Endophytes from Halotolerant Plants Aimed to Overcome Salinity and Draught
Abstract
:1. Introduction
2. Results
2.1. Plant Sampling and Measurement of Salt Content in the Soil
2.2. Isolation of Endophytic Bacteria
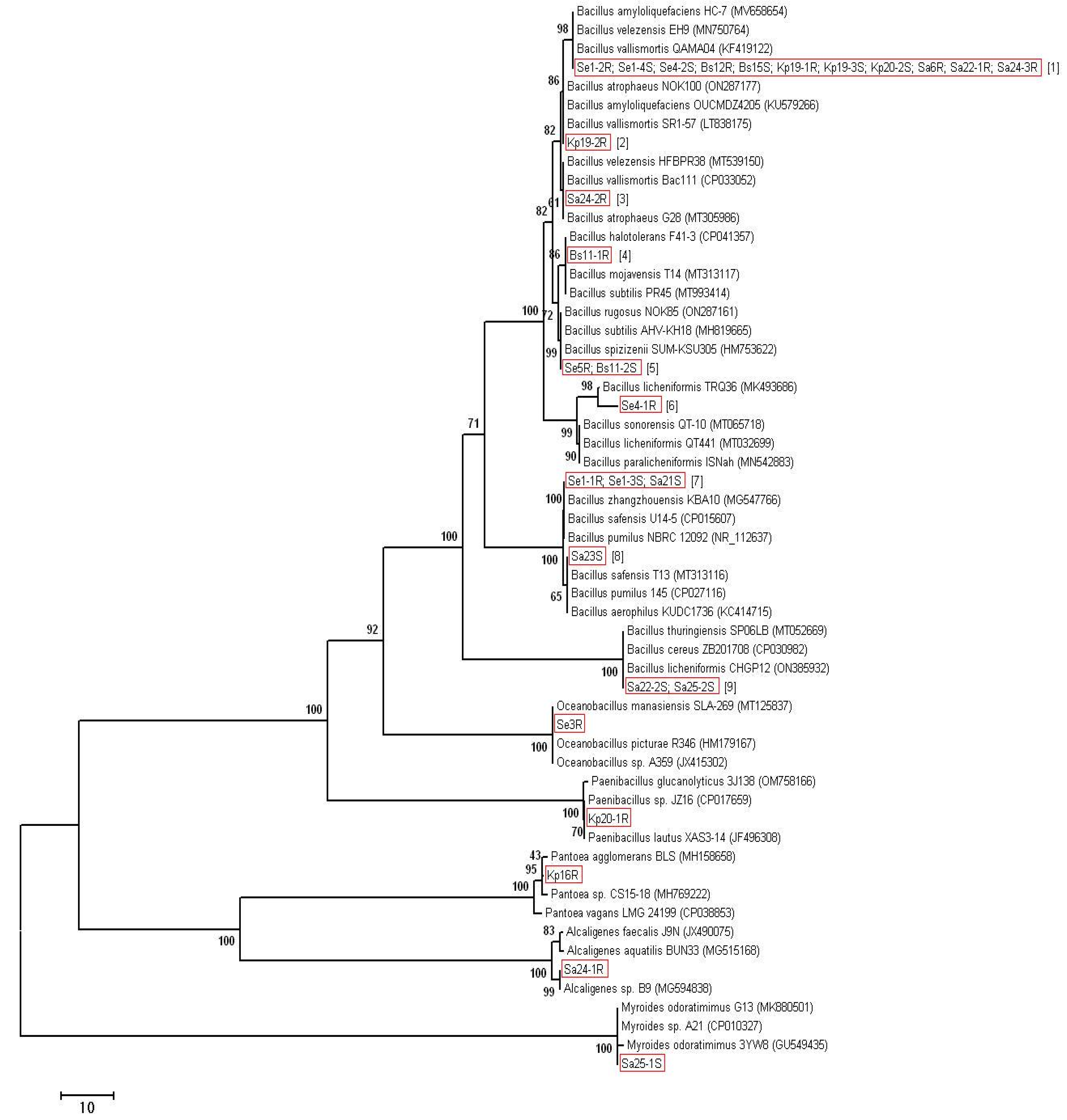
2.3. Molecular Identification of Endophytes
2.4. The Effect of Temperature on the Growth of Endophytes
2.5. The Effect of Different Concentrations of NaCl on the Growth of Endophytes
2.6. Properties of Endophytes
2.7. Plant Growth Promotion by Endophytes
2.8. Antifungal Activities of Endophytes
2.9. Polyphasic Characterisation of Endophytes
3. Discussion
4. Materials and Methods
4.1. Plant Sampling
4.2. Surface Sterilization of Plants and Isolation of Endophytic Bacteria
4.3. Measurement of Salt Content in the Soil
4.4. Molecular Identification of Endophytes
4.5. The Effect of Temperature on the Growth of Endophytes
4.6. The Effect of Different Concentrations of NaCl on the Growth of Endophytes
4.7. Nitrogen Fixing Ability
4.8. Estimation of Phosphorus Solubilization
4.9. Estimation of Cellulase Activity
4.10. Estimation of Amylase Activity
4.11. Estimation of Protease Activity
4.12. Estimation of Lipase Activity
4.13. Determination of Secretory Auxin
4.14. Estimation of Plant Growth Promotion Ability of Endophytes
4.15. Antifungal Activities of Endophytes
4.16. Statistical Analysis
4.17. Polyphasic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sgroy, V.; Cassán, F.; Masciarelli, O.; Del Papa, M.F.; Lagares, A.; Luna, V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 2009, 85, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Fujita, M. Plant response to salt stress and role of exogenous protectants to mitigate saltinduced damages. In Ecophysiology and Responses of Plants Under Salt Stress; Ahmad, P., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2013; pp. 25–87. [Google Scholar]
- Hasanuzzaman, M.; Nahar, K.; Fujita, M.; Ahmad, P.; Chandna, R.; Prasad, M.N.V.; Ozturk, M. Enhancing plant productivity under salt stress-relevance of poly-omics. In Salt Stress in Plants: Omics, Signaling and Responses; Ahmad, P., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 113–156. [Google Scholar]
- Khan, M.; Rolly, N.K.; Al Azzawi, T.N.I.; Imran, M.; Mun, B.-G.; Lee, I.-J.; Yun, B.-W. Lead (Pb)-induced oxi-dative stress alters the morphological and physio-biochemical properties of rice (Oryza sativa L.). Agronomy 2021, 11, 409. [Google Scholar] [CrossRef]
- FAO. Global Network on Integrated Soil Management for Sustain-Able Use of Salt-Affected Soils. Available online: http://www.fao.org/ag/agl/agll/spush (accessed on 14 September 2022).
- Pitmanand, M.G.; Läuchli, A. Global impact of salinity and agricultural ecosystem. In Salinity: Environment-Plants-Molecules; Läuchli, A., Lüttge, U., Eds.; Kluwer Academic: Dodrecht, The Netherlands, 2002; pp. 3–20. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Breckle, S.W.; Scheffer, A.; Wucherer, W. Halophytes on the dry sea floor of the aral sea. In Sustainable Land-Use in Deserts; Breckle, S.W., Veste, M., Wucherer, W., Eds.; Springer: New York, NY, USA, 2001; pp. 139–146. [Google Scholar]
- El Shaer, H.M. Halophytes and salt-tolerant plants as potential forage for ruminants in the near east region. Small Rumin. Res. 2010, 91, 3–12. [Google Scholar] [CrossRef]
- Redman, R.S.; Kim, Y.O.; Woodward, C.J.; Greer, C.; Espino, L.; Doty, S.L.; Rodriguez, R.J. Increased fitness of rice plants to abiotic stress via habitat adapted symbiosis: A strategy for mitigating impacts of climate change. PLoS ONE 2011, 6, e14823. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.J.; Henson, J.; Van Volkenburgh, E.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.O.; Redman, R.S. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008, 2, 404–416. [Google Scholar] [CrossRef] [Green Version]
- Chebotar, V.K.; Malfanova, N.V.; Scherbakov, A.V.; Ahtemova, G.A.; Borisov, A.Y.; Lugtenberg, B.; Tikhonovich, I.A. Endophytic bacteria in microbial preparations that improve plant development. Appl. Biochem. Microbiol. 2015, 51, 271–277. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Tkacz, A.; Poole, P. Role of root microbiota in plant productivity. J. Exp. Bot. 2015, 66, 2167–2175. [Google Scholar] [CrossRef]
- Vaishnav, A.; Shukla, A.K.; Sharma, A.; Kumar, R.; Choudhary, D.K. Endophytic bacteria in plant salt stress tolerance: Current and future prospects. J. Plant Growth Regul. 2019, 38, 650–668. [Google Scholar] [CrossRef]
- Naveed, M.; Hafeez, S.; Rafique, M.; Mumtaz, M.Z.; Subhani, Z.; Holatko, J.; Hammerschmiedt, T.; Malicek, O.; Mustafa, A.; Kintl, A.; et al. Plant-endophyte mediated improvement in physiological and bio-protective abilities of marigold (Tagetes patula). Front. Plant Sci. 2022, 13, 993130. [Google Scholar] [CrossRef] [PubMed]
- Ruppel, S.; Franken, P.; Witzel, K. Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct. Plant Biol. 2013, 40, 940–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Yuan, Z.; Druzhinina, I.S.; Labbé, J.; Redman, R.; Qin, Y.; Rodriguez, R.; Zhang, C.; Tuskan, G.A.; Lin, F. Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci. Rep. 2016, 6, 32467. [Google Scholar] [CrossRef]
- Etesami, H.; Beattie, G.A. Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 2018, 9, 148. [Google Scholar] [CrossRef] [Green Version]
- Dod, I.C.; Pérez-Alfocea, F. Microbial amelioration of crop salinity stress. J. Exp. Bot. 2012, 63, 3415–3428. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Ullah, I.; Ali, S.; Kang, S.-M.; Lee, I.-J. Alleviation of salt stress response in soybean plants with the endophytic bacterial isolate Curtobacterium sp. SAK1. Ann. Microbiol. 2019, 69, 797–808. [Google Scholar] [CrossRef]
- Berg, G. Plant-microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef]
- Lucero, M.E.; Unc, A.; Cooke, P.; Dowd, S.; Sun, S. Endophyte microbiome diversity in micropropa-gated Atriplex canescens and Atriplex torreyi var griffithsii. PLoS ONE 2011, 6, e17693. [Google Scholar] [CrossRef]
- Zhao, S.; Zhou, N.; Zhao, Z.-Y.; Zhang, K.; Wu, G.-H.; Tian, C.-Y. Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr. Microbiol. 2016, 73, 574–581. [Google Scholar] [CrossRef]
- Piernik, A.; Hrynkiewicz, K.; Wojciechowska, A.; Szymańska, S.; Lis, M.I.; Muscolo, A. Effect of halotolerant endophytic bacteria isolated from Salicornia europaea L. on the growth of fodder beet (Beta vulgaris L.) under salt stress. Arch. Agron. Soil Sci. 2017, 63, 1404–1418. [Google Scholar] [CrossRef]
- El-Tarabily, K.A.; AlKhajeh, A.S.; Ayyash, M.M.; Alnuaimi, L.H.; Sham, A.; ElBaghdady, K.Z.; Tariq, S.; AbuQamar, S.F. Growth promotion of Salicornia bigelovii by Micromonospora chalcea UAE1, an endophytic 1-aminocyclopropane-1-carboxylic acid deaminase-producing actinobacterial isolate. Front. Microbiol. 2019, 24, 1694. [Google Scholar] [CrossRef]
- Qin, S.; Feng, W.-W.; Zhang, Y.-J.; Wang, T.-T.; Xiong, Y.-W.; Xing, K. Diversity of bacterial microbiota of coastal halophyte Limonium sinense and amelioration of salinity stress damage by symbiotic plant growth-promoting actinobacterium Glutamicibacter halophytocola KLBMP 5180. Appl. Environ. Micrbiol. 2018, 84, e01533-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokhari, A.; Essack, M.; Lafi, F.F.; Andres-Barrao, C.; Jalal, R.; Alamoudi, S.; Razali, R.; Alzubaidy, H.; Shah, K.H.; Siddique, S.; et al. Bioprospecting desert plant Bacillus endophytic strains for their potential to enhance plant stress tolerance. Sci. Rep. 2019, 9, 18154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, K.; Oguri, S.; Chiba, S.; Momonoki, Y.S. Molecular cloning of acetylcholinesterase gene from Salicornia europaea L. Plant Signal. Behav. 2009, 4, 361–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S. Salicornia: Evaluating the halophytic extremophile as a food and a pharmaceutical candidate. 3 Biotech 2016, 6, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malfanova, N.; Kamilova, F.; Validov, S.; Shcherbakov, A.; Chebotar, V.; Tikhonovich, I.; Lugtenberg, B. Charac-terization of Bacillus subtilis HC8, a novel plant-beneficial endophytic strain from giant hogweed. Microb. Biotechnol. 2011, 4, 523–532. [Google Scholar] [CrossRef] [Green Version]
- Schulz, B.; Wanke, U.; Draeger, S.; Aust, H.J. Endophytes from herbaceous plants and shrubs, effectiveness of surface sterilization methods. Mycol. Res. 1993, 97, 1447–1450. [Google Scholar] [CrossRef]
- Szymańska, S.; Borruso, L.; Brusetti, L.; Hulisz, P.; Furtado, B.; Hrynkiewicz, K. Bacterial microbiome of root-associated endophytes of Salicornia europaea in correspondence to different levels of salinity. Environ. Sci. Pollut. Res. Int. 2018, 25, 25420–25431. [Google Scholar] [CrossRef]
- Chen, C.; Bauske, E.M.; Musson, G.; Rodriguez-Kibana, R.; Kloepper, J.W. Biological control of Fusarium wilt on cotton by use of endophytic bacteria. Biol. Control 1995, 5, 83–91. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Nisar, M.; Ali, H.; Hazrat, A.; Hayat, K.; Keerio, A.A.; Ihsan, M.; Laiq, M.; Ullah, S.; Fahad, S.; et al. Drought tolerance improvement in plants: An endophytic bacterial approach. Appl. Microbiol. Biotechnol. 2019, 103, 7385–7397. [Google Scholar] [CrossRef] [PubMed]
- Yasseen, B.T.; Al-Thani, R.F. Endophytes and halophytes to remediate industrial wastewater and saline soils: Perspectives from Qatar. Plants 2022, 11, 1497. [Google Scholar] [CrossRef] [PubMed]
- Shurigin, V.; Alikulov, B.; Davranov, K.; Ismailov, Z. Bacterial endophytes from halophyte black saxaul (Haloxylon aphyllum Minkw.) and their plant growth-promoting properties. J. Appl. Biol. Biotechnol. 2022, 10, 45–53. [Google Scholar]
- Arora, S.A.; Patel, P.N.; Vanza, M.J.; Rao, G.G. Isolation and characterization of endophytic bacteria colonizing halophyte and other salt tolerant plant species from coastal Gujarat. Afr. J. Microbiol. Res. 2014, 8, 1779–1788. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.-L.; Tang, S.-K.; Chu, X.; Jiang, Z.; Xu, L.-H.; Zhi, X.-Y. Oceanobacillus endoradicis sp. nov., an endophytic bacterial species isolated from the root of Paris polyphylla Smith var. yunnanensis. Antonie Van Leeuwenhoek 2016, 109, 957–964. [Google Scholar] [CrossRef]
- Sorty, A.M.; Meena, K.K.; Choudhary, K.; Bitla, U.M.; Minhas, P.S.; Krishnani, K.K. Effect of plant growth promoting bacteria associated with halophytic weed (Psoralea corylifolia L.) on germination and seedling growth of wheat under saline conditions. Appl. Biochem. Biotechnol. 2016, 180, 872–882. [Google Scholar] [CrossRef]
- Zhang, C.; Cai, K.; Li, M.; Zheng, J.; Han, Y. Plant-growth-promoting potential of PGPE isolated from Dactylis glomerata L. Microorganisms 2022, 10, 731. [Google Scholar] [CrossRef]
- Chernin, L.; Ismailov, Z.; Haran, S.; Chet, I. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl. Environ. Microbiol. 1995, 61, 1720–1726. [Google Scholar] [CrossRef] [Green Version]
- Vaddepalli, P.; Fulton, L.; Wieland, J.; Wassmer, K.; Schaeffer, M.; Ranf, S.; Schneitz, K. The cell wall-localized atypical β-1,3 glucanase ZERZAUST controls tissue morphogenesis in Arabidopsis thaliana. Development 2017, 144, 2259–2269. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kulkarni, J.; Jha, B. Halotolerant rhizobacteria promote growth and enhance salinity tolerance in peanut. Front. Microbiol. 2016, 7, 1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, A.H. Bacterial solubilization of mineral phosphates: Historical perspective and future prospects. Am. J. Altern. Agric. 1986, 1, 51–57. [Google Scholar] [CrossRef]
- Bianco, C.; Imperlini, E.; Calogero, R.; Senatore, B.; Amoresano, A.; Carpentieri, A.; Pucci, P.; Defez, R. Indole-3-acetic acid improves Escherichia coli’s defences to stress. Arch. Microbiol. 2006, 185, 373–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahir, Z.A.; Shah, M.K.; Naveed, M.; Akhter, M.J. Substrate dependent auxin production by Rhizobium phaseoli improves the growth and yield of Vigna radiata L. under salt stress conditions. J. Microbiol. Biotechnol. 2010, 20, 1288–1294. [Google Scholar] [CrossRef]
- Ilangumaran, G.; Smith, D.L. Plant growth promoting rhizobacteria in amelioration of salinity stress: A systems biology perspective. Front. Plant Sci. 2017, 8, 1768. [Google Scholar] [CrossRef] [Green Version]
- Albacete, A.; Ghanem, M.E.; Martínez-Andújar, C.; Acosta, M.; Sánchez-Bravo, J.; Martínez, V.; Lutts, S.; Dodd, I.C.; Pérez-Alfocea, F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008, 59, 4119–4131. [Google Scholar] [CrossRef]
- Park, J.M.; Radhakrishnan, R.; Kang, S.M.; Lee, I.J. IAA producing Enterobacter sp. I-3 as a potent bio-herbicide candidate for weed control: A special reference with lettuce growth inhibition. Indian J. Microbiol. 2015, 55, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Tsukanova, K.A.; Chebotar, V.K.; Meyer, J.J.M.; Bibikova, T.N. Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. S. Afr. J. Bot. 2017, 113, 91–102. [Google Scholar] [CrossRef]
- Plonka, P.M.; Grabacka, M. Melanin synthesis in microorganisms—biotechnological and medical aspects. Acta Biochim. Pol. 2006, 53, 429–443. [Google Scholar] [CrossRef] [Green Version]
- Claus, H.; Decker, H. Bacterial tyrosinases. Syst. Appl. Microbiol. 2006, 29, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zerrad, A.; Anissi, J.; Ghanam, J.; Sendide, K.; El Hassouni, M. Antioxidant and antimicrobial activities of melanin produced by a Pseudomonas balearica strain. J. Biotechnol. Lett. 2014, 5, 87–94. [Google Scholar]
- Ghadge, V.; Kumar, P.; Singh, S.; Mathew, D.E.; Bhattacharya, S.; Nimse, S.B.; Shinde, P.B. Natural melanin produced by the endophytic Bacillus subtilis 4NP-BL associated with the halophyte Salicornia brachiate. J. Agric. Food Chem. 2020, 68, 6854–6863. [Google Scholar] [CrossRef] [PubMed]
- Chizhevskaya, E.P.; Naidenova, E.A.; Onishchuk, O.P.; Andronov, E.E.; Simarov, B.V. The melanin biosynthesis gene from the CA15-1 strain of alfalfa nodule bacteria: Molecular analysis and phylogeny. Rus. J. Genet. 2018, 54, 925–932. [Google Scholar] [CrossRef]
- Piñero, S.; Rivera, J.; Romero, D.; Cevallos, M.A.; Martínez, A.; Bolívar, F.; Gosset, G. Tyrosinase from Rhizobium etli is involved in nodulation efficiency and symbiosis-associated stress resistance. J. Mol. Microbiol. Biotechnol. 2007, 13, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Lee, S.Y.; Kong, H.G.; Jo, E.J.; Choi, H.K.; Khan, R.; Lee, S.-W. Genetic determinants for pyomelanin production and its protective effect against oxidative stress in Ralstonia solanacearum. PLoS ONE 2016, 11, e0160845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttila, A.M.; Compant, S.; Campisano, A.; Doring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Prieto, P.; Schiliro, E.; Maldonado-Gonzalez, M.M.; Valderrama, R.; Barroso-Albarracm, J.B.; Mercado-Blanco, J. Root hairs play a key role in the endophytic colonization of olive roots by Pseudomonas spp. with biocontrol activity. Microb. Ecol. 2011, 62, 435–445. [Google Scholar] [CrossRef] [Green Version]
- de Souza, A.L.S.R.; De Souza, S.A.; De Oliveira, M.V.V.; Ferraz, T.M.; Figueiredo, F.A.M.M.A.; Da Silva, N.D.; Rangel, P.L.; Panisset, C.R.S.; Olivares, F.L.; Campostrini, E.; et al. Endophytic colonization of Arabidopsis thaliana by Gluconacetobacter diazotrophicus and its effect on plant growth promotion, plant physiology, and activation of plant defense. Plant Soil 2016, 399, 257–270. [Google Scholar] [CrossRef]
- Ali, S.; Duan, J.; Charles, T.C.; Glick, B.R. A bioinformatics approach to the determination of genes involved in endophytic behavior in Burkholderia spp. J. Theor. Biol. 2014, 343, 193–198. [Google Scholar] [CrossRef]
- Bado, S.; Forster, B.P.; Ghanim, A.M.A.; Jankowicz-Cieslak, J.; Berthold, G.; Luxiang, L. Protocol for measuring soil salinity. In Protocols for Pre-Field Screening of Mutants for Salt Tolerance in Rice; Wheat and Barley; Springer: Cham, Switzerland, 2016; pp. 13–19. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Egamberdieva, D.; Kucharova, Z.; Davranov, K.; Berg, G.; Makarova, N.; Azarova, T.; Chebotar, V.; Tikhonovich, I.; Kamilova, F.; Validov, S.Z.; et al. Bacteria able to control foot and root rot and to promote growth of cucumber in salinated soils. Biol. Fertil. Soils 2011, 47, 197–205. [Google Scholar] [CrossRef]
- Shcherbakov, A.V.; Bragina, A.; Kuzmina, E.Y.; Berg, C.; Muntyan, A.N.; Makarova, N.V.; Malfanova, N.V.; Cardinale, M.; Berg, G.; Chebotar, V.K.; et al. Endophytic bacteria of sphagnum mosses as promising objects of agricultural microbiology. Microbiology 2013, 82, 306–315. [Google Scholar] [CrossRef]
- Kamilova, F.; Validov, S.; Azarova, T.; Mulders, I.; Lugtenberg, B. Enrichment for enhanced competitive root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 2005, 7, 1809–1817. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indole acetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Chrastil, J. Colorimetric estimation of indole-3-acetic acid. Anal. Biochem. 1976, 72, 134–138. [Google Scholar] [CrossRef]





| Host Plant | Number Plant Sample | Isolation Site | Isolate |
|---|---|---|---|
| Neftekumsky district, Stavropol territory, 44.5453 N 45.1907 E | |||
| Salicornia europaea L. | 1 | Root | Se1-1R; Se1-2R |
| Stem | Se1-3S; Se1-4S | ||
| 2 | - | - | |
| 3 | Root | Se3R | |
| 4 | Root | Se4-1R | |
| Stem | Se4-2S | ||
| 5 | Root | Se5R | |
| Salsola australis (R.Br.) | 6 | Root | Sa6R |
| 7 | - | - | |
| 8 | - | - | |
| 9 | - | - | |
| 10 | - | - | |
| Bassia sedoides (Pall.) | 11 | Root | Bs11-1R |
| Stem | Bs11-2S | ||
| 12 | Root | Bs12R | |
| 13 | - | - | |
| 14 | - | - | |
| 15 | Stem | Bs15S | |
| Levokumsky district, Stavropol Territory, 44.5017 N 44.2901 E | |||
| Kochia prostrata (L.) Schrad. | 16 | Root | Kp16R |
| 17 | - | - | |
| 18 | - | - | |
| 19 | Root | Kp19-1R; Kp19-2R | |
| Stem | Kp19-3S | ||
| 20 | Root | Kp20-1R | |
| Stem | Kp20-2S | ||
| Salsola australis (R.Br.) | 21 | Stem | Sa21S |
| 22 | Root | Sa22-1R | |
| Stem | Sa22-2S | ||
| 23 | Stem | Sa23S | |
| 24 | Root | Sa24-1R; Sa24-2R; Sa24-3R | |
| 25 | Stem | Sa25-1S; Sa25-2S | |
| Group | Isolate | Identity | Nearest Homolog Sequences (Accession Number) |
|---|---|---|---|
| 1 | Se1-2R; Se1-4S; Se4-2S; Bs12R; Bs15S; Kp19-1R; Kp19-3S; Kp20-2S; Sa6R; Sa22-1R; Sa24-3R | 100% | Bacillus amyloliquefaciens strain HC-7 (MW658654) Bacillus velezensis strain EH9 (MN750764) Bacillus vallismortis strain QAMA04 (KF419122) |
| 2 | Kp19-2R | 100% | Bacillus amyloliquefaciens strain OUCMDZ4205 (KU579266) Bacillus atrophaeus strain NOK100 (ON287177) Bacillus vallismortis isolate SR1-57 (LT838175) |
| 3 | Sa24-2R | 100% | Bacillus atrophaeus strain G28 (MT305986) Bacillus vallismortis strain Bac111 (CP033052) Bacillus velezensis strain HFBPR38 (MT539150) |
| 4 | Bs11-1R | 100% | Bacillus subtilis strain PR45 (MT993414) Bacillus halotolerans strain F41-3 (CP041357) Bacillus mojavensis strain T14 (MT313117) |
| 5 | Se5R; Bs11-2S | 100% | Bacillus subtilis strain AHV-KH18 (MH819665) Bacillus spizizenii strain SUM-KSU305 (HM753622) Bacillus rugosus strain NOK85 (ON287161) |
| 6 | Se4-1R | 99% | Bacillus licheniformis strain TRQ36 (MK493686) Bacillus licheniformis strain QT441 (MT032699) Bacillus paralicheniformis strain ISNah (MN542883) Bacillus sonorensis strain QT-10 (MT065718) |
| 7 | Se1-1R; Se1-3S; Sa21S | 100% | Bacillus pumilus strain NBRC 12092 (NR_112637) Bacillus safensis strain U14-5 (CP015607) Bacillus zhangzhouensis strain KBA10 (MG547766) |
| 8 | Sa23S | 100% | Bacillus pumilus strain 145 (CP027116) Bacillus safensis strain T13 (MT313116) Bacillus aerophilus strain KUDC1736 (KC414715) |
| 9 | Sa22-2S; Sa25-2S | 100% | Bacillus thuringiensis strain SP06LB (MT052669) Bacillus licheniformis strain CHGP12 (ON385932) Bacillus cereus strain ZB201708 (CP030982) |
| - | Se3R | 100% | Oceanobacillus manasiensis strain SLA-269 (MT125837) Oceanobacillus picturae strain R346(HM179167) Oceanobacillus sp. A359 (JX415302) |
| - | Kp20-1R | 100% 100% 99% | Paenibacillus lautus strain XAS3-14 (JF496308) Paenibacillus sp. strain JZ16 (CP017659) Paenibacillus glucanolyticus strain 3J138 (OM758166) |
| - | Kp16R | 99% | Pantoea agglomerans strain BLS (MH158658) Pantoea sp. strain CS15-18 (MH769222) Pantoea vagans strain LMG 24199 (CP038853) |
| - | Sa24-1R | 100% 99% 99% | Alcaligenes sp. strain B9 (MG594838) Alcaligenes aquatilis strain BUN33 (MG515168) Alcaligenes faecalis strain J9N (JX490075) |
| - | Sa25-1S | 100% 100% 99% | Myroides odoratimimus strain G13 (MK880501) Myroides sp. strain A21 (CP010327) Myroides odoratimimus strain 3YW8 (GU549435) |
| Genus | Group | Strain | 10 °C | 15 °C | 20 °C | 28 °C | 37 °C | 45 °C | 55 °C |
|---|---|---|---|---|---|---|---|---|---|
| Bacillus sp. | [1] | Se1-2R | - | - | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [1] | Se1-4S | - | - | +++ | +++ | +++ | ++ | ++ |
| Bacillus sp. | [1] | Se4-2S | - | ++ | +++ | +++ | +++ | +++ | + |
| Bacillus sp. | [1] | Bs12R | - | + | +++ | +++ | +++ | +++ | + |
| Bacillus sp. | [1] | Bs15S | - | ++ | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [1] | Kp19-1R | - | + | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [1] | Kp19-3S | - | ++ | +++ | +++ | +++ | +++ | ++ |
| Bacillus sp. | [1] | Kp20-2S | - | - | +++ | +++ | +++ | ++ | - |
| Bacillus sp. | [1] | Sa6R | - | - | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [1] | Sa22-1R | - | + | +++ | +++ | +++ | +++ | + |
| Bacillus sp. | [1] | Sa24-3R | - | - | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [2] | Kp19-2R | - | ++ | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [3] | Sa24-2R | - | ++ | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [4] | Bs11-1R | - | + | +++ | +++ | +++ | ++ | + |
| Bacillus sp. | [5] | Se5R | - | ++ | +++ | +++ | +++ | + | - |
| Bacillus sp. | [5] | Bs11-2S | - | ++ | +++ | +++ | +++ | +++ | - |
| Bacillus sp. | [6] | Se4-1R | - | - | +++ | +++ | +++ | ++ | ++ |
| Bacillus sp. | [7] | Se1-1R | - | ++ | +++ | +++ | +++ | +++ | + |
| Bacillus sp. | [7] | Se1-3S | + | ++ | +++ | +++ | +++ | ++ | + |
| Bacillus sp. | [7] | Sa21S | - | - | +++ | +++ | ++ | ++ | - |
| Bacillus sp. | [8] | Sa23S | - | - | +++ | +++ | ++ | ++ | - |
| Bacillus sp. | [9] | Sa22-2S | + | +++ | +++ | +++ | +++ | + | - |
| Bacillus sp. | [9] | Sa25-2S | + | +++ | +++ | +++ | ++ | - | - |
| Oceanobacillus sp. | Se3R | - | + | +++ | +++ | +++ | - | - | |
| Paenibacillus sp. | Kp20-1R | - | - | +++ | ++ | ++ | + | - | |
| Pantoea sp. | Kp16R | + | ++ | +++ | +++ | +++ | - | - | |
| Alcaligenes sp. | Sa24-1R | - | ++ | +++ | +++ | ++ | - | - | |
| Myroides sp. | Sa25-1S | - | + | +++ | +++ | - | - | - |
| Genus | Group | Strain | CA | AA | PA | LA | PSA | NF | IAA |
|---|---|---|---|---|---|---|---|---|---|
| Bacillus sp. | [1] | Se1-2R | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Se1-4S | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Se4-2S | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Bs12R | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Bs15S | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Kp19-1R | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Kp19-3S | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Kp20-2S | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Sa6R | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Sa22-1R | + | + | + | - | - | + | + |
| Bacillus sp. | [1] | Sa24-3R | + | + | + | - | - | - | + |
| Bacillus sp. | [2] | Kp19-2R | + | + | + | - | - | - | + |
| Bacillus sp. | [3] | Sa24-2R | + | + | + | - | - | - | + |
| Bacillus sp. | [4] | Bs11-1R | + | + | + | - | - | + | + |
| Bacillus sp. | [5] | Se5R | + | + | + | - | - | + | + |
| Bacillus sp. | [5] | Bs11-2S | + | + | + | - | - | + | + |
| Bacillus sp. | [6] | Se4-1R | + | + | + | - | - | + | + |
| Bacillus sp. | [7] | Se1-1R | - | - | - | - | - | - | + |
| Bacillus sp. | [7] | Se1-3S | - | - | + | - | - | - | + |
| Bacillus sp. | [7] | Sa21S | - | - | - | - | - | - | + |
| Bacillus sp. | [8] | Sa23S | - | - | + | - | - | - | + |
| Bacillus sp. | [9] | Sa22-2S | + | + | + | - | - | - | + |
| Bacillus sp. | [9] | Sa25-2S | + | + | + | - | - | - | + |
| Oceanobacillus sp. | Se3R | - | - | - | - | - | - | + | |
| Paenibacillus sp. | Kp20-1R | + | + | + | + | + | - | ++ | |
| Pantoea sp. | Kp16R | - | - | - | - | + | + | ++ | |
| Alcaligenes sp. | Sa24-1R | - | - | - | - | - | - | - | |
| Myroides sp. | Sa25-1S | - | - | + | - | - | - | ++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chebotar, V.K.; Chizhevskaya, E.P.; Baganova, M.E.; Keleinikova, O.V.; Yuzikhin, O.S.; Zaplatkin, A.N.; Khonina, O.V.; Kostitsin, R.D.; Lapenko, N.G. Endophytes from Halotolerant Plants Aimed to Overcome Salinity and Draught. Plants 2022, 11, 2992. https://doi.org/10.3390/plants11212992
Chebotar VK, Chizhevskaya EP, Baganova ME, Keleinikova OV, Yuzikhin OS, Zaplatkin AN, Khonina OV, Kostitsin RD, Lapenko NG. Endophytes from Halotolerant Plants Aimed to Overcome Salinity and Draught. Plants. 2022; 11(21):2992. https://doi.org/10.3390/plants11212992
Chicago/Turabian StyleChebotar, Vladimir K., Elena P. Chizhevskaya, Maria E. Baganova, Oksana V. Keleinikova, Oleg S. Yuzikhin, Alexander N. Zaplatkin, Olesya V. Khonina, Roman D. Kostitsin, and Nina G. Lapenko. 2022. "Endophytes from Halotolerant Plants Aimed to Overcome Salinity and Draught" Plants 11, no. 21: 2992. https://doi.org/10.3390/plants11212992







