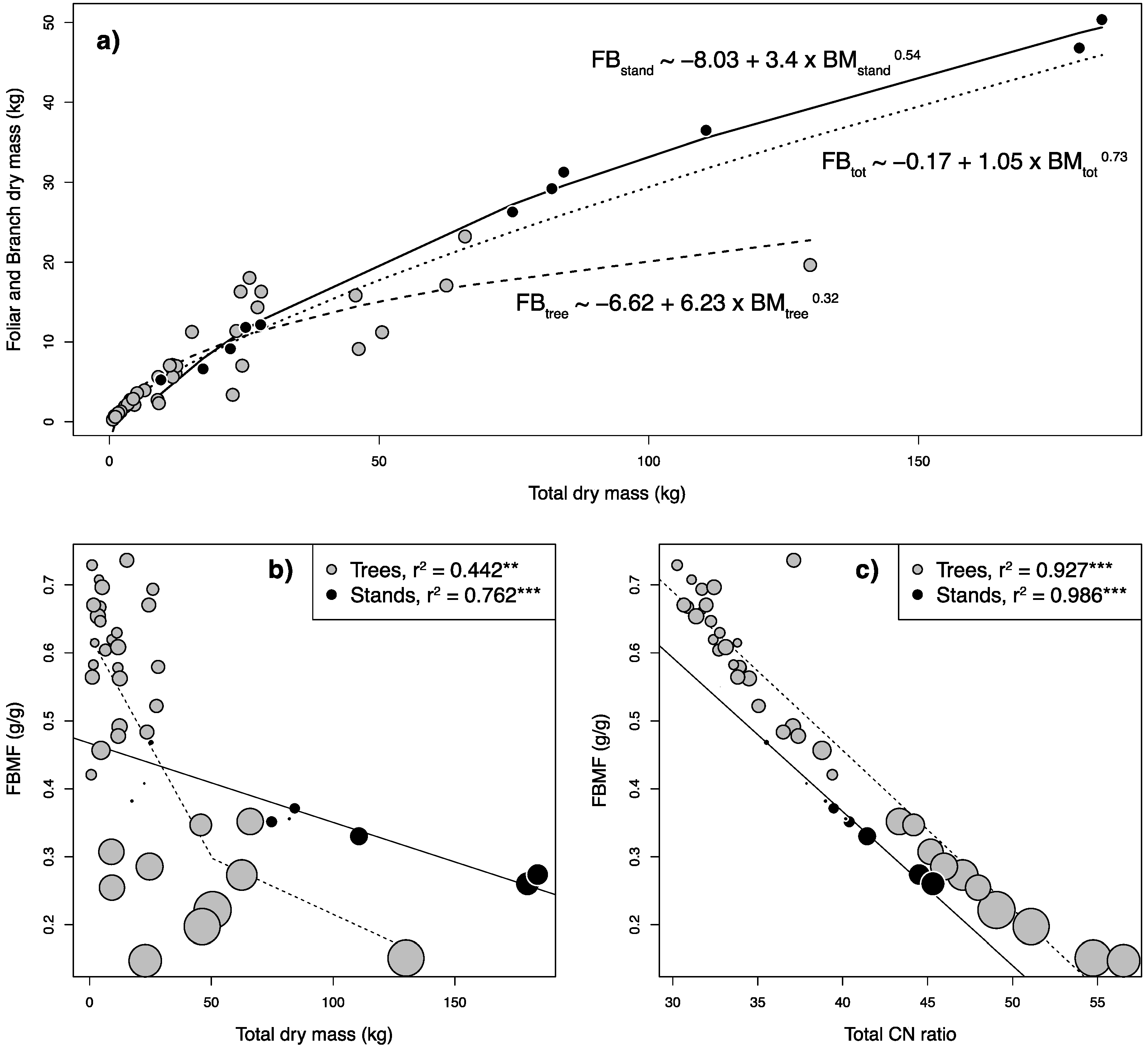
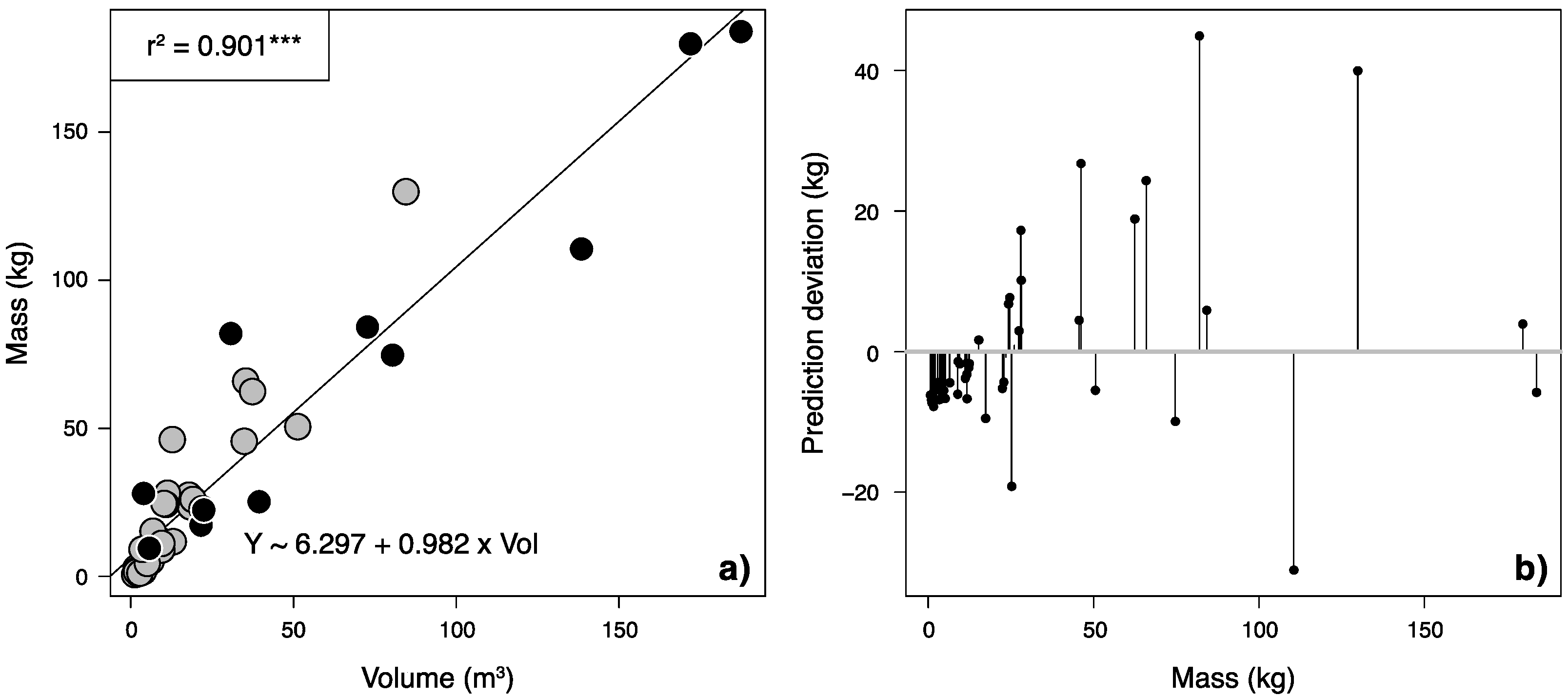
2.3. Allometric Equations
2.4. Model Simplifications towards In Situ Application
3. Materials and Methods
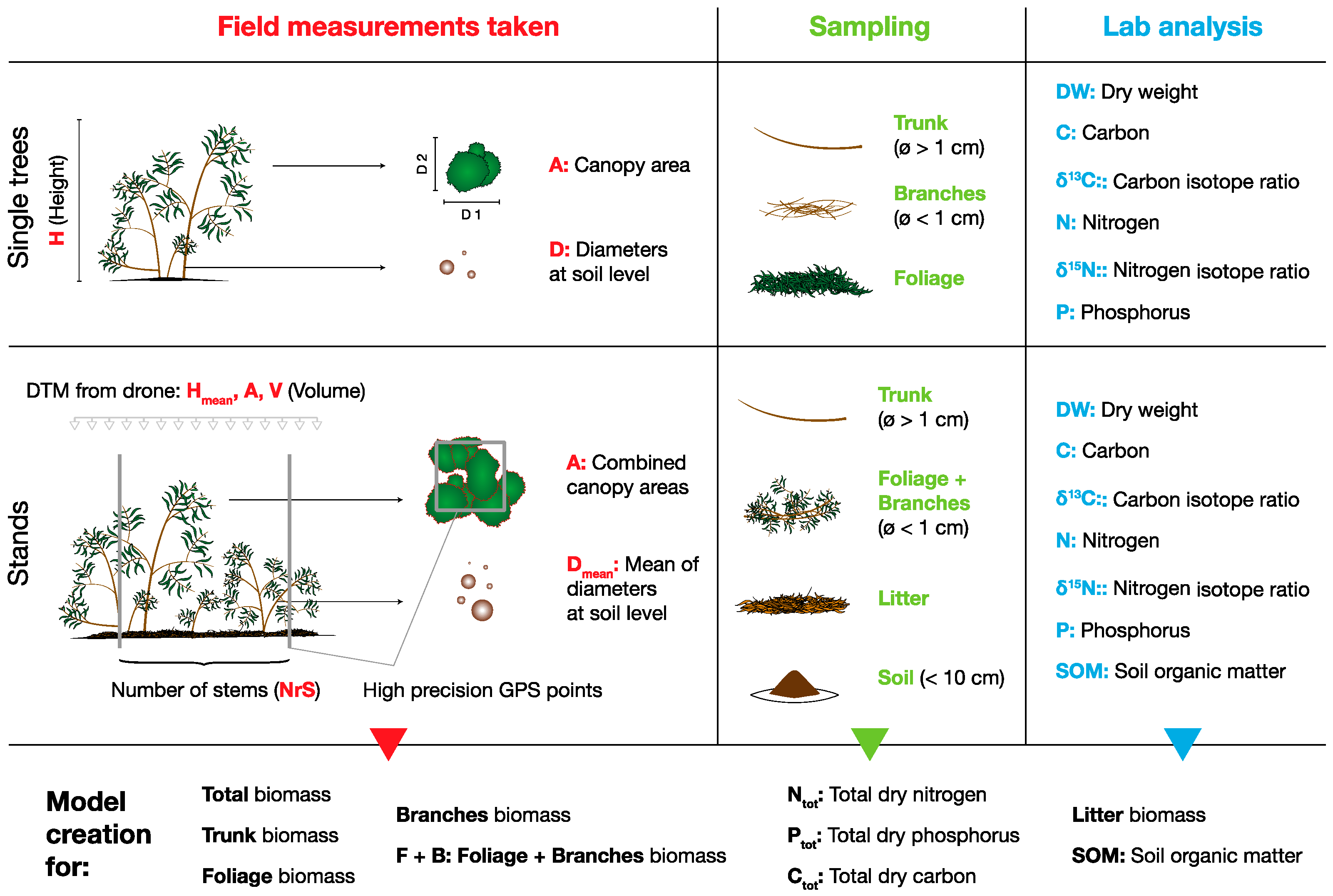
3.1. Field Measurements
3.2. Drone Flight, GPS Points and Data Analysis
3.3. Sampling, Dry Weight Determination and Total P
3.4. C/N Analysis
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sage, R.F. Global change biology: A primer. Glob. Change Biol. 2020, 26, 3–30. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.M.; Hui, C.; Nuñez, M.A.; Pauchard, A. Tree invasions: Patterns, processes, challenges and opportunities. Biol. Invasions 2014, 16, 473–481. [Google Scholar] [CrossRef]
- Hastings, A.; Byers, J.E.; Crooks, J.A.; Cuddington, K.; Jones, C.G.; Lambrinos, J.G.; Talley, T.S.; Wilson, W.G. Ecosystem engineering in space and time. Ecol. Lett. 2007, 10, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Fei, S.; Dukes, J.S.; Oswalt, C.M.; Iannone, B.V., III; Potter, K.M. A unified approach for quantifying invasibility and degree of invasion. Ecology 2015, 96, 2613–2621. [Google Scholar] [CrossRef]
- Lauri, P.; Havlík, P.; Kindermann, G.; Forsell, N.; Böttcher, H.; Obersteiner, M. Woody biomass energy potential in 2050. Energy Policy 2014, 66, 19–31. [Google Scholar] [CrossRef]
- Parikka, M. Global biomass fuel resources. Biomass Bioenergy 2004, 27, 613–620. [Google Scholar] [CrossRef]
- Kranert, M.; Gottschall, R.; Bruns, C.; Hafner, G. Energy or compost from green waste?—A CO2—Based assessment. Waste Manag. 2010, 30, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.B.; Lourenço, P.; Teixeira, A.; Azevedo, E.B.; Alves, M.; Elias, R.B.; Silva, L. Biomass valorization in the management of woody plant invaders: The case of Pittosporum undulatum in the Azores. Biomass Bioenergy 2018, 109, 155–165. [Google Scholar] [CrossRef]
- Van Meerbeek, K.; Appels, L.; Dewil, R.; Calmeyn, A.; Lemmens, P.; Muys, B.; Hermy, M. Biomass of invasive plant species as a potential feedstock for bioenergy production. Biofuels Bioprod. Biorefining 2015, 9, 273–282. [Google Scholar] [CrossRef]
- Souza-Alonso, P.; Rodríguez, J.; González, L.; Lorenzo, P. Here to stay. Recent advances and perspectives about Acacia invasion in Mediterranean areas. Ann. For. Sci. 2017, 74, 55. [Google Scholar] [CrossRef]
- Carneiro, M.; Moreira, R.; Gominho, J.; Fabião, A. Could control of invasive Acacias be a source of biomass for energy under Mediterranean conditions? Chem. Eng. 2014, 37, 187–192. [Google Scholar]
- Ferreira, T.; Paiva, J.M.; Pinho, C. Performance assessment of invasive Acacia dealbata as a fuel for a domestic pellet boiler. Chem. Eng. Trans. 2014, 42, 73–78. [Google Scholar]
- Correia, R.; Quintela, J.C.; Duarte, M.P.; Gonçalves, M. Insights for the valorization of biomass from portuguese invasive Acacia spp. in a biorefinery perspective. Forests 2020, 11, 1342. [Google Scholar] [CrossRef]
- Brito, L.M.; Mourão, I.; Coutinho, J.; Smith, S. Composting for management and resource recovery of invasive Acacia species. Waste Manag. Res. 2013, 31, 1125–1132. [Google Scholar] [CrossRef]
- Ulm, F.; Avelar, D.; Hobson, P.; Penha-Lopes, G.; Dias, T.; Máguas, C.; Cruz, C. Sustainable urban agriculture using compost and an open-pollinated maize variety. J. Clean. Prod. 2019, 212, 622–629. [Google Scholar] [CrossRef]
- Brown, S. Measuring carbon in forests: Current status and Future challenges. Environ. Pollut. 2002, 116, 363–372. [Google Scholar] [CrossRef]
- Van Breugel, M.; Ransijn, J.; Craven, D.; Bongers, F.; Hall, J.S. Estimating carbon stock in secondary forests: Decisions and uncertainties associated with allometric biomass models. For. Ecol. Manag. 2011, 262, 1648–1657. [Google Scholar] [CrossRef]
- Lovell, H.; MacKenzie, D. Allometric Equations and Timber Markets: An Important Forerunner of REDD+? In The Politics of Carbon Markets, 1st ed.; Stephan, B., Lane, R., Eds.; Taylor & Francis Group: London, UK, 2014; pp. 69–90. [Google Scholar]
- Picard, N.; Saint-André, L.; Henry, M. Manual for Building Tree Volume and Biomass Allometric Equations: From Field Measurement to Prediction; Food and Agricultural Organization of the United Nations: Rome, Italy; Centre de Coopération Internationale en Recherche Agronomique pour le Développement: Montpellier, France, 2012; p. 215. [Google Scholar]
- Van Laar, A.; Theron, J.M. Equations for predicting the biomass of Acacia cyclops and Acacia saligna in the western and eastern Cape regions of South Africa Part 1: Tree-level models. South. Afr. For. J. 2004, 201, 25–34. [Google Scholar]
- Bi, H.; Turner, J.; Lambert, M.J. Additive biomass equations for native eucalypt forest trees of temperate Australia. Trees 2004, 18, 467–479. [Google Scholar] [CrossRef]
- Jonson, J.H.; Freudenberger, D. Restore and sequester: Estimating biomass in native Australian woodland ecosystems for their carbon-Funded restoration. Aust. J. Bot. 2011, 59, 640–653. [Google Scholar] [CrossRef] [Green Version]
- Vega, J.A.; Arellano-Pérez, S.; Álvarez-González, J.G.; Fernández, C.; Jiménez, E.; Fernández-Alonso, J.M.; Vega-Nieva, D.J.; Briones-Herrera, C.; Alonso-Rego, C.; Fontúrbel, T.; et al. Modelling aboveground biomass and fuel load components at stand level in shrub communities in NW Spain. For. Ecol. Manag. 2022, 505, 119926. [Google Scholar] [CrossRef]
- Bonham, C.D. Measurements for Terrestrial Vegetation, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013. [Google Scholar]
- Fonseca, F.; de Figueiredo, T.; Ramos, B. Carbon storage in the Mediterranean upland shrub communities of Montesinho Natural Park, northeast of Portugal. Agrofor. Syst. 2012, 86, 463–475. [Google Scholar] [CrossRef]
- Ruiz-Peinado, R.; Moreno, G.; Juarez, E.; Montero, G.; Roig, S. The contribution of two common shrub species to aboveground and belowground carbon stock in Iberian dehesas. J. Arid. Environ. 2013, 91, 22–30. [Google Scholar] [CrossRef]
- Jayathunga, S.; Owari, T.; Tsuyuki, S. The use of fixed–wing UAV photogrammetry with LiDAR DTM to estimate merchantable volume and carbon stock in living biomass over a mixed conifer-broadleaf forest. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 767–777. [Google Scholar] [CrossRef]
- Guimarães, N.; Pádua, L.; Marques, P.; Silva, N.; Peres, E.; Sousa, J.J. Forestry remote sensing from unmanned aerial vehicles: A review focusing on the data, processing and potentialities. Remote Sens. 2020, 12, 1046. [Google Scholar] [CrossRef] [Green Version]
- Dainelli, R.; Toscano, P.; Di Gennaro, S.F.; Matese, A. Recent advances in Unmanned Aerial Vehicles forest remote sensing—A systematic review. Part II: Research applications. Forests 2021, 12, 397. [Google Scholar] [CrossRef]
- Puliti, S.; Breidenbach, J.; Astrup, R. Estimation of forest growing stock volume with UAV laser scanning data: Can it be done without field data? Remote Sens. 2020, 12, 1245. [Google Scholar] [CrossRef] [Green Version]
- Muukkonen, P. Generalized allometric volume and biomass equations for some tree species in Europe. Eur. J. For. Res. 2007, 126, 157–166. [Google Scholar]
- Ravindranath, N.H.; Ostwald, M. Methods for Estimating Above-Ground Biomass; Springer: Dordrecht, The Netherlands, 2008; pp. 113–147. [Google Scholar]
- Henry, M.; Bombelli, A.; Trotta, C.; Alessandrini, A.; Birigazzi, L.; Sola, G.; Vieilledent, G.; Santenoise, P.; Longuetaud, F.; Valentini, R.; et al. GlobAllomeTree: International platform for tree allometric equations to support volume, biomass and carbon assessment. Iforest Biogeosci. For. 2013, 6, 326. [Google Scholar] [CrossRef] [Green Version]
- White, J.F.; Gould, S.J. Interpretation of the coefficient in the allometric equation. Am. Nat. 1965, 99, 5–18. [Google Scholar] [CrossRef]
- Niklas, K.J. Plant allometry: Is there a grand unifying theory? Biol. Rev. 2004, 79, 871–889. [Google Scholar] [CrossRef]
- McCarthy, M.C.; Enquist, B.J. Consistency between an allometric approach and optimal partitioning theory in global patterns of plant biomass allocation. Funct. Ecol. 2007, 21, 713–720. [Google Scholar] [CrossRef]
- Poorter, H.; Jagodzinski, A.M.; Ruiz-Peinado, R.; Kuyah, S.; Luo, Y.; Oleksyn, J.; Usoltsev, V.A.; Buckley, T.N.; Reich, P.B.; Sack, L. How does biomass distribution change with size and differ among species? An analysis for 1200 plant species from five continents. New Phytol. 2015, 208, 736–749. [Google Scholar] [CrossRef] [Green Version]
- Poorter, H.; Niklas, K.J.; Reich, P.B.; Oleksyn, J.; Poot, P.; Mommer, L. Biomass allocation to leaves, stems and roots: Meta-analyses of interspecific variation and environmental control. New Phytol. 2012, 193, 30–50. [Google Scholar] [CrossRef] [PubMed]
- Forrester, D.I.; Tachauer, I.H.H.; Annighoefer, P.; Barbeito, I.; Pretzsch, H.; Ruiz-Peinado, R.; Stark, H.; Vacchiano, G.; Zlatanov, T.; Chakraborty, T.; et al. Generalized biomass and leaf area allometric equations for European tree species incorporating stand structure, tree age and climate. For. Ecol. Manag. 2017, 396, 160–175. [Google Scholar] [CrossRef]
- Reyes-Torres, M.; Oviedo-Ocaña, E.R.; Dominguez, I.; Komilis, D.; Sánchez, A. A systematic review on the composting of green waste: Feedstock quality and optimization strategies. Waste Manag. 2018, 77, 486–499. [Google Scholar] [CrossRef]
- Obernberger, I.; Brunner, T.; Bärnthaler, G. Chemical properties of solid biofuels—Significance and impact. Biomass Bioenergy 2006, 30, 973–982. [Google Scholar] [CrossRef]
- Farquhar, G.D.; Hubick, K.T.; Condon, A.G.; Richards, R.A. Carbon Isotope Fractionation and Plant Water-Use-Efficiency. In Stable Isotopes in Ecological Research; Rundel, P.W., Ehleringer, J.R., Nagy, K.H., Eds.; Springer: Berlin, Germany, 1989; pp. 21–40. [Google Scholar]
- Große-Stoltenberg, A.; Hellmann, C.; Thiele, J.; Oldeland, J.; Werner, C. Invasive acacias differ from native dune species in the hyperspectral/biochemical trait space. J. Veg. Sci. 2018, 29, 325–335. [Google Scholar] [CrossRef]
- Ulm, F.; Hellmann, C.; Cruz, C.; Máguas, C. N/P imbalance as a key driver for the invasion of oligotrophic dune systems by a woody legume. Oikos 2017, 126, 231–240. [Google Scholar] [CrossRef]
- Craine, J.M.; Elmore, A.J.; Aidar, M.P.; Bustamante, M.; Dawson, T.E.; Hobbie, E.A.; Kahmen, A.; Mack, M.C.; McLauchlan, K.K.; Michelsen, A.; et al. Global patterns of foliar nitrogen isotopes and their relationships with climate, mycorrhizal fungi, foliar nutrient concentrations, and nitrogen availability. New Phytol. 2009, 183, 980–992. [Google Scholar] [CrossRef]
- Goodman, R.C.; Phillips, O.L.; Baker, T.R. The importance of crown dimensions to improve tropical tree biomass estimates. Ecol. Appl. 2014, 24, 680–698. [Google Scholar] [CrossRef] [Green Version]
- Wertz, B.; Bembenek, M.; Karaszewski, Z.; Ochał, W.; Skorupski, M.; Strzeliński, P.; Węgiel, A.; Mederski, P.S. Impact of Stand Density and Tree Social Status on Aboveground Biomass Allocation of Scots Pine Pinus sylvestris L. Forests 2020, 11, 765. [Google Scholar] [CrossRef]
- Holmes, P. A comparison of the impacts of winter versus summer burning of slash fuel in alien-invaded fynbos areas in the Western Cape. South. Afr. For. J. 2001, 192, 41–49. [Google Scholar] [CrossRef]
- Jucker, T.; Caspersen, J.; Chave, J.; Antin, C.; Barbier, N.; Bongers, F.; Dalponte, M.; van Ewijk, K.Y.; Forrester, D.I.; Haeni, M.; et al. Allometric equations for integrating remote sensing imagery into forest monitoring programmes. Glob. Change Biol. 2017, 23, 177–190. [Google Scholar] [CrossRef]
- De Sa, N.C.; Castro, P.; Carvalho, S.; Marchante, E.; López-Núñez, F.A.; Marchante, H. Mapping the flowering of an invasive plant using unmanned aerial vehicles: Is there potential for biocontrol monitoring? Front. Plant Sci. 2018, 9, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, C.; Santana, P.; Brandão, T.; Guedes, M. Automatic detection of Acacia longifolia invasive species based on UAV-acquired aerial imagery. Inf. Process. Agric. 2022, 9, 276–287. [Google Scholar] [CrossRef]
- Eichhorn, F. Beziehungen zwischen Bestandshöhe und Bestandsmasse. Allg. Forst Jagdztg. 1904, 80, 45–49. [Google Scholar]
- Snowdon, P.; Raison, R.J.; Keith, H.; Ritson, P.; Grierson, P.; Adams, M.; Montagu, K.; Bi, H.Q.; Burrows, W.; Eamus, D. Protocol for Sampling Tree and Stand Biomass; Australian Greenhouse Office: Canberra, Australia, 2002. [Google Scholar]
- QGIS Development Team. QGIS Geographic Information System; Open Source Geospatial Foundation: Beaverton, OR, USA, 2021; Available online: http://qgis.org (accessed on 20 October 2022).
- D’Angelo, E.; Crutchfield, J.; Vandiviere, M. Rapid, sensitive, microscale determination of phosphate in water and soil. J. Environ. Qual. 2001, 30, 2206–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preston, T.; Owens, N.J.P. Interfacing an automatic elemental analyser with an isotope ratio mass spectrometer: The potential for fully automated total nitrogen and nitrogen-15 analysis. Analyst 1983, 108, 971–977. [Google Scholar] [CrossRef]
- Coleman, M.; Meier-Augenstein, W. Ignoring IUPAC guidelines for measurement and reporting of stable isotope abundance values affects us all. Rapid Commun. Mass Spectrom. 2014, 28, 1953–1955. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 20 October 2022).
- Muggeo, V.M.R. Segmented: An R Package to Fit Regression Models with Broken-Line Relationships. R News 2008, 8, 20–25. Available online: https://cran.r-project.org/doc/Rnews/ (accessed on 20 October 2022).
- Elzhov, T.V.; Mullen, K.M.; Spiess, A.N.; Bolker, B. minpack.lm: R Interface to the Levenberg-Marquardt Nonlinear Least-Squares Algorithm Found in MINPACK, Plus Support for Bounds. R package version 1.2-1. Available online: https://CRAN.R-project.org/package=minpack.lm (accessed on 20 October 2022).

| Variable | Min | Max | Mmean | ∆ | |
|---|---|---|---|---|---|
| Trees | Volume (m3) | 2.4 | 169.1 | 26.9 | 166.7 |
| Area (m2) | 1.2 | 24.6 | 7.2 | 23.4 | |
| H (m) | 2.1 | 10.6 | 4.9 | 8.5 | |
| D (cm) | 1.9 | 25.5 | 9.8 | 23.6 | |
| Trunks dry (kg) | 0.3 | 110.3 | 12.5 | 110 | |
| Branches dry (kg) | 0.1 | 11.2 | 3.3 | 11.1 | |
| Foliage dry (kg) | 0.2 | 12 | 4.1 | 11.8 | |
| F + B dry (kg) | 0.3 | 23.2 | 7.3 | 22.9 | |
| Total dry biomass (kg) | 0.7 | 129.9 | 19.8 | 129.2 | |
| Stands | Volume (m3) | 3.9 | 187.5 | 70.5 | 183.6 |
| Area (m2) | 1.2 | 29.2 | 15 | 28 | |
| Number of stems | 3 | 45 | 10.8 | 42 | |
| Dmean (cm) | 7.2 | 34.1 | 19.9 | 26.9 | |
| Hmean (m) | 0.4 | 7 | 3 | 6.6 | |
| Trunks dry (kg) | 4.3 | 133.6 | 50.2 | 129.3 | |
| F + B dry (kg) | 5.2 | 50.4 | 24.1 | 45.2 | |
| Total dry biomass (kg) | 9.5 | 184 | 74.4 | 174.5 | |
| SOM (%) | 0.3 | 1.4 | 0.7 | 1.1 | |
| Litter (kg/m2) | <0.01 | 1.9 | 0.7 | 1.9 |
| Trees | Stand | |||||
|---|---|---|---|---|---|---|
| Trunks | Branches | Foliage | F + B | Trunks | F + B | |
| C (%) | 43.2 (0.1) a | 45.1 (0.2) b | 48.1 (0.2) c | 47.7 (0.2) c | 44.1 (0.7) ab | 47.2 (0.5) c |
| N (%) | 0.6 (0.1) a | 1.1 (0.1) b | 2.3 (0.1) c | 2.2 (0.1) c | 0.6 (0) a | 2 (0.1) c |
| P (‰) | 0.8 (0.1) a | - | 2.4 (0.2) b | - | - | - |
| C/N ratio | 97.2 (10.5) ab | 42.3 (2.3) a | 21.7 (0.8) c | 24.2 (1.1) c | 72.2 (3.4) b | 23.5 (0.7) c |
| NP ratio | 11.3 (0.8) a | - | 11.4 (0.7) a | - | - | - |
| δ15N (‰) | −1.2 (0.1) a | −1.7 (0.1) ab | −1.1 (0.1) a | −1.2 (0.1) a | −2.2 (0.1) c | −2.1 (0.2) bc |
| δ13C (‰) | −27.5 (0.2) a | −25.9 (0.4) b | −28.4 (0.2) ac | −28.1 (0.3) ac | −27.5 (0.2) ab | −29 (0.4) c |
| Most Parsimonious Model | Best Model | ||||
|---|---|---|---|---|---|
| Y (Dry Mass) | Equation | RMSE | Equation | RMSE | ΔΡΜΣΕ (%) |
| Total | Y ~ 0.25 + 1.46 × V | 7.93 | Y ~ −7.56 + 1.3 × D + 1.12 × V | 7.21 | 9.9 |
| Trunk | Y ~ 0.73 + 0.34 × V1.29 | 6.28 | Y ~ −4.5 + 1.4 × 10−05 × D4.78 + 0.06 × H2.61 + 1.92 × A0.64 | 3.5 | 79.6 |
| Foliage | Y ~ −2.48 + 3.83 × ln(A) | 1.9 | Y ~ −2.48 + | 1.9 | 0 |
| 3.83 × ln(A) | |||||
| Branches | Y ~ −1.03 + 0.23 × D + 0.29 × A | 1.55 | Y ~ −1.03 + 0.23 × D + 0.29 × A | 1.55 | 0 |
| F + B | Y ~ 0.2 + 0.99 × A | 3.28 | Y ~ 0.2 + 0.99 × A | 3.28 | 0 |
| Ctot | Y ~ 0.25 + 0.64 × V | 3.51 | Y ~ −3.27 + 0.57 × D + 0.49 × V | 3.18 | 10.3 |
| Ntot | Y ~ −0.07 + 0.06 × V0.64 | 0.08 | Y ~ −0.05 + 0.02 × D + 0.01 × V | 0.08 | 2.5 |
| Ptot | Y ~ 0.003 + 0.001 × V | 0.01 | Y ~ −0.01 + 0.0013 × D + 0.001 × V | 0.007 | 14.3 |
| Most Parsimonious Model | Best Model | ||||
|---|---|---|---|---|---|
| Y (Dry Mass) | Equation | RMSE | Equation | RMSE | ΔΡΜΣΕ (%) |
| Total | Y ~ −31.01 + 0.22 × V1.28 + 19.96 × NrS0.41 | 7.23 | Y ~ −190.97 + 29.28 × NrS0.41 + 0.0008 × A3.38 + 0.013 × Hmean4.51 + 78.16 × Dmean0.24 | 2.13 | 239.5 |
| Trunk | Y ~ −28.52 + 0.07 × V1.43 + 18.15 × NrS0.36 | 5.78 | Y ~ −109.35 + 21.64 × NrS0.4 + 7.7 × 10−05 × A4 + 0.001 × Hmean5.52 + 30.95 × Dmean0.34 | 0.9 | 539.9 |
| F + B | Y ~ 2.13 + 0.24 × V + 0.46 × NrS | 2.67 | Y ~ 2.13 + 0.24 × V + 0.46 × NrS | 2.67 | 0 |
| Litter | Y ~ −0.28 + 0.02 × V + 0.06 × NrS | 0.63 | Y ~ −0.81 + 0.001 × Hmean4.13 + 0.58 × NrS0.48 | 0.32 | 97.5 |
| Ctot | Y ~ −13.79 + 0.1 × V1.27 + 8.89 × NrS0.41 | 3.24 | Y ~ −107.24 + 12.9 × NrS0.42 + 0.0004 × A3.36 + 0.007 × Hmean4.38 + 54.39 × Dmean0.18 | 0.94 | 244 |
| Ntot | Y ~ −0.02 + 0.01 × V+ 0.02 × NrS | 0.09 | Y ~ −2.94 + 0.0002 × V1.67 + 1.29 × Dmean0.2 + 0.51 × NrS0.34 | 0.04 | 154.3 |
| SOM | Y ~ 0.42 + 0.003 × V | 0.16 | Y ~ 0.13 + 0.0144 × NrS − 0.038 × A + 0.18 × Hmean + 0.019 × Dmean | 0.09 | 67.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulm, F.; Estorninho, M.; de Jesus, J.G.; de Sousa Prado, M.G.; Cruz, C.; Máguas, C. From a Lose–Lose to a Win–Win Situation: User-Friendly Biomass Models for Acacia longifolia to Aid Research, Management and Valorisation. Plants 2022, 11, 2865. https://doi.org/10.3390/plants11212865
Ulm F, Estorninho M, de Jesus JG, de Sousa Prado MG, Cruz C, Máguas C. From a Lose–Lose to a Win–Win Situation: User-Friendly Biomass Models for Acacia longifolia to Aid Research, Management and Valorisation. Plants. 2022; 11(21):2865. https://doi.org/10.3390/plants11212865
Chicago/Turabian StyleUlm, Florian, Mariana Estorninho, Joana Guedes de Jesus, Miguel Goden de Sousa Prado, Cristina Cruz, and Cristina Máguas. 2022. "From a Lose–Lose to a Win–Win Situation: User-Friendly Biomass Models for Acacia longifolia to Aid Research, Management and Valorisation" Plants 11, no. 21: 2865. https://doi.org/10.3390/plants11212865
APA StyleUlm, F., Estorninho, M., de Jesus, J. G., de Sousa Prado, M. G., Cruz, C., & Máguas, C. (2022). From a Lose–Lose to a Win–Win Situation: User-Friendly Biomass Models for Acacia longifolia to Aid Research, Management and Valorisation. Plants, 11(21), 2865. https://doi.org/10.3390/plants11212865










