Legume Pangenome: Status and Scope for Crop Improvement
Abstract
1. Introduction
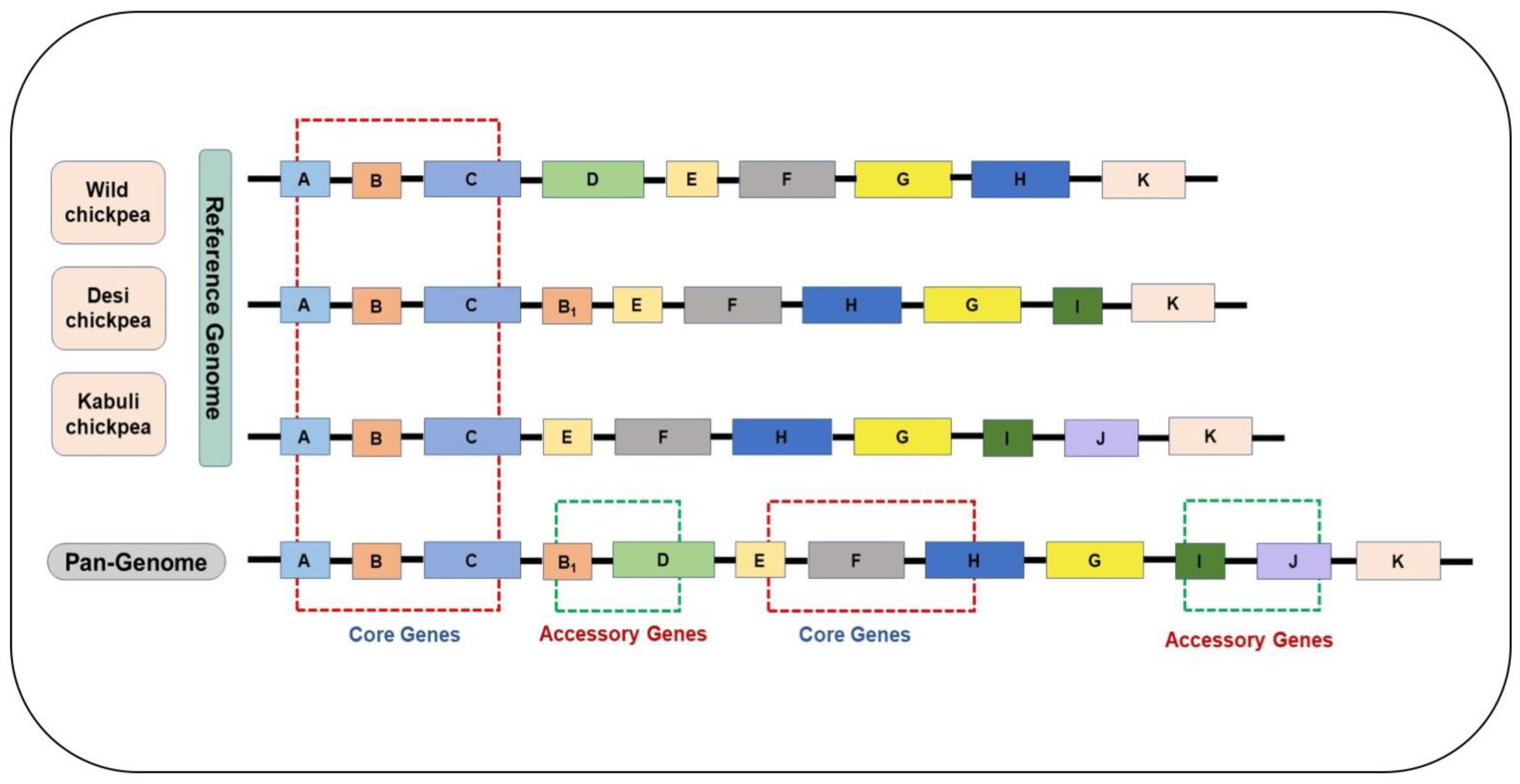
2. Pangenome Capturing Structural Variations Not Present in Single Reference Genomes
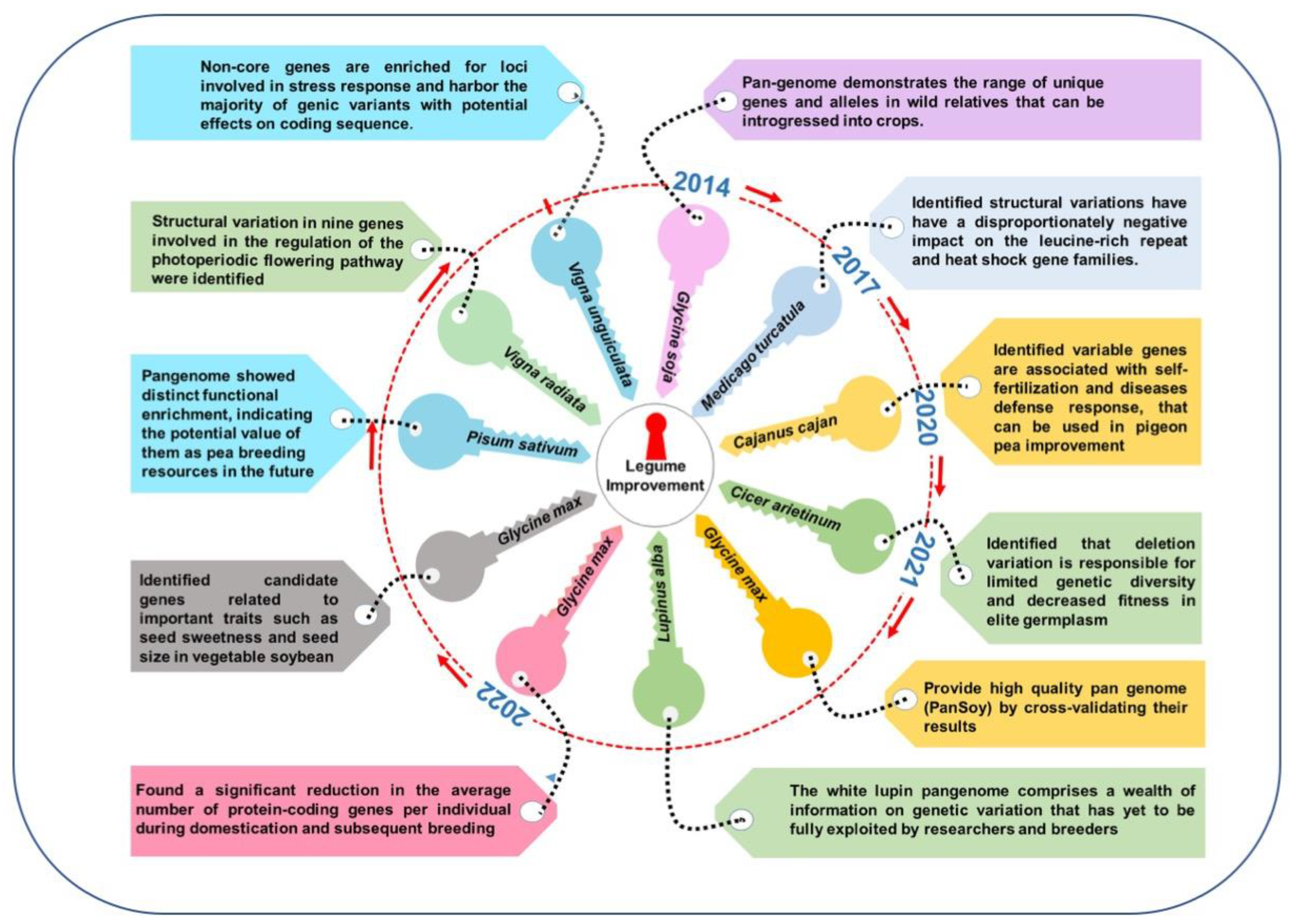
3. Soybean (Glycine max L.)
4. Chickpea (Cicer arietinum L.) Pangenome
5. Cowpea (Vigna ungiculata L.) Pangenome
6. Pigeonpea (Cajanus cajan L.) Pangenome
7. Mungbean (Vigna radiata L.) Pangenome
8. White Lupin (Lupinus alba L.) Pangenome
9. Barrel Medic (Medicago truncatula) Pangenome
10. Lentil (Lens culinaris) Pangenome
11. Pea (Pisum sativum L.) Pangenome
12. Pangenome-Derived PAVs for GWAS Analysis to Explore Novel Genes
13. Pangenome and Scope of Crop Domestication for New Species
14. Prospects and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmidhuber, J.; Tubiello, F.N. Global food security under climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19703–19708. [Google Scholar] [CrossRef] [PubMed]
- Leisner, C.P. Climate change impacts on food security-focus on perennial cropping systems and nutritional value. Plant Sci. 2020, 293, 110412. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, B.; Collard, B.C.; Demont, M. Improving global food security through accelerated plant breeding. Plant Sci. 2019, 287, 110207. [Google Scholar] [CrossRef] [PubMed]
- Rivers, J.; Warthmann, N.; Pogson, B.J.; Borevitz, J.O. Genomic breeding for food, environment and livelihoods. Food Secur. 2015, 7, 375–382. [Google Scholar] [CrossRef]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trend Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef]
- Saxena, R.K.; Edwards, D.; Varshney, R.K. Structural variations in plant genomes. Brief. Funct. Genom. 2014, 13, 296–307. [Google Scholar] [CrossRef]
- Yuan, Y.; Bayer, P.E.; Batley, J.; Edwards, D. Current status of structural variation studies in plants. Plant Biotechnol. J. 2021, 19, 2153–2163. [Google Scholar] [CrossRef]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef]
- Tao, Y.; Jordan, D.R.; Mace, E.S. A graph-based pan-genome guides biological discovery. Mol. Plant 2020, 13, 1247–1249. [Google Scholar] [CrossRef] [PubMed]
- Danilevicz, M.F.; Fernandez, C.G.T.; Marsh, J.I.; Bayer, P.E.; Edwards, D. Plant pangenomics: Approaches, applications and advancements. Curr. Opin. Plant Biol. 2020, 54, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Bayer, P.E.; Barker, G.C.; Edger, P.P.; Kim, H.; Martinez, P.A.; Chan, C.K.; Severn-Ellis, A.; McCombie, W.R.; Parkin, I.A.; et al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 2016, 7, 13390. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Bayer, P.E.; Bhalla, P.L.; Batley, J.; Edwards, D. Pangenomics comes of age: From bacteria to plant and animal appli- cations. Trends Genet. 2020, 36, 132–145. [Google Scholar] [CrossRef]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef]
- Torkamaneh, D.; Lemay, M.A.; Belzile, F. The pan-genome of the cultivated soybean (PanSoy) reveals an extraordinarily conserved gene content. Plant Biotechnol. J. 2021, 19, 1852–1862. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.G.; Zhang, Z.; Guo, Y.; Zhang, J.; Sui, Y.; Zheng, L.; et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014, 32, 1045–1052. [Google Scholar] [CrossRef]
- Morgante, M.; De Paoli, E.; Radovic, S. Transposable elements and the plant pan-genomes. Curr. Opin. Plant Biol. 2007, 10, 149–155. [Google Scholar] [CrossRef]
- Jiao, W.B.; Schneeberger, K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat. Commun. 2020, 11, 989. [Google Scholar] [CrossRef]
- Schatz, M.C.; Maron, L.G.; Stein, J.C.; Wences, A.H.; Gurtowski, J.; Biggers, E.; Lee, H.; Kramer, M.; Antoniou, E.; Ghiban, E.; et al. Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 2014, 15, 506. [Google Scholar]
- Montenegro, J.D.; Golicz, A.A.; Bayer, P.E.; Hurgobin, B.; Lee, H.; Chan, C.K.; Visendi, P.; Lai, K.; Doležel, J.; Batley, J.; et al. The pangenome of hexaploid bread wheat. Plant J. 2017, 90, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Franco, J.A.V.; Gage, J.L.; Bradbury, P.J.; Johnson, L.C.; Miller, Z.R.; Buckler, E.S.; Romay, M.C. A Maize practical haplotype graph leverages diverse NAM assemblies. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gordon, S.P.; Contreras-Moreira, B.; Woods, D.P.; Des Marais, D.L.; Burgess, D.; Shu, S.; Stritt, C.; Roulin, A.C.; Schackwitz, W.; Tyler, L.; et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017, 8, 2184. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Contreras-Moreira, B.; Levy, J.J.; Djamei, A.; Czedik-Eysenberg, A.; Tartaglio, V.S.; Session, A.; Martin, J.; Cartwright, A.; Katz, A.; et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat. Commun. 2020, 11, 3670. [Google Scholar] [CrossRef]
- Ruperao, P.; Thirunavukkarasu, N.; Gandham, P.; Selvanayagam, S.; Govindaraj, M.; Nebie, B.; Manyasa, E.; Gupta, R.; Das, R.R.; Odeny, D.A.; et al. Sorghum pan-genome explores the functional utility for genomic-assisted breeding to accelerate the genetic gain. Front. Plant Sci. 2021, 12, 963. [Google Scholar] [CrossRef]
- Hurgobin, B.; Edwards, D. SNP discovery using a pangenome: Has the single reference approach become obsolete? Biology 2017, 6, 21. [Google Scholar] [CrossRef]
- Bayer, P.E.; Scheben, A.; Golicz, A.A.; Yuan, Y.; Faure, S.; Lee, H.; Chawla, H.S.; Anderson, R.; Bancroft, I.; Raman, H.; et al. Modelling of gene loss propensity in the pangenomes of three Brassica species suggests different mechanisms between polyploids and diploids. Plant Biotechnol. J. 2021, 19, 2488–2500. [Google Scholar] [CrossRef]
- Hübner, S.; Bercovich, N.; Todesco, M.; Mandel, J.R.; Odenheimer, J.; Ziegler, E.; Lee, J.S.; Baute, G.J.; Owens, G.L.; Grassa, C.J.; et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 2019, 5, 54–62. [Google Scholar] [CrossRef]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Ou, L.; Li, D.; Lv, J.; Chen, W.; Zhang, Z.; Li, X.; Yang, B.; Zhou, S.; Yang, S.; Li, W.; et al. Pan-genome of cultivated pepper (Capsicum) and its use in gene presence–absence variation analyses. New Phytol. 2018, 220, 360–363. [Google Scholar] [CrossRef]
- Li, J.; Yuan, D.; Wang, P.; Wang, Q.; Sun, M.; Liu, Z.; Si, H.; Xu, Z.; Ma, Y.; Zhang, B.; et al. Cotton pan-genome retrieves the lost sequences and genes during domestication and selection. Genome Biol. 2021, 22, 119. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Song, C.; Saxena, R.K.; Azam, S.; Yu, S.; Sharpe, A.G.; Cannon, S.; Baek, J.; Rosen, B.D.; Tar’an, B.; et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013, 31, 240–246. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Chen, W.; Li, Y.; Bharti, A.K.; Saxena, R.K.; Schlueter, J.A.; Donoghue, M.T.; Azam, S.; Fan, G.; Whaley, A.M.; et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012, 30, 83. [Google Scholar] [CrossRef] [PubMed]
- Kreplak, J.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Yang, T.; Liu, R.; Luo, Y.; Hu, S.; Wang, D. Improved pea reference genome and pan-genome highlight genomic features and evolutionary characteristics. Nat. Genet. 2022, 54, 1553–1563. [Google Scholar] [CrossRef]
- Hane, J.K.; Ming, Y.; Kamphuis, L.G.; Nelson, M.N.; Garg, G.; Atkins, C.A.; Bayer, P.E.; Bravo, A.; Bringans, S.; Cannon, S.; et al. A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: Insights into plant–microbe interactions and legume evolution. Plant Biotechnol. J. 2017, 15, 318–330. [Google Scholar] [CrossRef]
- Hufnagel, B.; Marques, A.; Soriano, A.; Marquès, L.; Divol, F.; Doumas, P.; Sallet, E.; Mancinotti, D.; Carrere, S.; Marande, W.; et al. High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nat. Commun. 2020, 11, 492. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.C.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J.; et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443. [Google Scholar] [CrossRef] [PubMed]
- Gaut, B.S.; Seymour, D.K.; Liu, Q.; Zhou, Y. Demography and its effects on genomic variation in crop domestication. Nat. Plants 2018, 4, 512–520. [Google Scholar] [CrossRef]
- Della Coletta, R.; Qiu, Y.; Ou, S.; Hufford, M.B.; Hirsch, C.N. How the pan-genome is changing crop genomics and improvement. Genome Biol. 2021, 22, 1–9. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Zhang, H.; Liu, Z.; Wang, Y.; Xing, L.; He, Q.; Du, H. Plant pan-genomics: Recent advances, new challenges, and roads ahead. J. Genet. Genom. 2022, 49, 833–846. [Google Scholar] [CrossRef]
- Jiang, N.; Bao, Z.; Zhang, X.; Eddy, S.R.; Wessler, S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature 2004, 431, 569–573. [Google Scholar] [CrossRef]
- Fedoroff, N.V. Transposable elements, epigenetics, and genome evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef]
- Wei, L.; Cao, X. The effect of transposable elements on phenotypic variation: Insights from plants to humans. Sci. China Life Sci. 2016, 59, 24–37. [Google Scholar] [CrossRef]
- Yandeau-Nelson, M.D.; Xia, Y.; Li, J.; Neuffer, M.G.; Schnable, P.S. Unequal sister chromatid and homolog recombination at a tandem duplication of the A1 locus in maize. Genetics 2006, 173, 2211–2226. [Google Scholar] [CrossRef][Green Version]
- Lei, L.; Goltsman, E.; Goodstein, D.; Wu, G.A.; Rokhsar, D.S.; Vogel, J.P. Plant pan-genomics comes of age. Annu. Rev. Plant Biol. 2021, 72, 411–435. [Google Scholar] [CrossRef] [PubMed]
- Jayakodi, M.; Schreiber, M.; Stein, N.; Mascher, M. Building pan-genome infrastructures for crop plants and their use in association genetics. DNA Res. 2021, 28, dsaa030. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book/Am. Soc. Plant Biol. 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-genome of wild and cultivated soybeans. Cell 2020, 182, 162–176. [Google Scholar] [CrossRef]
- Bayer, P.E.; Valliyodan, B.; Hu, H.; Marsh, J.I.; Yuan, Y.; Vuong, T.D.; Patil, G.; Song, Q.; Batley, J.; Varshney, R.K.; et al. Sequencing the USDA core soybean collection reveals gene loss during domestication and breeding. Plant Genome 2022, 15, e20109. [Google Scholar] [CrossRef]
- Liu, C.; Wang, Y.; Peng, J.; Fan, B.; Xu, D.; Wu, J.; Cao, Z.; Gao, Y.; Wang, X.; Li, S.; et al. High-quality genome assembly and pan-genome studies facilitate genetic discovery in mung bean and its improvement. Plant Commun. 2022, 26, 100352. [Google Scholar] [CrossRef]
- Liang, Q.; Munoz-Amatriain, M.; Shu, S.; Lo, S.; Wu, X.; Carlson, J.W.; Davidson, P.; Goodstein, D.M.; Phillips, J.; Janis, N.M.; et al. A view of the pan-genome of domesticated cowpea (Vigna unguiculata [L.] Walp.). bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhao, J.; Bayer, P.E.; Ruperao, P.; Saxena, R.K.; Khan, A.W.; Golicz, A.A.; Nguyen, H.T.; Batley, J.; Edwards, D.; Varshney, R.K. Trait associations in the pangenome of pigeon pea (Cajanus cajan). Plant Biotechnol. J. 2020, 18, 1946–1954. [Google Scholar] [CrossRef]
- Varshney, R.K.; Roorkiwal, M.; Sun, S.; Bajaj, P.; Chitikineni, A.; Thudi, M.; Singh, N.P.; Du, X.; Upadhyaya, H.D.; Khan, A.W.; et al. A chickpea genetic variation map based on the sequencing of 3,366 genomes. Nature 2021, 599, 622–627. [Google Scholar] [CrossRef]
- Zhou, P.; Silverstein, K.A.; Ramaraj, T.; Guhlin, J.; Denny, R.; Liu, J.; Farmer, A.D.; Steele, K.P.; Stupar, R.M.; Miller, J.R.; et al. Exploring structural variation and gene family architecture with De Novo assemblies of 15 Medicago genomes. BMC Genom. 2017, 18, 261. [Google Scholar] [CrossRef]
- Hufnagel, B.; Soriano, A.; Taylor, J.; Divol, F.; Kroc, M.; Sanders, H.; Yeheyis, L.; Nelson, M.; Péret, B. Pangenome of white lupin provides insights into the diversity of the species. Plant Biotechnol. J. 2021, 19, 2532–2543. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Gonzalez, J.J.; García, P.; Polanco, C.; González, A.I.; Vaquero, F.; Vences, F.J.; Pérez de la Vega, M.; Sáenz de Miera, L.E. Multi-Species Transcriptome Assemblies of Cultivated and Wild Lentils (Lens sp.) Provide a First Glimpse at the Lentil Pangenome. Agronomy 2022, 12, 1619. [Google Scholar] [CrossRef]
- Shen, Y.; Liu, J.; Geng, H.; Zhang, J.; Liu, Y.; Zhang, H.; Xing, S.; Du, J.; Ma, S.; Tian, Z. De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 2018, 61, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Du, H.; Liu, Y.; Ni, L.; Wang, Z.; Liang, C.; Tian, Z. Update soybean Zhonghuang 13 genome to a golden reference. Sci. China Life Sci. 2019, 62, 1257–1260. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chung, C.Y.; Li, M.W.; Wong, F.L.; Wang, X.; Liu, A.; Wang, Z.; Leung, A.K.; Wong, T.H.; Tong, S.W.; et al. A reference-grade wild soybean genome. Nat. Commun. 2019, 10, 1216. [Google Scholar] [CrossRef]
- Hu, Z.; Sun, C.; Lu, K.C.; Chu, X.; Zhao, Y.; Lu, J.; Shi, J.; Wei, C. EUPAN enables pan-genome studies of a large number of eukaryotic genomes. Bioinformatics 2017, 33, 2408–2409. [Google Scholar] [CrossRef]
- Jain, M.; Misra, G.; Patel, R.K.; Priya, P.; Jhanwar, S.; Khan, A.W.; Shah, N.; Singh, V.K.; Garg, R.; Jeena, G.; et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 2013, 74, 715–729. [Google Scholar] [CrossRef]
- Timko, M.P.; Singh, B.B. Cowpea, a multifunctional legume. In Genomics of Tropical Crop Plants; Moore, P.H., Ming, R., Eds.; Springer: New York, NY, USA, 2008; pp. 227–258. [Google Scholar]
- Medina, C.A.; Samac, D.A.; Yu, L.X. Pan-transcriptome identifying master genes and regulation network in response to drought and salt stresses in Alfalfa (Medicago sativa L.). Sci. Rep. 2021, 11, 17203. [Google Scholar] [CrossRef]
- Varshney, R.; Saxena, R.; Upadhyaya, H.; Khan, A.; Yu, Y.; Kim, C.; Rathore, A.; Kim, D.; Kim, J.; An, S.; et al. Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat. Genet. 2017, 49, 1082–1088. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Parida, S.K.; Bakır, M.; von Wettberg, E.J.; Siddique, K.H.M. Progress of Genomics-Driven Approaches for Sustaining Underutilized Legume Crops in the Post-Genomic Era. Front. Genet. 2022, 13, 831656. [Google Scholar] [CrossRef]
- Wolko, B.; Clements, J.C.; Naganowska, B.; Nelson, M.; Huaan, Y. Lupinus. In Wild Crop Relatives: Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 153–206. [Google Scholar]
- Taylor, J.L.; De Angelis, G.; Nelson, M.N. How have narrow-leafed lupin genomic resources enhanced our understanding of lupin domestication? In The Lupin Genome; Singh, K.B., Kamphuis, L.G., Nelson, M.N., Eds.; Springer: Cham, Switzerland, 2020; pp. 95–108. [Google Scholar]
- Xu, W.; Zhang, Q.; Yuan, W.; Xu, F.; Muhammad Aslam, M.; Miao, R.; Li, Y.; Wang, Q.; Li, X.; Zhang, X.; et al. The genome evolution and low-phosphorus adaptation in white lupin. Nat. Commun. 2020, 11, 1069. [Google Scholar] [CrossRef] [PubMed]
- Ksiazkiewicz, M.; Nazzicari, N.; Yang, H.; Nelson, M.N.; Renshaw, D.; Rychel, S.; Ferrari, B.; Carelli, M.; Tomaszewska, M.; Stawiński, S.; et al. A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci. Rep. 2017, 7, 15335. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Renshaw, D.; Luckett, D.; Clements, J.; Yan, G.; Adhikari, K.; Buirchell, B.; Sweetingham, M.; Yang, H. Development of a sequence-specific PCR marker linked to the gene “pauper” conferring low-alkaloids in white lupin (Lupinus albus L.) for marker assisted selection. Mol. Breed. 2009, 23, 153–161. [Google Scholar] [CrossRef]
- Young, N.D.; Udvardi, M. Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 2009, 12, 193–201. [Google Scholar] [CrossRef]
- von Wettberg, E.J.; Porter, S.S.; Moriuchi, K.S.; Mukherjee, J.R. Chapter 2: Medicago as a model forage and ecological model. In The Model Legume Medicago Truncatula; de Bruijn, F.J., Ed.; Wiley Publishers: Hoboken, NJ, USA, 2020. [Google Scholar]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef]
- de la Vega, M.P.; Torres, A.M.; Cubero, J.I.; Kole, C. (Eds.) Genetics, Genomics and Breeding of Cool Season Grain Legumes; CRC Press: Boca Raton, FL, USA, 2011; pp. 98–150. [Google Scholar]
- Ramsay, L.; Koh, C.; Konkin, D.; Cook, D.; Penmetsa, V.; Dongying, G.; Coyne, C.; Humann, J.; Kaur, S.; Dolezel, J.; et al. Lens Culinaris CDC Redberry Genome Assembly v2.0. Available online: https://knowpulse.usask.ca/genome-assembly/Lcu.2RBY (accessed on 7 April 2021).
- Ramsay, L.; Koh, C.S.; Kagale, S.; Gao, D.; Kaur, S.; Haile, T.; Gela, T.S.; Chen, L.A.; Cao, Z.; Konkin, D.J.; et al. Genomic rearrangements have consequences for introgression breeding as revealed by genome assemblies of wild and cultivated lentil species. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hirsch, C.N.; Foerster, J.M.; Johnson, J.M.; Sekhon, R.S.; Muttoni, G.; Vaillancourt, B.; Peñagaricano, F.; Lindquist, E.; Pedraza, M.A.; Barry, K.; et al. Insights into the maize pan-genome and pan-transcriptome. Plant Cell 2014, 26, 121–135. [Google Scholar] [CrossRef]
- Guerra-Garcia, A.; Haile, T.; Ogutcen, E.; Bett, K.E.; von Wettberg, E.J. An evolutionary look into the history of lentil reveals unexpected diversity. Evol. Appl. 2022, 15, 1313–1325. [Google Scholar] [CrossRef]
- Ogutcen, E.; Ramsay, L.; von Wettberg, E.B.; Bett, K.E. Capturing variation in Lens (Fabaceae): Development and utility of an exome capture array for lentil. Appl. Plant Sci. 2018, 6, e01165. [Google Scholar] [CrossRef]
- Dahl, W.J.; Foster, L.M.; Tyler, R.T. Review of the health benefits of peas (Pisum sativum L.). Br. J. Nutr. 2012, 108, S3–S10. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M. Domestication of pulses in the Old World: Legumes were companions of wheat and barley when agriculture began in the Near East. Science 1973, 182, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Gizlice, Z.; Carter, T.E., Jr.; Burton, J.W. Genetic diversity patterns in North American public soybean cultivars based on coefficient of parentage. Crop Sci. 1996, 36, 753–765. [Google Scholar] [CrossRef]
- Hyten, D.L.; Song, Q.; Zhu, Y.; Choi, I.Y.; Nelson, R.L.; Costa, J.M.; Specht, J.E.; Shoemaker, R.C.; Cregan, P.B. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 2006, 103, 16666–16671. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K. The domestication syndrome. Kulturpflanze 1984, 32, 11–34. (In German) [Google Scholar] [CrossRef]
- Gepts, P. Crop domestication as a long-term selection experiment. Plant Breed. Rev. 2010, 24, 1–44. [Google Scholar]
- Milla, R.; Osborne, C.P.; Turcotte, M.M.; Violle, C. Plant domestication through an ecological lens. Trends Ecol. Evol. 2015, 30, 463–469. [Google Scholar] [CrossRef]
- Smýkal, P.; Nelson, M.N.; Berger, J.D.; Von Wettberg, E.J. The impact of genetic changes during crop domestication. Agronomy 2018, 8, 119. [Google Scholar] [CrossRef]
- Hufford, M.B.; Teran, J.C.B.M.Y.; Gepts, P. Crop Biodiversity: An Unfinished Magnum Opus of Nature. Annu. Rev. Plant Biol. 2019, 70, 727–751. [Google Scholar] [CrossRef]
- Ladizinsky, G. Seed dispersal in relation to the domestication of Middle East legumes. Econ. Bot. 1979, 33, 284–289. [Google Scholar] [CrossRef]
- Ogutcen, E.; Pandey, A.; Khan, M.K.; Marques, E.; Penmetsa, R.V.; Kahraman, A.; Von Wettberg, E.J. Pod shattering: A homologous series of variation underlying domestication and an avenue for crop improvement. Agronomy 2018, 8, 137. [Google Scholar] [CrossRef]
- Balarynová, J.; Klčová, B.; Sekaninová, J.; Kobrlová, L.; Cechová, M.Z.; Krejčí, P.; Leonova, T.; Gorbach, D.; Ihling, C.; Smržová, L.; et al. The loss of polyphenol oxidase function is associated with hilum pigmentation and has been selected during pea domestication. New Phytol. 2022, 235, 1807–1821. [Google Scholar] [CrossRef] [PubMed]
- Kluyver, T.A.; Charles, M.; Jones, G.; Rees, M.; Osborne, C.P. Did greater burial depth increase the seed size of domesticated legumes? J. Expt. Bot. 2013, 64, 4101–4108. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.D.; Shrestha, D.; Ludwig, C. Reproductive strategies in Mediterranean legumes: Trade-offs between phenology, seed size and vigor within and between wild and domesticated Lupinus species collected along aridity gradients. Front. Plant Sci. 2017, 8, 548. [Google Scholar] [CrossRef] [PubMed]
- Salinas, A.D.; Bonet, A.; Gepts, P. The Wild Relative of PhaseolusVulgaris in Middle America. In Genetic Resources of Phaseolus Beans; Springer: Dordrecht, The Netherlands, 1988; pp. 163–184. [Google Scholar]
- Fernandez, A.R.; Sáez, A.; Quintero, C.; Gleiser, G.; Aizen, M.A. Intentional and unintentional selection during plant domestication: Herbivore damage, plant defensive traits and nutritional quality of fruit and seed crops. New Phytol. 2021, 231, 1586–1598. [Google Scholar] [CrossRef]
- Weeden, N.F. Genetic changes accompanying the domestication of Pisum sativum: Is there a common genetic basis to the ‘domestication syndrome’ for legumes? Ann. Bot. 2007, 100, 1017–1025. [Google Scholar] [CrossRef]
- Abbo, S.; van-Oss, R.P.; Gopher, A.; Saranga, Y.; Ofner, I.; Peleg, Z. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant Sci. 2014, 19, 351–360. [Google Scholar] [CrossRef]
- Tay Fernandez, C.G.; Nestor, B.J.; Danilevicz, M.F.; Gill, M.; Petereit, J.; Bayer, P.E.; Finnegan, P.M.; Batley, J.; Edwards, D. Pangenomes as a Resource to Accelerate Breeding of Under-Utilised Crop Species. Int. J. Mol. Sci. 2022, 23, 2671. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]

| Crop | Size of Pangenome | Number of Core and Dispensable Genes | Accessions Used | Reference |
|---|---|---|---|---|
| Wild soybean (Glycine soja) | 889.33–1118.34 Mb | 48.6% of the gene families | 7 | [17] |
| were core genomic units and 51.4% of gene families (30,364) were non-core | ||||
| Soybean (Glycine max) | 108 Mb | 49 431 (90.6%) hard-core genes, 7.2% variable genes | 204 | [16] |
| Soybean (Glycine max) | 992.3 Mb to 1059.8 Mb | 20,623 core genes and 28,679 dispensable genes | 26 | [54] |
| Soybean (Glycine max) | 1213 Mbp | 86.8% of genes were core and 13.2% were dispensable | 1110 | [55] |
| Mungbean (Vigna radiata) | 762.92 Mb | 83.1% were core genes and 16.9% were variable | 217 | [56] |
| Cowpea (Vigna ungiculata) | 449.91 Mb | 21,330 core and 23,531 non-core genes | 6 | [57] |
| Pigeonpea (Cajanus cajan) | 622 Mbp | 48 067 (86.6%) core genes and 7445 (13.4%) non-core | 89 | [58] |
| Chickpea (Cicer arietinum) | 592.58 Mbp | - | 3366 | [59] |
| Barrel medic (Medicago turcatula) | 388 Mbp to 428 Mbp | 250 Mbp core sequence and 180 Mbp dispensable | 15 | [60] |
| sequence | ||||
| White lupin (Lupinus alba) | 462.7 Mbp | 32,068 core genes, 14,822 non-core genes | 39 | [61] |
| Lentil (Lens culinaris) | - | 15,910 core genes, 24,226 accessory genes | 8 | [62] |
| Pea (Pisum sativum) | 15,470 core genes, 6170 softcore genes, 41,028 shell genes, and 50,108 cloud genes | 116 | [38] |
| Crop | Salient Features | Reference |
|---|---|---|
| Wild soybean (Glycine soja) | 2.3–3.9 Mbp of G. soja specific PAV related to defence response, cell growth, and photosynthesis. Pangenome analysis | [17] |
| informed variation for protein, oil, flowering time, and organ-size traits in the wild and cultivated soybean species | ||
| Soybean (Glycine max) | PanSoy sheds novel insights into the intraspecific variation in G. max | [16] |
| Soybean (Glycine max) | The function of core genes were related to growth, immune system, reproductive, cellular, and cellular-component organization or biogenesis. | [54] |
| The function of non-core genes were related to abiotic and biotic response genes, such as different NBS gene families. Structural variation related to | ||
| domestication traits and agronomic trait, viz., iron-deficiency chlorosis | ||
| Soybean (Glycine max) | Domestication selection sweeps on chromosome Gm20, breeding-related selective sweep region on Gm20, | [55] |
| 110 genomic regions with signatures of domestication-selective sweeps harboring 1266 protein-coding genes, | ||
| 86 genomic regions with signatures of breeding-selective sweeps harboring 1434 protein-coding genes | ||
| Mungbean (Vigna radiata) | A total of nine presence/absence variations controlling flowering regulation. Bruchid-resistant genes Pang34265, Pang44622, Pang57772, Pang58608, and Pang64254 | [56] |
| Cowpea (Vigna ungiculata) | PAVs contributing to black seed-coat color in cowpeas | [57] |
| Pigeonpea (Cajanus cajan) | PAVs attributing phenotypic diversity, including various phenotypic traits, viz., seed weight, days to 50% flowering, and plant height | [58] |
| Chickpea (Cicer arietinum) | 643 gene-gain and 247 gene-loss CNVs in C. reticulatum accessions, | [59] |
| insertions (139,483), deletions (47,882), inversions (61,171), | ||
| intra-chromosomal translocations (417) and inter-chromosomal translocations (2410) in cultivated and 287,854 insertions, | ||
| 67,351 deletions, 58,070 inversions, 446 intra-chromosomal translocations and 2066 inter-chromosomal translocations among C. reticulatum accessions | ||
| Barrel medic (Medicago truncatula) | 500,000–1,500,000 short indels (<50 bp), 27,000–110,000 large indels, 49,000–169,000 copy number variants (CNVs), | [60] |
| and 2700–12,700 translocations; NBS-LRRs showed high SNP diversity; | ||
| Medicago sativa | circRNA, lncRNA responsive to salinity and drought | [69] |
| White lupin (Lupinus alba) | Alkaloid-related genes/QTLs/genomic region, 1195 PAVs | [61] |
| Lentil (Lens culinaris) | Comparative analysis and evolutionary analysis, transcriptome of L. culinaris contained genes 58,375 | [62] |
| L. nigricans transcriptome contained the minimum number of 46,742 genes | ||
| Pea (Pisum sativum) | Two seed-dormancy genes Psat02G0081200 and Psat02G0507900 were elucidated; pan genes of P. abyssinicum were unique in response to chemicals and stimuli; | [38] |
| pan genes of P. fulvum showed their role in development, cytoskeleton, and tropism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jha, U.C.; Nayyar, H.; von Wettberg, E.J.B.; Naik, Y.D.; Thudi, M.; Siddique, K.H.M. Legume Pangenome: Status and Scope for Crop Improvement. Plants 2022, 11, 3041. https://doi.org/10.3390/plants11223041
Jha UC, Nayyar H, von Wettberg EJB, Naik YD, Thudi M, Siddique KHM. Legume Pangenome: Status and Scope for Crop Improvement. Plants. 2022; 11(22):3041. https://doi.org/10.3390/plants11223041
Chicago/Turabian StyleJha, Uday Chand, Harsh Nayyar, Eric J. B. von Wettberg, Yogesh Dashrath Naik, Mahendar Thudi, and Kadambot H. M. Siddique. 2022. "Legume Pangenome: Status and Scope for Crop Improvement" Plants 11, no. 22: 3041. https://doi.org/10.3390/plants11223041
APA StyleJha, U. C., Nayyar, H., von Wettberg, E. J. B., Naik, Y. D., Thudi, M., & Siddique, K. H. M. (2022). Legume Pangenome: Status and Scope for Crop Improvement. Plants, 11(22), 3041. https://doi.org/10.3390/plants11223041










