Mixed-Ploidy and Dysploidy in Hypericum perforatum: A Karyomorphological and Genome Size Study
Abstract
:1. Introduction
2. Results
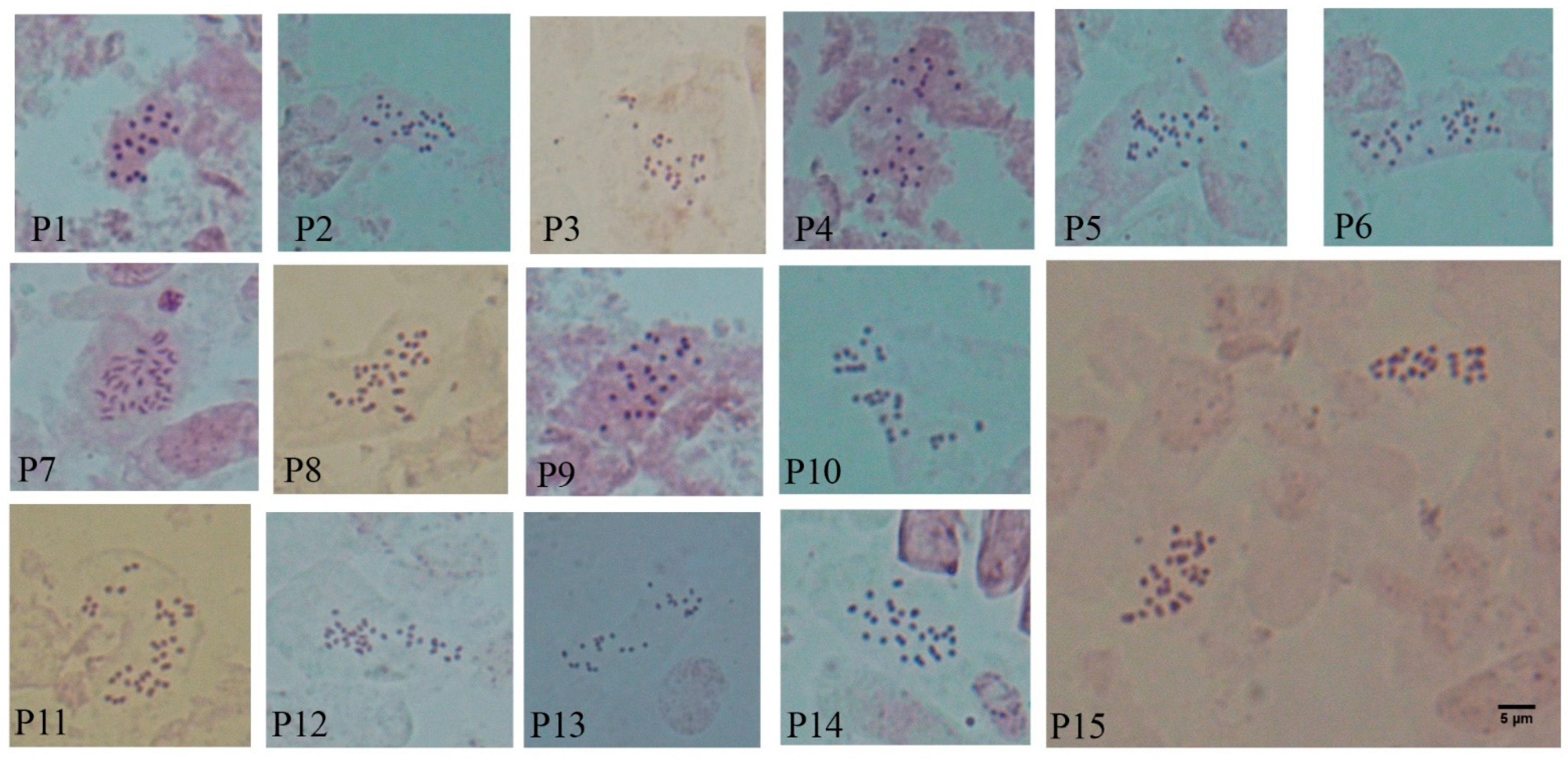
2.1. Chromosome Counts, Basic Numbers and Ploidy Levels
2.2. Karyotypes
2.3. Regression Analysis
2.4. Principal Coordinate and Cluster Analysis
2.5. Flow Cytometry
3. Discussion
3.1. Variation of Chromosome Numbers in H. perforatum
3.2. Dysploidy Caused the Considerable Chromosome Number Variation
3.3. Chromosome Structure
3.4. Genome Size Differentiation
4. Materials and Methods
4.1. Plant Materials
4.2. Karyologic Analysis
4.3. Flow Cytometric (FCM) Analyses
4.4. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pellestor, F.; Gatinois, V. Chromoanagenesis: A piece of the macroevolution scenario. Mol. Cytogenet. 2020, 13, 3. [Google Scholar] [PubMed]
- Mehravi, S.; Ranjbar, G.A.; Najafi-Zarrini, H.; Mirzaghaderi, G.; Hanifei, M.; Severn-Ellis, A.A.; Edwards, D.; Batley, J. Karyology and genome size analyses of Iranian endemic Pimpinella (Apiaceae) species. Front. Plant Sci. 2022, 13, 898881. [Google Scholar]
- Winterfeld, G.; Ley, A.; Hoffmann, M.H.; Paule, J.; Röser, M. Dysploidy and polyploidy trigger strong variation of chromosome numbers in the prayer-plant family (Marantaceae). Plant Syst. Evol. 2020, 306, 36. [Google Scholar]
- Peruzzi, L.; Góralski, G.; Joachimiak, A.J.; Bedini, G. Does actually mean chromosome number increase with latitude in vascular plants? An answer from the comparison of Italian, Slovak and Polish floras. Comp. Cytogenet. 2012, 6, 371–377. [Google Scholar]
- Carta, A.; Bedini, G.; Peruzzi, L. Unscrambling phylogenetic effects and ecological determinants of chromosome number in major angiosperm clades. Sci. Rep. 2018, 8, 14258. [Google Scholar]
- Claude, S.J.; Park, S.; Park, S. Gene loss, genome rearrangement, and accelerated substitution rates in plastid genome of Hypericum ascyron (Hypericaceae). BMC Plant Biol. 2022, 22, 135. [Google Scholar]
- Stevens, P.F. Hpericaceae Jussieu, (‘Hyperica’). In The Families and Genera of Vascular Plants; Kubitzki, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 9, pp. 194–201. [Google Scholar]
- Nürk, N.M.; Crockett, S.L. Morphological and phytochemical diversity among Hypericum species of the Mediterranean Basin. Med. Aromat. Plant Sci. Biotechnol. 2011, 5, 14. [Google Scholar]
- Hammer, K. Guttiferae (Clusiaceae). In Mansfeld’s Encyclopedia of Agricultural and Horticultural Crops, 3rd ed.; Hanelt, P., Institut für Pflanzengenetik und Kulturpflanzenforschung, Eds.; Spinger: Berlin/Heidelberg, Germany; New York, NY, USA, 2001; pp. 1345–1360. [Google Scholar]
- Koch, M.A.; Scheriau, C.; Betzin, A.; Hohmann, N.; Sharbel, T.F. Evolution of cryptic gene pools in Hypericum perforatum: The influence of reproductive system and gene flow. Ann. Bot. 2013, 111, 1083–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robson, N.K.B. Studies in the genus Hypericum L. (Guttiferae) 4 (2). Section 9. Hypericum sensu lato (part 2): Subsection 1. Hypericum series 1. Hypericum. Bull. Nat. Hist. Mus. Bot. Ser. 2002, 32, 61–123. [Google Scholar]
- Robson, N. Hypericum botany. In Hypericum: The Genus Hypericum; Ernst, E., Ed.; Taylor and Francis: London, UK, 2003; pp. 1–22. [Google Scholar]
- Gagnieu, A.; Wilhelm, J. Genre Hypericum. In Les Chromosomes Dans La Cellule, La Plante, L’esp`Ece; Gagnieu, A., Ed.; Travaux biologiques d’edit `a Prof. Plantefol: Paris, France, 1965; pp. 472–473. [Google Scholar]
- Bell, C.R. Documented plant chromosome numbers. SIDA 1965, 2, 168–170. [Google Scholar]
- Kogi, M. A karyomorphological study of the genus Hypericum (Hypericaceae) in Japan. Bot. Mag. 1984, 97, 333–343. [Google Scholar]
- Nielsen, N. Chromosome numbers in the genus Hypericum. Hereditas 1924, 5, 378–382. [Google Scholar]
- Robson, N.K.B.; Adams, P. Chromosome Numbers in Hypericum and Related Genera. Brittonia 1968, 20, 95–106. [Google Scholar]
- Matzk, F.; Meister, A.; Brutovská, R.; Schubert, I. Reconstruction of reproductive diversity in Hypericum perforatum L. opens novel strategies to manage apomixis. Plant J. 2001, 26, 275–282. [Google Scholar]
- Qu, L.; Widrlechner, M.P.; Rigby, S.M. Analysis of breeding systems, ploidy, and the role of hexaploids in three Hypericum perforatum L. populations. Ind. Crop. Prod. 2010, 32, 1–6. [Google Scholar]
- Bruňáková, K.; Čellárová, E. Conservation Strategies in the Genus Hypericum via Cryogenic Treatment. Front. Plant Sci. 2016, 7, 558. [Google Scholar]
- Brutovská, R.; Čellárová, E.; Schubert, I. Cytogenetic characterization of three Hypericum species by in situ hybridization. Theor. Appl. Genet. 2000, 101, 46–50. [Google Scholar]
- Campbell, M.; Delfosse, E. Biology of Australian weeds. 13. Hypericum perforatum L. J. Aust. Institut. Agric. Sci. 1984, 50, 63–73. [Google Scholar]
- Brutovská, R.; Kušniriková, P.; Bogyiová, E.; Čellárová, E. Karyotype analysis of Hypericum perforatum L. Biol. Plant. 2000, 43, 133–136. [Google Scholar]
- Molins, M.P.; Corral, J.M.; Aliyu, O.M.; Koch, M.A.; Betzin, A.; Maron, J.L.; Sharbel, T.F. Biogeographic variation in genetic variability, apomixis expression and ploidy of St. John’s wort (Hypericum perforatum) across its native and introduced range. Ann. Bot. 2014, 113, 417–427. [Google Scholar] [CrossRef] [Green Version]
- Brutovská, R.; Čellárová, E.; Doležel, J. Cytogenetic variability of in vitro regenerated Hypericum perforatum L. plants and their seed progenies. Plant Sci. 1998, 133, 221–229. [Google Scholar]
- Matzk, F.; Hammer, K.; Schubert, I. Coevolution of apomixis and genome size within the genus Hypericum. Sex. Plant Reprod. 2003, 16, 51–58. [Google Scholar]
- Mayo, G.M.; Langridge, P. Modes of reproduction in Australian populations of Hypericum perforatum L. (St. John’s wort) revealed by DNA fingerprinting and cytological methods. Genome 2003, 46, 573–579. [Google Scholar] [PubMed]
- Moraes, J.C.R. Caracterização Citogenética e da Biologia Reprodutiva de Três Espécies do Gênero Hypericum L. (Clusiaceae). Master’s Thesis, Instituto Agronômico, Campinas, Brazil, 2007. [Google Scholar]
- Greilhuber, J.; Doležel, J.; Lysák, M.A.; Bennett, M.D. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef]
- Knight, C.A.; Molinari, N.A.; Petrov, D.A. The large genome constraint hypothesis: Evolution, ecology and phenotype. Ann. Bot. 2005, 95, 177–190. [Google Scholar]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar]
- Nowicka, A.; Sliwinska, E.; Grzebelus, D.; Baranski, R.; Simon, P.W.; Nothnagel, T.; Grzebelus, E. Nuclear DNA content variation within the genus Daucus (Apiaceae) determined by flow cytometry. Sci. Hortic. 2016, 209, 132–138. [Google Scholar]
- Esfahani, S.T.; Karimzadeh, G.; Naghavi, M.R. 2C DNA value of Persian poppy (Papaver bracteatum Lindl.) medicinal plant as revealed by flow cytometry analysis; a quick effective criteria for distinguishing unidentified papaver species. Int. J. Adv. Biotechnol. Res. 2016, 7, 573–578. [Google Scholar]
- Bennett, M.D.; Price, H.J.; Johnston, J.S. Anthocyanin inhibits propidium iodide DNA fluorescence in Euphorbia pulcherrima: Implications for genome size variation and flow cytometry. Ann. Bot. 2008, 101, 777–790. [Google Scholar]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold Press: London, UK, 1971; p. 216. [Google Scholar]
- Armstrong, K.C. The evolution of Bromus inermis and related species of Bromus sect. Pnigma. Bot. Jahrb. Syst. 1981, 102, 427–443. [Google Scholar]
- Joachimiak, A.; Kula, A.; Sliwinska, E.; Sobieszczanska, A. C-banding and nuclear DNA amount in six Bromus species. Acta Biol. Cracov. Ser. Bot. 2001, 43, 105–115. [Google Scholar]
- Brochmann, C.; Brysting, A.K.; Alsos, I.G.; Borgen, L.; Grundt, H.H.; Scheen, A.C.; Elven, R. Polyploidy in arctic plants. Biol. J. Linn. Soc. 2004, 82, 521–536. [Google Scholar]
- Escudero, M.; Martín-Bravo, S.; Mayrose, I.; Fernández-Mazuecos, M.; Fiz-Palacios, O.; Hipp, A.L.; Pimentel, M.; Jiménez-Mejías, P.; Valcárcel, V.; Vargas, P.; et al. Karyotypic changes through dysploidy persist longer over evolutionary time than polyploid changes. PLoS ONE 2014, 9, e85266. [Google Scholar]
- Winterfeld, G.; Schneider, J.; Becher, H.; Dickie, J.; Roser, M. Karyosystematics of the Australasian stipoid grass Austrostipa and related genera: Chromosome sizes, ploidy, chromosome base numbers and phylogeny. Aust. Syst. Bot. 2015, 28, 145–159. [Google Scholar]
- Li, J.; Zhu, K.; Wang, Q.; Chen, X. Genome size variation and karyotype diversity in eight taxa of Sorbus sensu stricto (Rosaceae) from China. Comp. Cytogenet. 2021, 15, 137–148. [Google Scholar] [CrossRef]
- Mishiba, K.; Mii, M. Polysomaty analysis in diploid and tetraploid Portulaca grandiflora. Plant Sci. 2000, 156, 213–219. [Google Scholar]
- Ferraz, M.E.; Fonsêca, A.; Pedrosa-Harand, A. Multiple and independent rearrangements revealed by comparative cytogenetic mapping in the dysploid Leptostachyus group (Phaseolus L., Leguminosae). Chromosome Res. 2020, 28, 395–405. [Google Scholar]
- Baptista-Giacomelli, F.R.; Pagliarini, M.S.; de Almeida, J.L. Meiotic behavior in several Brazilian oat cultivars (Avena sativa L.). Cytologia 2000, 65, 371–378. [Google Scholar]
- Orr, B.; Godek, K.M.; Compton, D. Aneuploidy. Curr. Biol. 2015, 25, R538–R542. [Google Scholar] [PubMed] [Green Version]
- Baltisberger, M.; Hörandl, E. Karyotype evolution supports the molecular phylogeny in the genus Ranunculus (Ranunculaceae). Perspect. Plant Ecol. Evol. Syst. 2016, 18, 1–14. [Google Scholar]
- Ernst, E. Hypericum: The Genus Hypericum; CRC Press: Boca Raton, FL, USA, 2003; p. 284. [Google Scholar]
- Galla, G.; Barcaccia, G.; Schallau, A.; Puente Molins, M.; Bäumlein, H.; Sharbel, T.F. The cytohistological basis of apospory in Hypericum perforatum L. Sex. Plant Reprod. 2011, 24, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.N.; Merg, K.F. Evolution of polyploidy and the diversification of plant–pollinator interactions. Ecology 2008, 89, 2197–2206. [Google Scholar] [PubMed]
- Ramsey, J. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. USA 2011, 108, 7096–7101. [Google Scholar]
- Baack, E.; Melo, M.C.; Rieseberg, L.H.; Ortiz-Barrientos, D. The origins of reproductive isolation in plants. New Phytol. 2015, 207, 968–984. [Google Scholar] [PubMed]
- Porturas, L.D.; Segraves, K.A. Whole genome duplication does not promote common modes of reproductive isolation in Trifolium pratense. Am. J. Bot. 2020, 107, 833–841. [Google Scholar]
- Weiss-Schneeweiss, H.; Schneeweiss, G.M. Karyotype diversity and evolutionary trends in angiosperms. In Plant Genome Diversity; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: New York, NY, USA, 2013; pp. 209–230. [Google Scholar]
- Carta, A.; Bedini, G.; Peruzzi, L. A deep dive into the ancestral chromosome number and genome size of flowering plants. New Phytol. 2020, 228, 1097–1106. [Google Scholar]
- Soltis, D.E.; Tago-Nakazawa, M.; Xiang, Q.Y.; Kawano, S.; Murata, J.; Wakabayashi, M.; Hibsch-Jetter, C. Phylogenetic relationships and evolution in Chrysosplenium (Saxifragaceae) based on matK sequence data. Am. J. Bot. 2001, 88, 883–893. [Google Scholar]
- Church, S.A. Molecular phylogenetics of Houstonia (Rubiaceae): Descending aneuploidy and breeding system evolution in the radiation of the lineage across North America. Mol. Phylogenet. Evol. 2003, 27, 223–238. [Google Scholar] [PubMed]
- Fonsêca, A.; Ferraz, M.E.; Pedrosa-Harand, A. Speeding up chromosome evolution in Phaseolus: Multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 2016, 125, 413–421. [Google Scholar] [PubMed]
- Chen, G.F.; Sun, W.G.; Hong, D.Y.; Zhou, Z.; Niu, Y.; Nie, Z.L.; Sun, H.; Zhang, J.W.; Li, Z.M. Systematic significance of cytology in Cyananthus (Campanulaceae) endemic to the Sino-Himalayan region. J. Syst. Evol. 2014, 52, 260–270. [Google Scholar]
- Mandáková, T.; Pouch, M.; Harmanová, K.; Zhan, S.H.; Mayrose, I.; Lysak, M.A. Multispeed genome diploidization and diversification after an ancient allopolyploidization. Mol. Ecol. 2017, 26, 6445–6462. [Google Scholar]
- Winterfeld, G.; Becher, H.; Voshell, S.; Hilu, K.; Röser, M. Karyotype evolution in Phalaris (Poaceae): The role of reductional dysploidy, polyploidy and chromosome alteration in a wide-spread and diverse genus. PLoS ONE 2018, 13, e0192869. [Google Scholar]
- Aparicio, A.; Escudero, M.; Valdés-Florido, A.; Pachón, M.; Rubio, E.; Albaladejo, R.G.; Martín-Hernanz, S.; Pradillo, M. Karyotype evolution in Helianthemum (Cistaceae): Dysploidy, achiasmate meiosis and ecological specialization in H. squamatum, a true gypsophile. Bot. J. Linn. Soc. 2019, 191, 484–501. [Google Scholar] [CrossRef]
- Koprivý, L.; Fráková, V.; Kolarčik, V.; Mártonfiová, L.; Dudáš, M.; Mártonfi, P. Genome size and endoreplication in two pairs of cytogenetically contrasting species of Pulmonaria (Boraginaceae) in Central Europe. AoB Plants 2022, 14, plac036. [Google Scholar]
- Liu, L.; Astuti, G.; Coppi, A.; Peruzzi, L. Different chromosome numbers but slight morphological differentiation and genetic admixture among populations of the Pulmonaria hirta complex (Boraginaceae). Taxon 2022, 71, 1025–1043. [Google Scholar]
- Maletskii, S.I. Hierarchy of units of hereditary: Variability and inheritance of traits in species formation in plants. In Epigenetika Rasteniy: Sb. Nauch. Tr. In-Ta Tsitologii i Genetiki SO RAN; The Epigenetics of Plants; Institute of Cytology and Genetics, Siberian Division of the Russian Academy of Sciences: Novosibirsk, Russia, 2005; pp. 7–53. [Google Scholar]
- Kunakh, V.A.; Adonin, V.I.; Ozheredov, S.P.; Blyum, Y.B. Mixoploidy in wild and cultivated species of Cruciferae capable of hybridizing with rapeseed Brassica napus. Cytol. Genet. 2008, 42, 204–209. [Google Scholar]
- Mártonfiová, L.E.N.K.A.; Danova, K.A.L.I.N.A.; Toteva, V.K.; Čellárová, E. Karyotype analysis of Hypericum rumeliacum Boiss. Thaiszia. J. Bot. 2014, 24, 143–150. [Google Scholar]
- Zuo, L.; Yuan, Q. The difference between the heterogeneity of the centromeric index and intrachromosomal asymmetry. Plant Syst. Evol. 2011, 297, 141–145. [Google Scholar]
- Levin, D.A. The Role of Chromosomal Change in Plant Evolution; Oxford University Press: New York, NY, USA, 2002; p. 240. [Google Scholar]
- Peruzzi, L.; Leitch, I.J.; Caparelli, K.F. Chromosome diversity and evolution in Liliaceae. Ann. Bot. 2009, 103, 459–475. [Google Scholar] [PubMed]
- Mahdavi, S.; Karimzadeh, G. Karyological and nuclear DNA content variation in some Iranian endemic Thymus species (Lamiaceae). J. Agric. Sci. Technol. 2010, 12, 447–458. [Google Scholar]
- Peruzzi, L.; Altınordu, F. A proposal for a multivariate quantitative approach to infer karyological relationships among taxa. Comp. Cytogenet. 2014, 8, 337–349. [Google Scholar] [PubMed]
- Harpke, D.; Carta, A.; Tomović, G.; Ranđelović, V.; Ranđelović, N.; Blattner, F.R.; Peruzzi, L. Phylogeny, karyotype evolution and taxonomy of Crocus series Verni (Iridaceae). Plant Syst. Evol. 2015, 301, 309–325. [Google Scholar] [CrossRef]
- Siljak-Yakovlev, S.; Peruzzi, L. Cytogenetic characterization of endemics: Past and future. Plant Biosyst. 2012, 146, 694–702. [Google Scholar]
- Peruzzi, L.; Eroğlu, H.E. Karyotype asymmetry: Again, how to measure and what to measure? Comp. Cytogenet. 2013, 7, 1–9. [Google Scholar] [PubMed] [Green Version]
- Peruzzi, L. Chromosome diversity and evolution in the genus Gagea (Liliaceae). Bocconea 2012, 24, 147–158. [Google Scholar]
- Doležel, J.; Bartoš, J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar]
- Majdi, M.; Karimzadeh, G.; Malboobi, M.A.; Omidbaigi, R.; Mirzaghaderi, G. Induction of tetraploidy to feverfew (Tanacetum parthenium Schulz-Bip.): Morphological, physiological, cytological, and phytochemical changes. HortScience 2010, 45, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Karimzadeh, G.; Mousavi, S.H.; Jafarkhani-Kermani, M.; Jalali-Javaran, M. Karyological and nuclear DNA variation in Iranian endemic muskmelon (Cucumis melo var. inodorus). Cytologia 2010, 75, 451–461. [Google Scholar]
- Karimzadeh, G.; Danesh-Gilevaei, M.; Aghaalikhani, M. Karyotypic and nuclear DNA variations in Lathyrus sativus (Fabaceae). Caryologia 2011, 64, 42–54. [Google Scholar]
- Tavan, M.; Mirjalili, M.H.; Karimzadeh, G. In vitro polyploidy induction: Changes in morphological, anatomical and phytochemical characteristics of Thymus persicus (Lamiaceae). Plant Cell Tissue Organ. Cult. 2015, 122, 573–583. [Google Scholar]
- Javadian, N.; Karimzadeh, G.; Sharifi, M.; Moieni, A.; Behmanesh, M. In vitro polyploidy induction: Changes in morphology, podophyllotoxin biosynthesis, and expression of the related genes in Linum album (Linaceae). Planta 2017, 245, 1165–1178. [Google Scholar] [CrossRef]
- Hamidi, F.; Karimzadeh, G.; Monfared, S.R.; Salehi, M. Assessment of Iranian endemic Artemisia khorassanica: Karyological, genome size, and gene expressions involved in artemisinin production. Turk. J. Biol. 2018, 42, 329–340. [Google Scholar]
- Esfahani, S.T.; Karimzadeh, G.; Naghavi, M.R. In vitro polyploidy induction in Persian Poppy (Papaver bracteatum Lindl.). Caryologia 2020, 73, 133–144. [Google Scholar]
- Leitch, I.; Bennett, M. Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 2004, 82, 651–663. [Google Scholar]
- Ozkan, H.; Tuna, M.; Arumuganathan, K. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. Plant Syst. Evol. 2003, 94, 260–264. [Google Scholar]
- Naranjo, C.A.; Ferrari, M.R.; Palermo, A.M.; Poggio, L. Karyotype, DNA content and meiotic behaviour in five South American species of Vicia (Fabaceae). Ann. Bot. 1998, 82, 757–764. [Google Scholar]
- Abedi, R.; Babaei, A.; Karimzadeh, G. Karyological and flow cytometric studies of Tulipa (Liliaceae) species from Iran. Plant Syst. Evol. 2015, 301, 1473–1484. [Google Scholar]
- Paszko, B. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst. Evol. 2006, 258, 39–48. [Google Scholar] [CrossRef]
- Doležel, J.; Bartos, J.; Voglmayr, H.; Greilhuber, J. Nuclear DNA content and genome size of trout and human. Cytom. J. Int. Soc. Anal. Cytol. 2003, 51, 127–128. [Google Scholar]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Gower, J.C. A general coefficient of similarity and some of its properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Sneath, P.H.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; CABI: San Francisco, CA, USA, 1973; p. 588. [Google Scholar]







| Population | pl | 2n | x | S (µm) | L (µm) | CL (µm) | AR | r-Value | RL% | F% | TCV (µm3) | CI% | MCA | CVCL | CVCI | KF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 2x | 16 | 8 | 0.37 | 0.47 | 0.84 | 1.27 | 0.80 | 12.50 | 5.53 | 0.25 | 44.22 | 0.74 | 26.63 | 6.79 | 16m |
| P2 | 3x | 24 | 8 | 0.39 | 0.49 | 0.88 | 1.29 | 0.79 | 8.33 | 3.67 | 0.31 | 44.03 | 0.74 | 16.73 | 8.08 | 24m |
| P3 | 3x | 24 | 8 | 0.37 | 0.48 | 0.85 | 1.29 | 0.78 | 8.33 | 3.64 | 0.30 | 43.75 | 0.72 | 19.68 | 6.31 | 24m |
| P4 | 4x | 32 | 8 | 0.45 | 0.58 | 1.03 | 1.31 | 0.77 | 7.81 | 3.39 | 0.53 | 43.39 | 0.69 | 19.18 | 7.80 | 32m |
| P5 | 4x | 32 | 8 | 0.39 | 0.54 | 0.93 | 1.39 | 0.73 | 7.83 | 3.30 | 0.27 | 42.11 | 0.69 | 18.21 | 7.56 | 32m |
| P6 | 4x | 32 | 8 | 0.35 | 0.45 | 0.81 | 1.28 | 0.79 | 7.78 | 3.43 | 0.34 | 44.05 | 0.74 | 17.58 | 6.74 | 32m |
| P7 | 4x | 32 | 8 | 0.50 | 0.66 | 1.16 | 1.30 | 0.78 | 7.77 | 3.38 | 0.53 | 43.67 | 0.65 | 20.91 | 7.49 | 32m |
| P8 | 4x | 32 | 8 | 0.39 | 0.49 | 0.88 | 1.25 | 0.81 | 7.79 | 3.48 | 0.39 | 44.65 | 0.66 | 21.17 | 6.05 | 32m |
| P9 | 2x | 22 | 11 | 0.40 | 0.48 | 0.88 | 1.22 | 0.83 | 9.09 | 4.11 | 0.55 | 45.19 | 0.78 | 15.54 | 7.79 | 22m |
| P10 | 2x | 30 | 15 | 0.38 | 0.48 | 0.86 | 1.30 | 0.79 | 7.92 | 3.47 | 0.38 | 43.85 | 0.70 | 18.36 | 7.74 | 30m |
| P11 | 2x | 30 | 15 | 0.49 | 0.61 | 1.10 | 1.25 | 0.81 | 7.89 | 3.51 | 0.45 | 44.64 | 0.69 | 20.40 | 5.51 | 30m |
| P12 | 2x | 38 | 19 | 0.43 | 0.55 | 0.98 | 1.32 | 0.77 | 7.58 | 3.29 | 0.39 | 43.40 | 0.70 | 20.88 | 6.96 | 38m |
| P13 | 2x | 26 | 13 | 0.38 | 0.48 | 0.86 | 1.26 | 0.80 | 8.14 | 3.61 | 0.35 | 44.39 | 0.71 | 19.04 | 6.40 | 26m |
| P14 | 2x | 28 | 14 | 0.40 | 0.50 | 0.90 | 1.26 | 0.81 | 8.02 | 3.56 | 0.34 | 44.51 | 0.70 | 18.71 | 7.27 | 28m |
| P15 | 3x | 24 | 8 | 0.39 | 0.49 | 0.88 | 1.29 | 0.79 | 8.33 | 3.67 | 0.30 | 44.03 | 0.72 | 16.73 | 8.08 | 24m |
| 4x | 32 | 8 | 0.40 | 0.49 | 0.89 | 1.23 | 0.81 | 7.60 | 3.42 | 0.40 | 44.83 | 0.70 | 26.61 | 3.39 | 32m |
| Population | Ploidy Level | 2n | 2C-Value (pg ± SE) | 1C-Value (pg) | 1Cx-Value (pg) |
|---|---|---|---|---|---|
| P1 | 2x | 16 | 0.87 ± 0.021 | 0.435 | 0.435 |
| P2 | 3x | 24 | 1.40 ± 0.018 | 0.700 | 0.470 |
| P3 | 3x | 24 | 1.51 ± 0.018 | 0.755 | 0.503 |
| P4 | 4x | 32 | 1.69 ± 0.021 | 0.845 | 0.423 |
| P5 | 4x | 32 | 1.86 ± 0.020 | 0.930 | 0.465 |
| P6 | 4x | 32 | 1.61 ± 0.022 | 0.805 | 0.403 |
| P7 | 4x | 32 | 1.62 ± 0.022 | 0.810 | 0.405 |
| P8 | 4x | 32 | 1.76 ± 0.022 | 0.880 | 0.440 |
| P9 | 2x | 22 | 1.20 ± 0.020 | 0.600 | 0.600 |
| P10 | 2x | 30 | 1.15 ± 0.021 | 0.575 | 0.575 |
| P11 | 2x | 30 | 1.80 ± 0.032 | 0.900 | 0.900 |
| P12 | 2x | 38 | 2.02 ± 0.022 | 1.010 | 1.010 |
| P13 | 2x | 26 | 1.13 ± 0.025 | 0.565 | 0.565 |
| P14 | 2x | 28 | 1.42 ± 0.021 | 0.710 | 0.710 |
| P15 | 3x | 24 | 1.30 ± 0.030 | 0.650 | 0.433 |
| 4x | 32 | 1.60 ± 0.030 | 0.800 | 0.400 |
| Population Codes | Local Collection Sites | Latitude (N) | Longitude (E) | Altitude (m) | Mean Temp. (°C) | Mean Rainfall (mm) |
|---|---|---|---|---|---|---|
| P1 | Sari, Mazandaran, Iran | 36°33′ | 53°03′ | 43 | 15 | 2122 |
| P2 | Rasht, Gilan, Iran | 37°16′ | 49°34′ | −3 | 15.9 | 1359 |
| P3 | Aq Qala, Golestan, Iran | 37°00′ | 54°27′ | −14 | 17 | 675 |
| P4 | Astara, Gilan, Iran | 38°25′ | 48°51′ | −26 | 16 | 1500 |
| P5 | Lakan, Gilan, Iran | 37°12′ | 49°35′ | 19.5 | 15.9 | 1359 |
| P6 | Chalus, Mazandaran, Iran | 36°39′ | 51°25′ | 24 | 30 | 700 |
| P7 | Baneh, Kordestan, Iran | 36°14′ | 46°16′ | 1496 | 11 | 386 |
| P8 | Behshahr, Mazandaran, Iran | 36°41′ | 53°32′ | 23 | 20 | 977 |
| P9 | Javaherdeh, Mazandaran, Iran | 36°51′ | 50°28′ | 1772 | 19 | 1200 |
| P10 | Ramsar, Mazandaran, Iran | 36°54′ | 50°39′ | 36 | 21 | 1200 |
| P11 | Ziarat, Golestan, Iran | 36°42′ | 54°28′ | 918 | 17 | 649 |
| P12 | Ziarat, Golestan, Iran | 36°42′ | 54°28′ | 918 | 17 | 649 |
| P13 | Aq Qala, Golestan, Iran | 37°00′ | 54°27′ | −14 | 17 | 675 |
| P14 | Talesh, Gilan, Iran | 37°48′ | 48°54′ | 42 | 15 | 1446 |
| P15 | Ziarat, Golestan, Iran | 36°42′ | 54°28′ | 918 | 17 | 649 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehravi, S.; Karimzadeh, G.; Kordenaeej, A.; Hanifei, M. Mixed-Ploidy and Dysploidy in Hypericum perforatum: A Karyomorphological and Genome Size Study. Plants 2022, 11, 3068. https://doi.org/10.3390/plants11223068
Mehravi S, Karimzadeh G, Kordenaeej A, Hanifei M. Mixed-Ploidy and Dysploidy in Hypericum perforatum: A Karyomorphological and Genome Size Study. Plants. 2022; 11(22):3068. https://doi.org/10.3390/plants11223068
Chicago/Turabian StyleMehravi, Shaghayegh, Ghasem Karimzadeh, Alaeddin Kordenaeej, and Mehrdad Hanifei. 2022. "Mixed-Ploidy and Dysploidy in Hypericum perforatum: A Karyomorphological and Genome Size Study" Plants 11, no. 22: 3068. https://doi.org/10.3390/plants11223068
APA StyleMehravi, S., Karimzadeh, G., Kordenaeej, A., & Hanifei, M. (2022). Mixed-Ploidy and Dysploidy in Hypericum perforatum: A Karyomorphological and Genome Size Study. Plants, 11(22), 3068. https://doi.org/10.3390/plants11223068






