Split-Ubiquitin Two-Hybrid Screen for Proteins Interacting with slToc159-1 and slToc159-2, Two Chloroplast Preprotein Import Receptors in Tomato (Solanum lycopersicum)
Abstract
:1. Introduction
2. Results
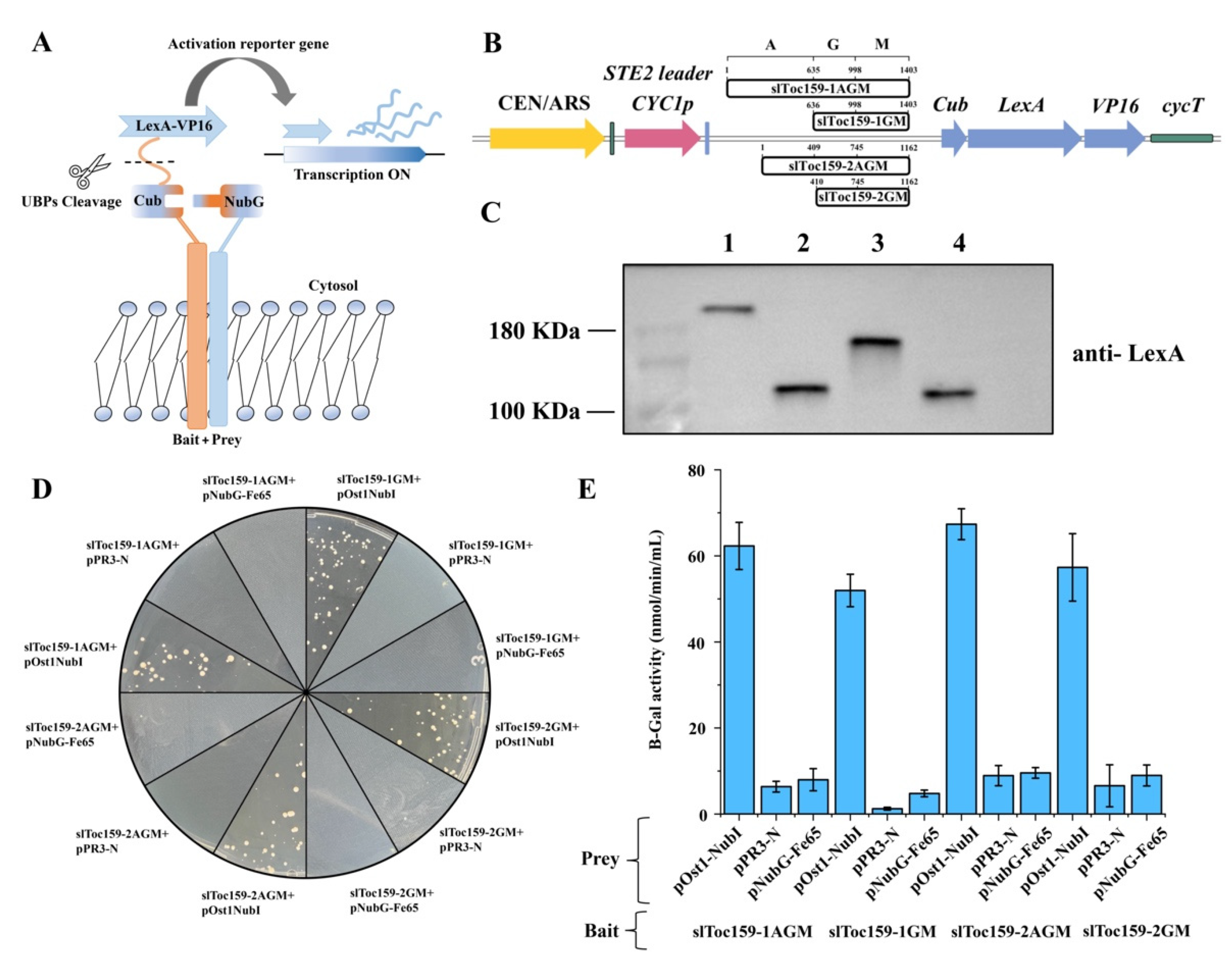
2.1. Generation of NMY51 Strains of S. cerevisiae Expressing Functional slToc159 Receptor Family Bait Proteins
2.2. Screening of Tomato cDNA Library with slToc159-1AGM and slToc159-2AGM Bait Proteins
2.3. Interactions of Toc34-1 and Toc34-2 Prey Proteins with Toc159-1AGM and Toc159-2AGM Bait Proteins
2.4. Interactions of Photosynthesis-Related Prey Proteins with Four Bait Proteins (slToc159-1AGM, slToc159-1GM, slToc159-2AGM, slToc159-2GM)
2.5. Interactions of Non-Photosynthesis-Related Prey Proteins with Four Bait Proteins (slToc159-1AGM, slToc159-1GM, slToc159-2AGM, slToc159-2GM)
2.6. BiFC Analysis of Preprotein Interactions between Different Domains (-A, -G, -M) of slToc159-1 and slToc159-2
3. Discussion
4. Materials and Methods
- All methods were performed according to the relevant guidelines and regulations
- Strains and growth media
- 3.
- Tomato cDNA library construction
- 4.
- Construction of bait vectors expressed in yeast
- 5.
- Split-ubiquitin Y2H screening
- 6.
- Immunoblot analysis
- 7.
- β-Galactosidase activity assay
- 8.
- O-nitrophenyl-ß-d-galactopyranoside (ONPG) assay
- 9.
- Pellet X-gal (PXG) assay
- 10.
- BiFC analysis
- 11.
- Subcellular localization analysis
- 12.
- Statistical analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Keeling, P.J. Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 2004, 91, 1481–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, N.; Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nature Reviews. Mol. Cell. Biol. 2004, 5, 971–982. [Google Scholar]
- McFadden, G.I. Chloroplast origin and integration. Plant Physiol. 2001, 125, 50–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, W.; Rujan, T.; Richly, E.; Hansen, A.; Cornelsen, S.; Lins, T.; Leister, D.; Stoebe, B.; Hasegawa, M.; Penny, D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 2002, 99, 12246–12251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Genetics 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Inaba, T.; Yazu, F.; Ito-Inaba, Y.; Kakizaki, T.; Nakayama, K. Retrograde signaling pathway from plastid to nucleus. Int. Rev. Cell Mol. Biol. 2011, 290, 167–204. [Google Scholar]
- Jarvis, P.; Lopez-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell. Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef]
- Chen, L.J.; Li, H.M. Stable megadalton TOC–TIC supercomplexes as major mediators of protein import into chloroplasts. Plant J. 2017, 92, 178–188. [Google Scholar] [CrossRef] [Green Version]
- Perry, S.E.; Keegstra, K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994, 6, 93–105. [Google Scholar]
- Hirsch, S.; Muckel, E.; Heemeyer, F.; von Heijne, G.; Soll, J. A receptor component of the chloroplast protein translocation machinery. Science 1994, 266, 1989–1992. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008, 179, 257–285. [Google Scholar] [CrossRef] [PubMed]
- Kessler, F.; Schnell, D. Chloroplast biogenesis: Diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 2009, 21, 494–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, L.G.L.; Paile, Y.D.; Siman, S.R.; Chen, Y.; Smith, M.D.; Schnell, D.J. Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 2014, 5, 269. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Kouranov, A.; LaSala, S.E.; Schnell, D.J. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 1996, 134, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Richardson, L.G.; Jelokhani-Niaraki, M.; Smith, M.D. The acidic domains of the Toc159 chloroplast preprotein receptor family are intrinsically disordered protein domains. BMC Biochem. 2009, 10, 35. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.; Hiltbrunner, A.; Weibel, P.; Vidi, P.A.; Alvarez-Huerta, M.; Smith, M.D.; Schnell, D.J.; Kessler, F. Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane. J. Cell Biol. 2002, 159, 845–854. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.D.; Hiltbrunner, A.; Kessler, F.; Schnell, D.J. The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J. Cell Biol. 2002, 159, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.; Teresinski, H.J.; Smith, M.D. A split-ubiquitin yeast twohybrid screen to examine the substrate specifificity of AtToc159 and AtToc132, two Arabidopsis chloroplast preprotein import receptors. PLoS ONE 2014, 9, e95026. [Google Scholar] [CrossRef] [Green Version]
- Jackson-Constan, D.; Keegstra, K. Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol. 2001, 125, 1567–1576. [Google Scholar] [CrossRef] [Green Version]
- Keegstra, K.; Cline, K. Protein import and routing systems of chloroplasts. Plant Cell 1999, 11, 557–570. [Google Scholar] [CrossRef] [Green Version]
- Afitlhile, M.; Duffield-Duncan, K.; Fry, M.; Workman, S.; Hum-Musser, S.; Hildebrand, D. The toc132toc120 heterozygote mutant of Arabidopsis thaliana accumulates reduced levels of hexadecatrienoic acid. Plant Physiol. Biochem. 2015, 96, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Kubis, S.; Patel, R.; Combe, J.; Bédard, J.; Kovacheva, S.; Lilley, K.; Biehl, A.; Leister, D.; Ríos, G.; Koncz, C.; et al. Function sepecialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 2004, 16, 2059–2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Campbell, J.H.; Glick, B.R.; Smith, M.D.; Liang, Y. Molecular characterization and expression analysis of chloroplast protein import components in tomato (Solanum lycopersicum). PLoS ONE 2014, 9, e95088. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, X.; Schnell, D.J. Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 2000, 122, 813–822. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, Y.; Smith, M.D.; Chen, K.; Schnell, D.J. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 2004, 15, 3379–3392. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.D.; Rounds, C.M.; Wang, F.; Chen, K.; Afitlhile, M.; Schnell, D.J. atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 2004, 165, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Fields, S.; Song, O. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Aronsson, H.; Combe, J.; Patel, R.; Agne, B.; Martin, M.; Kessler, F.; Jarvis, P. Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J. 2010, 63, 297–311. [Google Scholar] [CrossRef] [Green Version]
- Bedard, J.; Kubis, S.; Bimanadham, S.; Jarvis, P. Functional similarity between the chloroplast translocon component, Tic40, and the human co-chaperone, Hsp70-interacting protein (Hip). J. Biol. Chem. 2007, 282, 21404–21414. [Google Scholar] [CrossRef] [Green Version]
- Kuchler, M.; Decker, S.; Hormann, F.; Soll, J.; Heins, L. Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 2002, 21, 6136–6145. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Chiba, T.; Ozawa, R.; Yoshida, M.; Hattori, M.; Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 2001, 98, 4569–4574. [Google Scholar] [CrossRef] [Green Version]
- Uetz, P.; Giot, L.; Cagney, G.; Mansfield, T.A.; Judson, R.S.; Knight, J.R.; Lockshon, D.; Narayan, V.; Srinivasan, M.; Pochart, P.; et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 2000, 403, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Schulze, W.; Thaminy, S.; Stagljar, I.; Frommer, W.B.; Ward, J.M. Intra -and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure 2002, 10, 763–772. [Google Scholar] [CrossRef]
- Johnsson, N.; Varshavsky, A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA 1994, 91, 10340–10344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stagljar, I.; Korostensky, C.; Johnsson, N.; te Heesen, S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 1998, 95, 5187–5192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.; Assmann, S.M. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 2004, 16, 1616–1632. [Google Scholar] [CrossRef] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of webservers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Kessler, F.; Schnell, D.J. The function and diversity of plastid protein import pathways: A multilane GTPase highway into plastids. Traffic 2006, 7, 248–257. [Google Scholar] [CrossRef] [Green Version]
- Stengel, A.; Soll, J.; Bolter, B. Protein import into chloroplasts: New aspects of a well-known topic. Biol. Chem. 2007, 388, 765–772. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.; Chen, K.; Hiltbunner, A.; Wehrli, E.; Eugster, M.; Schnell, D.; Kessler, F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 2000, 403, 203–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniell, H.; Lin, C.; Yu, M.; Chang, W. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 1–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staehelin, L.A.; DeWit, M. Correlation of structure and function of chloroplast membranes at the supramolecular level. J. Cell. Biochem. 1984, 24, 261–269. [Google Scholar] [CrossRef]
- Inoue, H.; Li, M.; Schnell, D.J. An essential role for chloroplast heat shock protein 90(Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampinga, H.; Craig, E. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [Green Version]
- May, T.; Soll, J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 2000, 12, 53–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehlert, A.; Weltmeier, F.; Wang, X.; Mayer, C.S.; Smeekens, S.; Vicente-Carbajosa, J.; Dröge-Laser, W. Two hybrid protein-protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 2006, 46, 890–900. [Google Scholar] [CrossRef]
- Bischof, S.; Baerenfaller, K.; Wildhaber, T.; Troesch, R.; Vidi, P.A.; Roschitzki, B.; Hirsch-Hoffmann, M.; Hennig, L.; Kessler, F.; Gruissem, W.; et al. Plastid proteome assembly without Toc159: Photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 2011, 23, 3911–3928. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J.; et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013, 41, 348–352. [Google Scholar] [CrossRef] [Green Version]
- Fish, M.; Nash, D.; German, A.; Overton, A.; Jelokhani-Niaraki, M.; Chuong, S.; Smith, M.D. New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways. Int. J. Mol. Sci. 2022, 23, 1571. [Google Scholar] [CrossRef]
- Lung, S.C.; Smith, M.D.; Weston, J.K.; Gwynne, W.; Secord, N.; Chuong, S.D.X. The C-terminus of Bienertia sinuspersici Toc159 contains essential elements for its targeting and anchorage to the chloroplast outer membrane. Front. Plant Sci. 2014, 5, 722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Kim, S.J.; Lee, Y.J.; Jin, J.B.; Hwang, I. The M domain of atToc159 plays an essential role in the import of proteins into chloroplasts and chloroplast biogenesis. J. Biol. Chem. 2003, 278, 36794–36805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egea, I.; Barsan, C.; Bian, W.; Purgatto, E.; Latché, A.; Chervin, C.; Bouzayen, M.; Pech, J.-C. Chromoplast differentiation: Current status and perspectives. Plant Cell Physiol. 2010, 51, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Dhami, N. Chloroplast-to-chromoplast transition envisions provitamin A biofortification in green vegetables. Plant Cell Rep. 2021, 40, 799–804. [Google Scholar] [CrossRef]
- Ling, Q.; Sadali, N.M.; Soufi, Z.; Zhou, Y.; Huang, B.; Zeng, Y.; Rodriguez-Concepcion, M.; Jarvis, R.P. The chloroplast-associated protein degradation pathway controls chromoplast development and fruit ripening in tomato. Nat. Plants 2021, 7, 655–666. [Google Scholar] [CrossRef]
- Demarsy, E.; Lakshmanan, A.M.; Kessler, F. Border control: Selectivity of chloroplast protein import and regulation at the TOC-complex. Front. Plant Sci. 2014, 5, 483. [Google Scholar] [CrossRef] [Green Version]
- Teng, Y.S.; Chan, P.T.; Li, H.M. Diferential age-dependent import regulation by signal peptides. PLoS Biol. 2012, 10, e1001416. [Google Scholar] [CrossRef]
- Möckli, N.; Auerbach, D. Quantitative beta-galactosidase assay suitable for high-throughput applications in the yeast two-hybrid system. Biotechniques 2004, 36, 872–876. [Google Scholar] [CrossRef]







| Plasmid Combination | Usage |
|---|---|
| Bait + pOst1-NubI | Validating the correct expression of yeast proteins in the split-ubiquitin system |
| Bait + pPR3-N Bait + pNubG-Fe65 | Self-activation detection |
| Bait 2 | Speculative Function | Location | |||
|---|---|---|---|---|---|
| Gene Name | Gene Code 1 | Toc159-1AGM | Toc159-2AGM | ||
| RuBisCO small subunit 1(RBSC-1) | Solyc02g063150.2 | + | Carbon fixation | Chloroplast | |
| RuBisCO small subunit 3 (RBCS-3) | Solyc01g073930.3.1 | + | Carbon fixation | Chloroplast | |
| RuBisCO small subunit 2A (RBCS-2A) | Solyc03g034220.3.1 | + | Carbon fixation | Chloroplast | |
| Photosystem II 23 kDa protein (PSBP) | Solyc07g044860.3.1 | + | Photosystem-II-associated protein | Chloroplast | |
| Photosystem I subunit O (PSI-O) | Solyc06g074200.4.1 | + | + | Photosystem-I-associated protein | Chloroplast |
| RuBisCO small subunit 4 (RBCS-4) | Solyc02g085950.4.1 | + | + | Carbon fixation | Chloroplast |
| Oxygen-evolving enhancer protein 1 (PSBO) | Solyc02g065400.3.1 | + | Photosystem-II-associated protein | Chloroplast | |
| Photosystem I reaction center subunit IV A (PSI-A) | Solyc06g083680.3.1 | + | Photosystem-I-associated protein | Chloroplast | |
| Photosystem I reaction center subunit III (PSI-III) | Solyc02g069460.2 | + | Photosystem-I-associated protein | Chloroplast | |
| photosystem II subunit S (PSBS) | Solyc06g060340.3.1 | + | Photosystem-II-associated protein | Chloroplast | |
| Chlorophyll a-b binding protein CP24 (LHCP) | Solyc01g105030.2 | + | Photosystem-II-associated protein | Chloroplast | |
| Chlorophyll a-b binding protein 8 (CAB-8) | Solyc10g007690.2 | + | Photosystem-II-associated protein | Chloroplast | |
| Chlorophyll a/b binding protein Cab-3C (CAB-3C) | Solyc03g005780.1 | + | Photosystem-II-associated protein | Chloroplast | |
| Chlorophyll a-b binding protein (CAB11) | Solyc03g115900.2 | + | + | Photosystem-I-associated protein | Chloroplast |
| Chlorophyll a-b binding protein 7 (CAB-7) | Solyc10g006230.2 | + | Photosystem-II-associated protein | Chloroplast | |
| Photosystem I reaction center subunit II(PSI-D) | Solyc06g054260.1.1 | + | Photosystem-I-associated protein | Chloroplast | |
| ATP synthase subunit B (ASS-B) | Solyc06g066000.3.1 | + | + | ATP synthesis driven by the proton dynamic potential | Chloroplast |
| ATP synthase subunit delta chain (ASS-delta) | Solyc05g050500.1 | + | ATP synthesis driven by the proton dynamic potential | Chloroplast | |
| Bait 2 | Speculative Function | Location | |||
|---|---|---|---|---|---|
| Gene Name | Gene Code 1 | Toc159-1AGM | Toc159-2AGM | ||
| ATP-dependent Clp protease proteolytic 2 (ACP2) | Solyc08g079620.2 | + | A central component of the chloroplast protease network | Chloroplast | |
| ATP-dependent Clp protease proteolytic 4 (ACP4) | Solyc08g077890.2 | + | A central component of the chloroplast protease network | Chloroplast | |
| 60S ribosomal protein (L7a-2) | Solyc06g064470.4.1 | + | Ribosome biogenesis | Cytoplasm | |
| Glycine-rich RNA-binding protein (RBP1) | Solyc01g109660.2 | + | Development and stress adaptation | Chloroplast | |
| Glutamine synthetase (GS2) | Solyc01g080280.2 | + | Photorespiration and assimilation of ammonia from nitric acid reduction | Chloroplast | |
| Multiple organellar RNA editing factor 2 (MORF2) | Solyc06g008220.2 | + | Multiple RNA editing in plastids | Chloroplast | |
| Cell division protein FtsZ (CDP2-1) | Solyc09g009430.3.1 | + | Key cellular skeletal components in the mechanism of chloroplast division | Chloroplast | |
| Peroxiredoxin Q (PQ) | Solyc07g042440.2 | + | + | Redox reactions | Chloroplast |
| GTPase (HflX) | Solyc04g080770.3.1 | + | GTP enzymes that target chloroplasts | Chloroplast | |
| Hop-interacting protein (THI026) | Solyc04g018110.1.1 | + | Assemble the Hsp complex | Cytoplasm | |
| Heat shock cognate 70 kDa protein (HSP70) | Solyc11g066060.3.1 | + | + | Assists in targeted transport of preproteins to chloroplasts | Cytoplasm |
| Stearoyl-[acyl-carrier-protein] 9-desaturase (SAC9) | Solyc03g063110.2 | + | Fatty acid metabolism | Chloroplast | |
| GTP diphosphokinase (RSH1) | Solyc09g098580.3.1 | + | Transfer of high-energy phosphate groups and signal transduction | Chloroplast | |
| Calcium-binding protein (KIC) | Solyc10g009340.1.1 | + | Regulation of cell division and trichome morphogenesis | Mitochondrion | |
| Elongation factor 1-alpha (EF1) | Solyc06g005060.3.1 | + | Regulation of cell growth and division | Chloroplast | |
| thiamine biosynthesis protein (ThiC) | Solyc06g006080.3.1 | + | Vitamin B1 biosynthesis | Chloroplast | |
| RPM1-interacting protein 4 (RIP4) | Solyc06g083390.4.1 | + | Plant immunity | Chloroplast | |
| A/B barrel domain-containing protein (UP3) | Solyc07g041490.1 | + | + | Stress response | Chloroplast |
| SufE-like protein 1 (SLP1) | Solyc12g015910.1.1 | + | Formation of iron-sulfur clusters in chloroplasts | Chloroplast | |
| Serine hydroxymethyl transferase (SHT) | Solyc05g053810.2 | + | Catalyze the conversion of serine and glycine to each other | Mitochondrion | |
| 3-oxoacyl-[acyl-carrier-protein] reductase (FabG) | Solyc12g045030.1 | + | Fatty Acid Synthesis | Chloroplast | |
| NADH dehydrogenase [ubiquinone] flavoprotein 14 | Solyc02g087240.2 | + | Redox reactions | Mitochondrion | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Yue, J.; Zhang, C.; Yan, J. Split-Ubiquitin Two-Hybrid Screen for Proteins Interacting with slToc159-1 and slToc159-2, Two Chloroplast Preprotein Import Receptors in Tomato (Solanum lycopersicum). Plants 2022, 11, 2923. https://doi.org/10.3390/plants11212923
Wang Q, Yue J, Zhang C, Yan J. Split-Ubiquitin Two-Hybrid Screen for Proteins Interacting with slToc159-1 and slToc159-2, Two Chloroplast Preprotein Import Receptors in Tomato (Solanum lycopersicum). Plants. 2022; 11(21):2923. https://doi.org/10.3390/plants11212923
Chicago/Turabian StyleWang, Qi, Jiang Yue, Chaozhong Zhang, and Jianmin Yan. 2022. "Split-Ubiquitin Two-Hybrid Screen for Proteins Interacting with slToc159-1 and slToc159-2, Two Chloroplast Preprotein Import Receptors in Tomato (Solanum lycopersicum)" Plants 11, no. 21: 2923. https://doi.org/10.3390/plants11212923
APA StyleWang, Q., Yue, J., Zhang, C., & Yan, J. (2022). Split-Ubiquitin Two-Hybrid Screen for Proteins Interacting with slToc159-1 and slToc159-2, Two Chloroplast Preprotein Import Receptors in Tomato (Solanum lycopersicum). Plants, 11(21), 2923. https://doi.org/10.3390/plants11212923





