Camellia japonica Root Extract Increases Antioxidant Genes by Induction of NRF2 in HeLa Cells
Abstract
:1. Introduction
2. Results
2.1. The Chemical Profile of CJRE
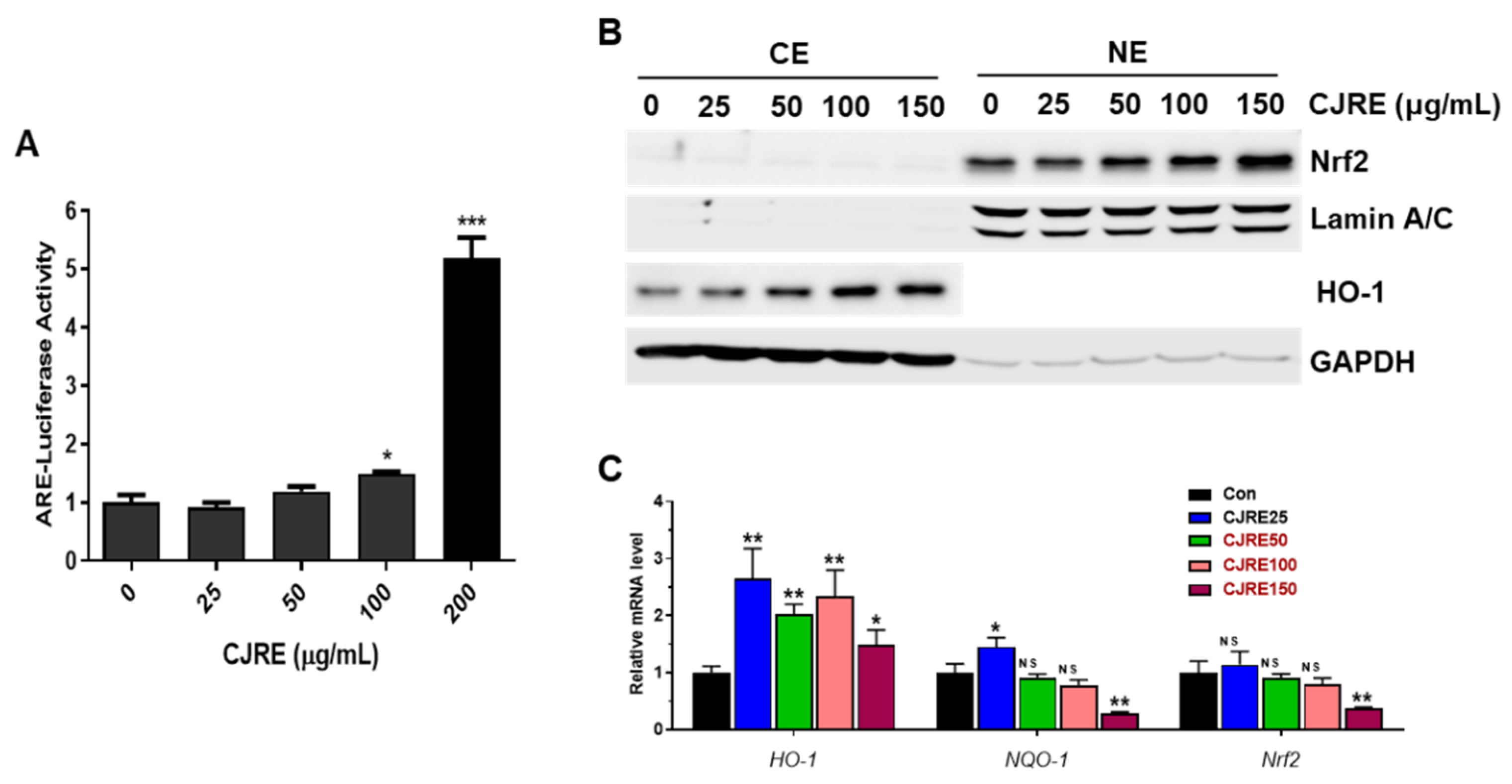
2.2. CJRE Increases NRF2 Activity through ARE System in HeLa Cells
2.3. CJRE Has a Radical Scavenging Activity
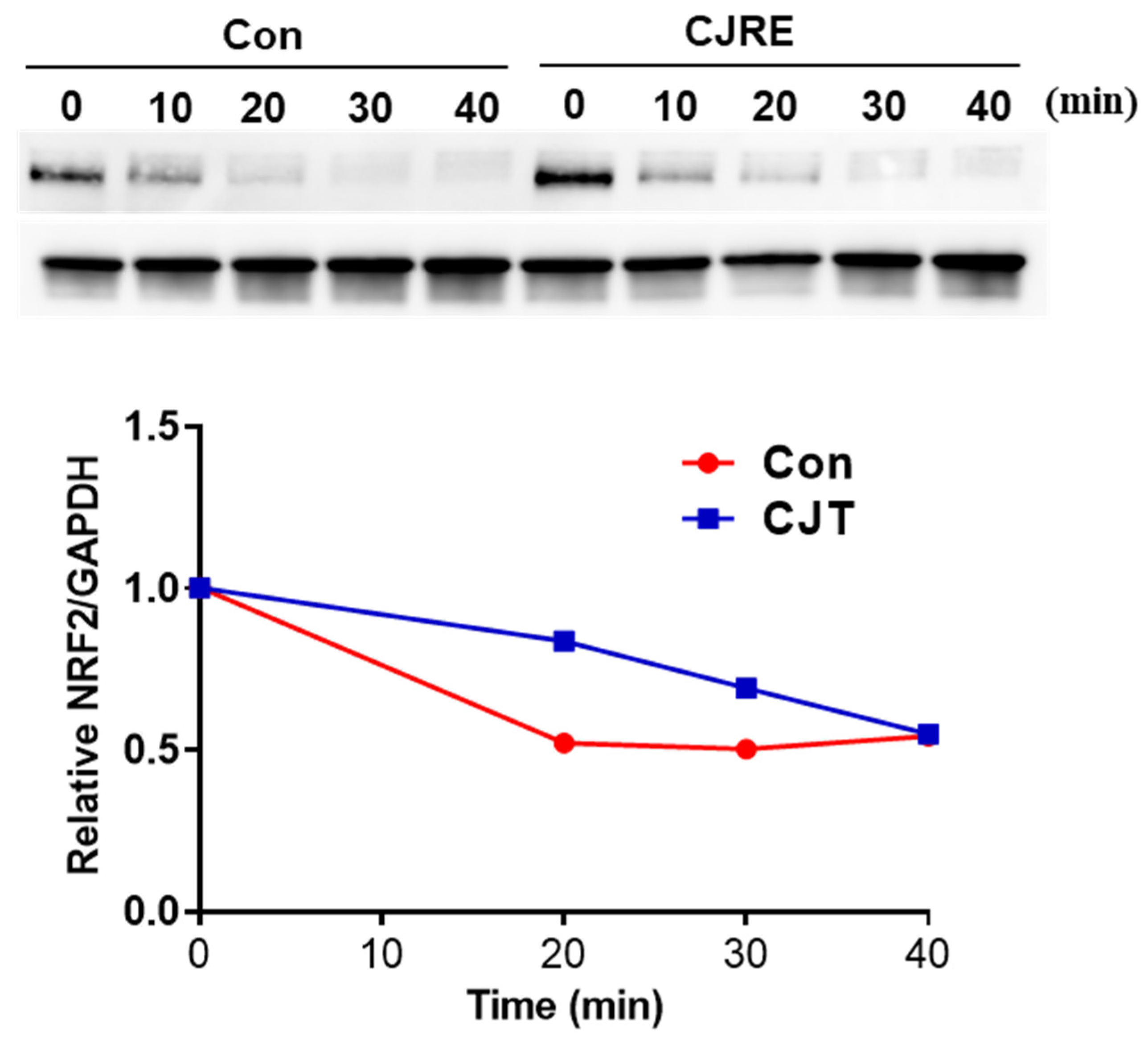
2.4. CJRE Potentiates the NRF2 Stability in HeLa Cells
2.5. CJRE Increases the Transcriptional Activity NRF2 Gene
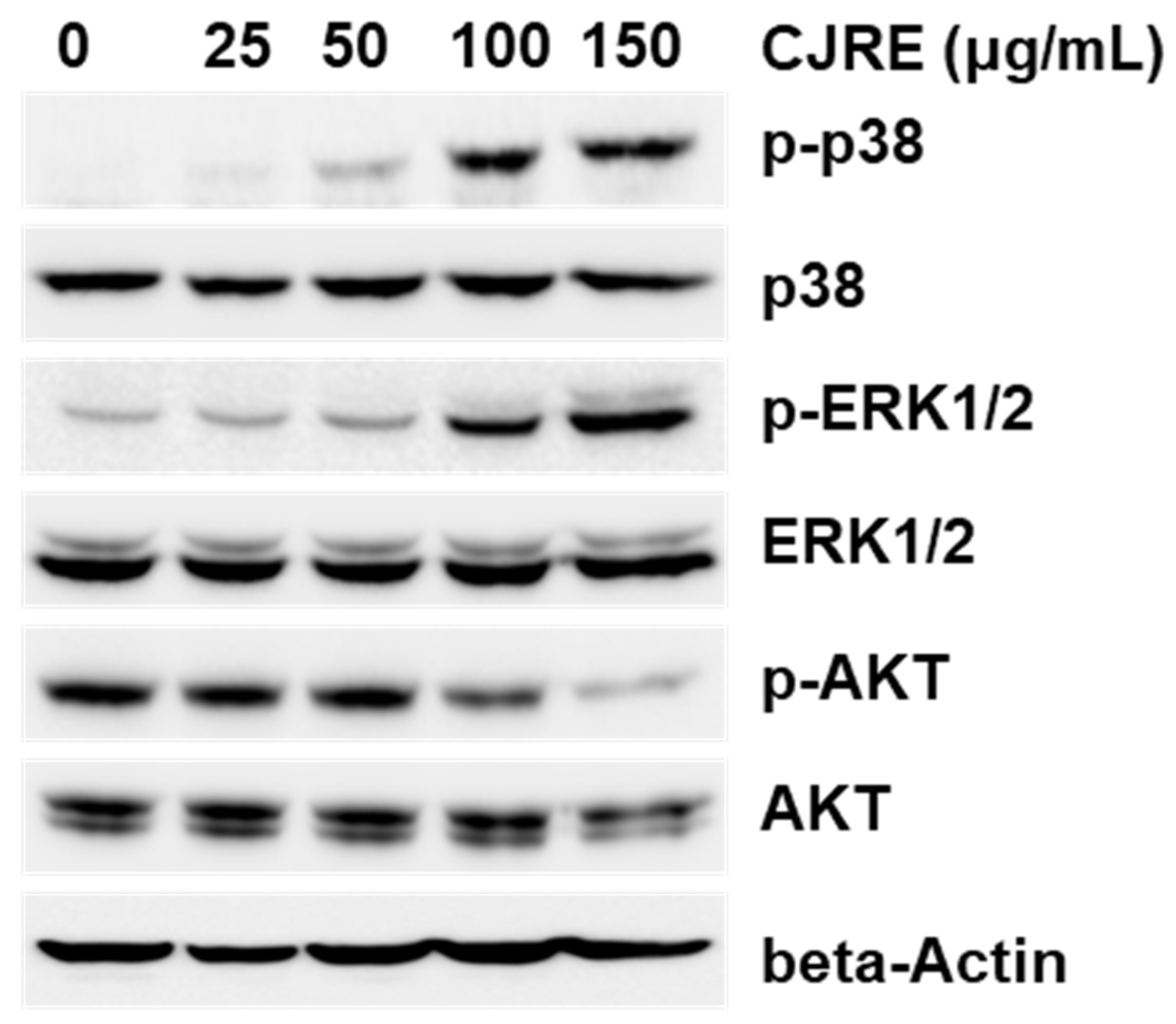
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. LC-qTOFMS Analysis
4.4. Cell Toxicity Assay
4.5. Radical Scavenging Test
4.6. ARE Luciferase Assay
4.7. Western Blot Analysis
4.8. Real-Time PCR Analysis
4.9. Protein Stability Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.L.; Kensler, T.W.; et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Sajadimajd, S.; Khazaei, M. Oxidative Stress and Cancer: The Role of Nrf2. Curr. Cancer Drug Targets 2018, 18, 538–557. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [Green Version]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Xu, E.Y.; Sacks, D.B.; Lee, J.; Shu, L.; Xia, B.; Kong, A.N. Identification and functional studies of a new Nrf2 partner IQGAP1: A critical role in the stability and transactivation of Nrf2. Antioxid. Redox Signal. 2013, 19, 89–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, K.S.; Kundu, J.; Kundu, J.K.; Surh, Y.J. Targeting Nrf2-Keap1 signaling for chemoprevention of skin carcinogenesis with bioactive phytochemicals. Toxicol. Lett. 2014, 229, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Kang, D.H.; Lee, D.H.; Bae, S.H. Fenofibrate activates Nrf2 through p62-dependent Keap1 degradation. Biochem. Biophys. Res. Commun. 2015, 465, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.K.; Wakabayashi, N.; Kensler, T.W. Chemoprevention through the Keap1-Nrf2 signaling pathway by phase 2 enzyme inducers. Mutat. Res. 2004, 555, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Johnson, J.A. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J. Biochem. Mol. Biol. 2004, 37, 139–143. [Google Scholar] [CrossRef]
- Nguyen, T.; Sherratt, P.J.; Pickett, C.B. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 233–260. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Ryter, S.W.; Alam, J.; Choi, A.M. Heme oxygenase-1/carbon monoxide: From basic science to therapeutic applications. Physiol. Rev. 2006, 86, 583–650. [Google Scholar] [CrossRef]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.H.; Sharrocks, A.D.; Whitmarsh, A.J. MAP kinase signalling cascades and transcriptional regulation. Gene 2013, 513, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.A.; Jaiswal, A.K. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 2003, 278, 44675–44682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.; Sherratt, P.J.; Huang, H.C.; Yang, C.S.; Pickett, C.B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 2003, 278, 4536–4541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef] [PubMed]
- Joo, M.S.; Kim, W.D.; Lee, K.Y.; Kim, J.H.; Koo, J.H.; Kim, S.G. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Mol. Cell. Biol. 2016, 36, 1931–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Khalil, A.A.K.; Kim, H.J.; Kim, S.E.; Ahn, M.J. 2-Methoxy-7-Acetonyljuglone Isolated from Reynoutria japonica Increases the Activity of Nuclear Factor Erythroid 2-Related Factor-2 through Inhibition of Ubiquitin Degradation in HeLa Cells. Antioxidants 2019, 8, 398. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.; Wahyudi, L.D.; Keum, Y.S.; Yang, H.; Kim, J.H. E-p-Methoxycinnamoyl-alpha-l-rhamnopyranosyl Ester, a Phenylpropanoid Isolated from Scrophularia buergeriana, Increases Nuclear Factor Erythroid-Derived 2-Related Factor 2 Stability by Inhibiting Ubiquitination in Human Keratinocytes. Molecules 2018, 23, 768. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, K.; Nakamura, S.; Nakashima, S.; Matsumoto, T.; Uno, K.; Ohta, T.; Miura, T.; Matsuda, H.; Yoshikawa, M. Medicinal flowers. XXXV. Nor-oleanane-type and acylated oleanane-type triterpene saponins from the flower buds of Chinese Camellia japonica and their inhibitory effects on melanogenesis. Chem. Pharm. Bull. 2012, 60, 1188–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, S.; Fujimoto, K.; Nakashima, S.; Matsumoto, T.; Miura, T.; Uno, K.; Matsuda, H.; Yoshikawa, M. Medicinal flowers. XXXVI.1) Acylated oleanane-type triterpene saponins with inhibitory effects on melanogenesis from the flower buds of Chinese Camellia japonica. Chem. Pharm. Bull. 2012, 60, 752–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, S.; Moriura, T.; Park, S.; Fujimoto, K.; Matsumoto, T.; Ohta, T.; Matsuda, H.; Yoshikawa, M. Melanogenesis inhibitory and fibroblast proliferation accelerating effects of noroleanane- and oleanane-type triterpene oligoglycosides from the flower buds of Camellia japonica. J. Nat. Prod. 2012, 75, 1425–1430. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Ha, T.K.; Dhodary, B.; Pyo, E.; Nguyen, N.H.; Cho, H.; Kim, E.; Oh, W.K. Oleanane triterpenes from the flowers of Camellia japonica inhibit porcine epidemic diarrhea virus (PEDV) replication. J. Med. Chem. 2015, 58, 1268–1280. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.T.; Hung, T.M.; Cuong, T.D.; Kim, J.C.; Kim, E.H.; Jin, S.E.; Na, M.; Lee, Y.M.; Kim, Y.H.; Choi, J.S.; et al. 28-nor-oleanane-type triterpene saponins from Camellia japonica and their inhibitory activity on LPS-induced NO production in macrophage RAW264.7 cells. Bioorg. Med. Chem. Lett. 2010, 20, 7435–7439. [Google Scholar] [CrossRef]
- Thao, N.T.; Hung, T.M.; Lee, M.K.; Kim, J.C.; Min, B.S.; Bae, K. Triterpenoids from Camellia japonica and their cytotoxic activity. Chem. Pharm. Bull. 2010, 58, 121–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, M.; Murakami, T.; Yoshizumi, S.; Murakami, N.; Yamahara, J.; Matsuda, H. Bioactive saponins and glycosides. V. Acylated polyhydroxyolean-12-ene triterpene oligoglycosides, camelliasaponins A1, A2, B1, B2, C1, and C2, from the seeds of Camellia japonica L.: Structures and inhibitory activity on alcohol absorption. Chem. Pharm. Bull. 1996, 44, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Harada, E.; Murakami, T.; Matsuda, H.; Yamahara, J.; Murakami, N. Camelliasaponins B1, B2, C1 and C2, new type inhibitors of ethanol absorption in rats from the seeds of Camellia japonica L. Chem Pharm Bull. 1994, 42, 742–744. [Google Scholar] [CrossRef] [Green Version]
- Akanda, M.R.; Park, B.Y. Involvement of MAPK/NF-kappaB signal transduction pathways: Camellia japonica mitigates inflammation and gastric ulcer. Biomed. Pharmacother. 2017, 95, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Sharma, G.; Yang, J.L.; Choi, H.S.; Lim, S.I.; Kang, K.W.; Oh, W.K. Oleanane triterpenes as protein tyrosine phosphatase 1B (PTP1B) inhibitors from Camellia japonica. Phytochemistry 2014, 103, 99–106. [Google Scholar] [CrossRef]
- Onodera, K.; Hanashiro, K.; Yasumoto, T. Camellianoside, a novel antioxidant glycoside from the leaves of Camellia japonica. Biosci. Biotechnol. Biochem. 2006, 70, 1995–1998. [Google Scholar] [CrossRef] [Green Version]
- Ko, K.; Wahyudi, L.D.; Kwon, Y.S.; Kim, J.H.; Yang, H. Nuclear Factor Erythroid 2-Related Factor 2 Activating Triterpenoid Saponins from Camellia japonica Roots. J. Nat. Prod. 2018, 81, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Chen, C.; Tony Kong, A.N. Resveratrol inhibits genistein-induced multi-drug resistance protein 2 (MRP2) expression in HepG2 cells. Arch. Biochem. Biophys. 2011, 512, 160–166. [Google Scholar] [CrossRef]
- Kim, Y.E.; Kim, J.W.; Cheon, S.; Nam, M.S.; Kim, K.K. Alpha-Casein and Beta-Lactoglobulin from Cow Milk Exhibit Antioxidant Activity: A Plausible Link to Antiaging Effects. J. Food Sci. 2019, 84, 3083–3090. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yu, S.; Chen, J.D.; Kong, A.N. The nuclear cofactor RAC3/AIB1/SRC-3 enhances Nrf2 signaling by interacting with transactivation domains. Oncogene 2013, 32, 514–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahyudi, L.D.; Jeong, J.; Yang, H.; Kim, J.H. Amentoflavone-induced oxidative stress activates NF-E2-related factor 2 via the p38 MAP kinase-AKT pathway in human keratinocytes. Int. J. Biochem. Cell. Biol. 2018, 99, 100–108. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-H.; Yang, H.; Kim, K.K. Camellia japonica Root Extract Increases Antioxidant Genes by Induction of NRF2 in HeLa Cells. Plants 2022, 11, 2914. https://doi.org/10.3390/plants11212914
Kim J-H, Yang H, Kim KK. Camellia japonica Root Extract Increases Antioxidant Genes by Induction of NRF2 in HeLa Cells. Plants. 2022; 11(21):2914. https://doi.org/10.3390/plants11212914
Chicago/Turabian StyleKim, Jung-Hwan, Heejung Yang, and Kee K. Kim. 2022. "Camellia japonica Root Extract Increases Antioxidant Genes by Induction of NRF2 in HeLa Cells" Plants 11, no. 21: 2914. https://doi.org/10.3390/plants11212914
APA StyleKim, J.-H., Yang, H., & Kim, K. K. (2022). Camellia japonica Root Extract Increases Antioxidant Genes by Induction of NRF2 in HeLa Cells. Plants, 11(21), 2914. https://doi.org/10.3390/plants11212914







