Functional Analysis of V2 Protein of Beet Curly Top Iran Virus
Abstract
1. Introduction
2. Results
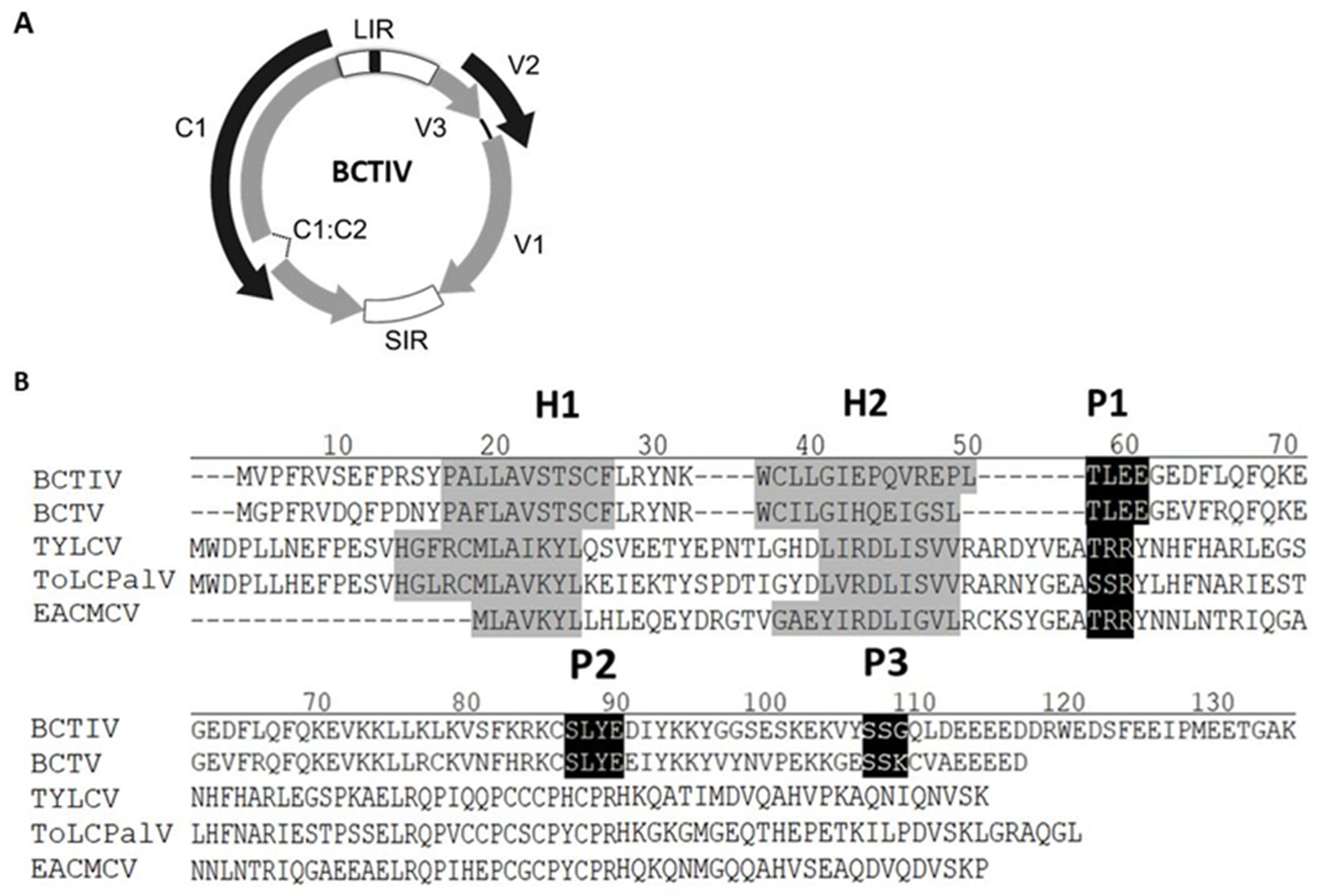
2.1. BCTIV V2 Aminoacidic Sequence Analysis
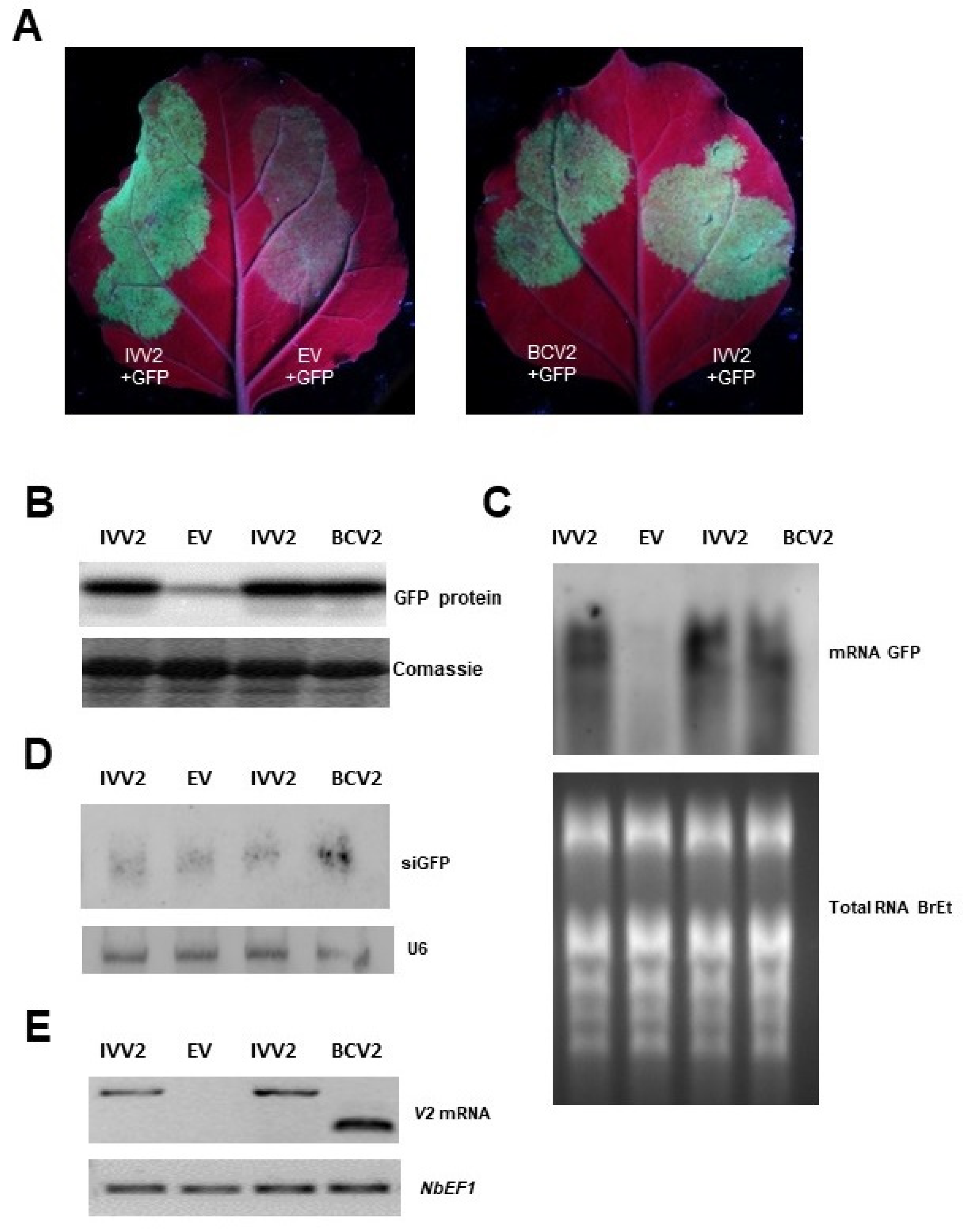
2.2. BCTIV V2 Is a Local PTGS Suppressor
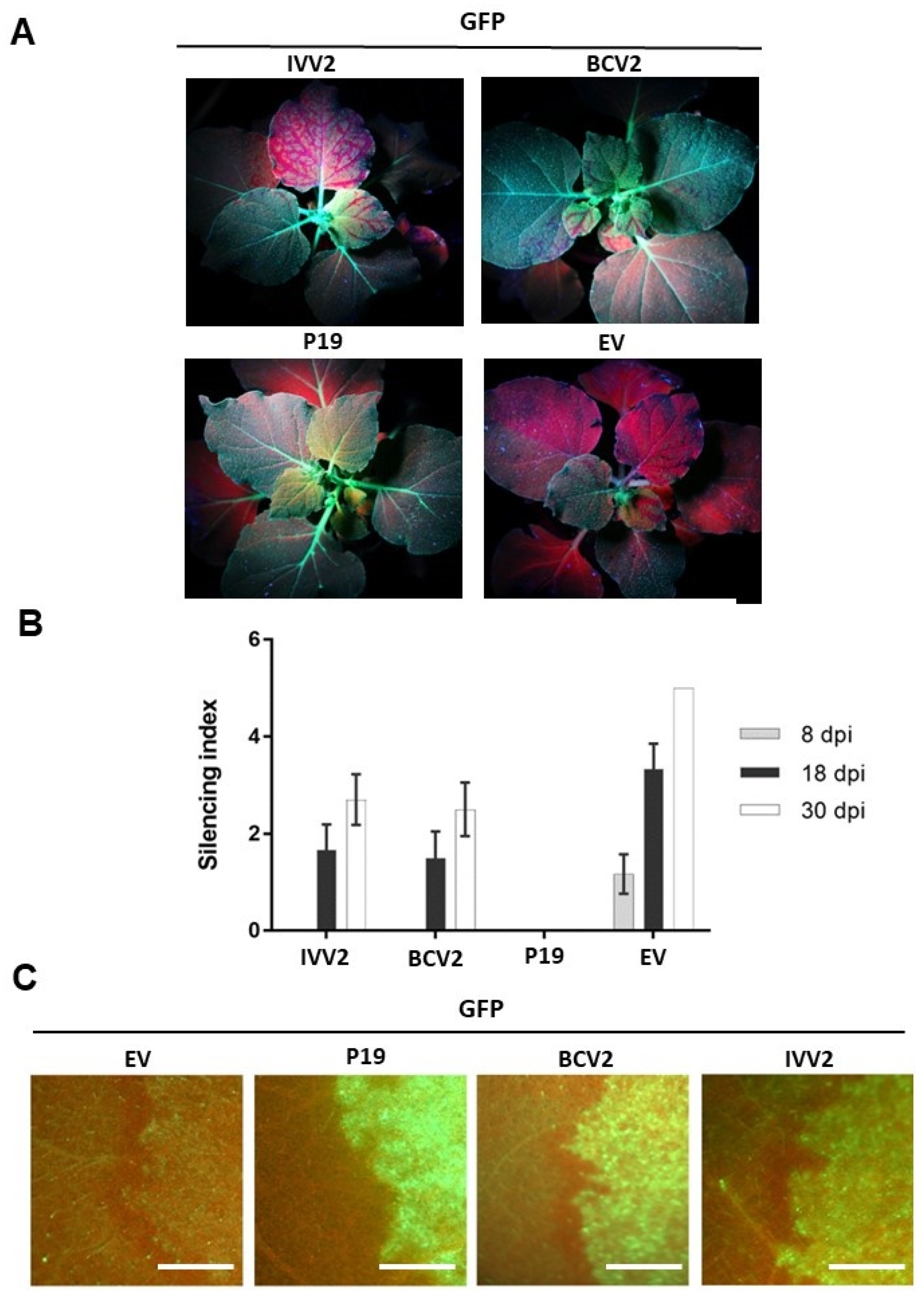
2.3. BCTIV V2 Causes a Delay in the Long-Distance Spread of RNA Silencing
2.4. BCTIV V2 Cannot Suppress Short-Range (Cell-to-Cell) Spread of Gene Silencing
2.5. BCTIV V2 Triggers HR-like Response When Expressed from a PVX Derived Vector
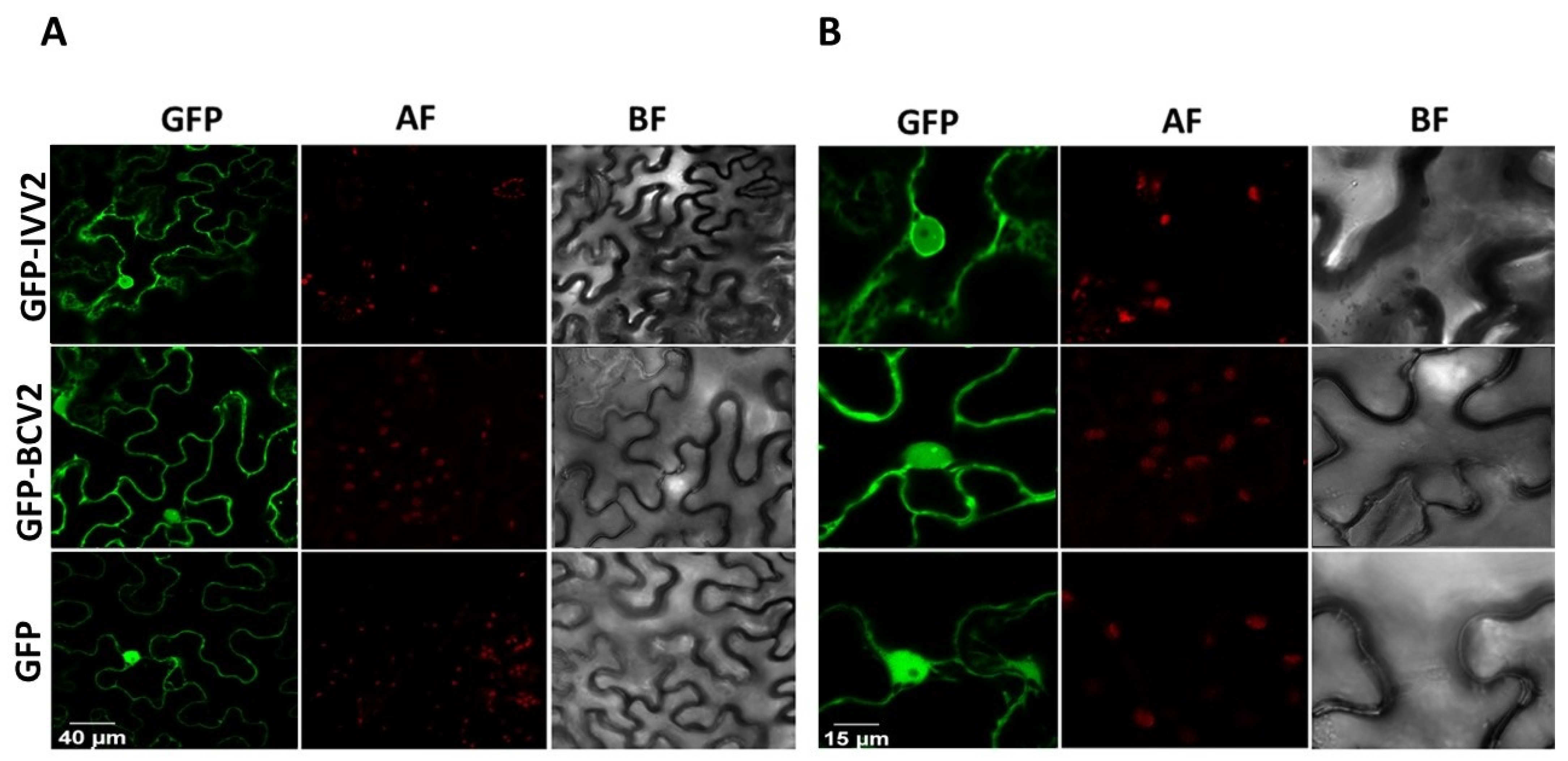
2.6. BCTIV V2 Localizes in the Nucleus and Perinuclear Region
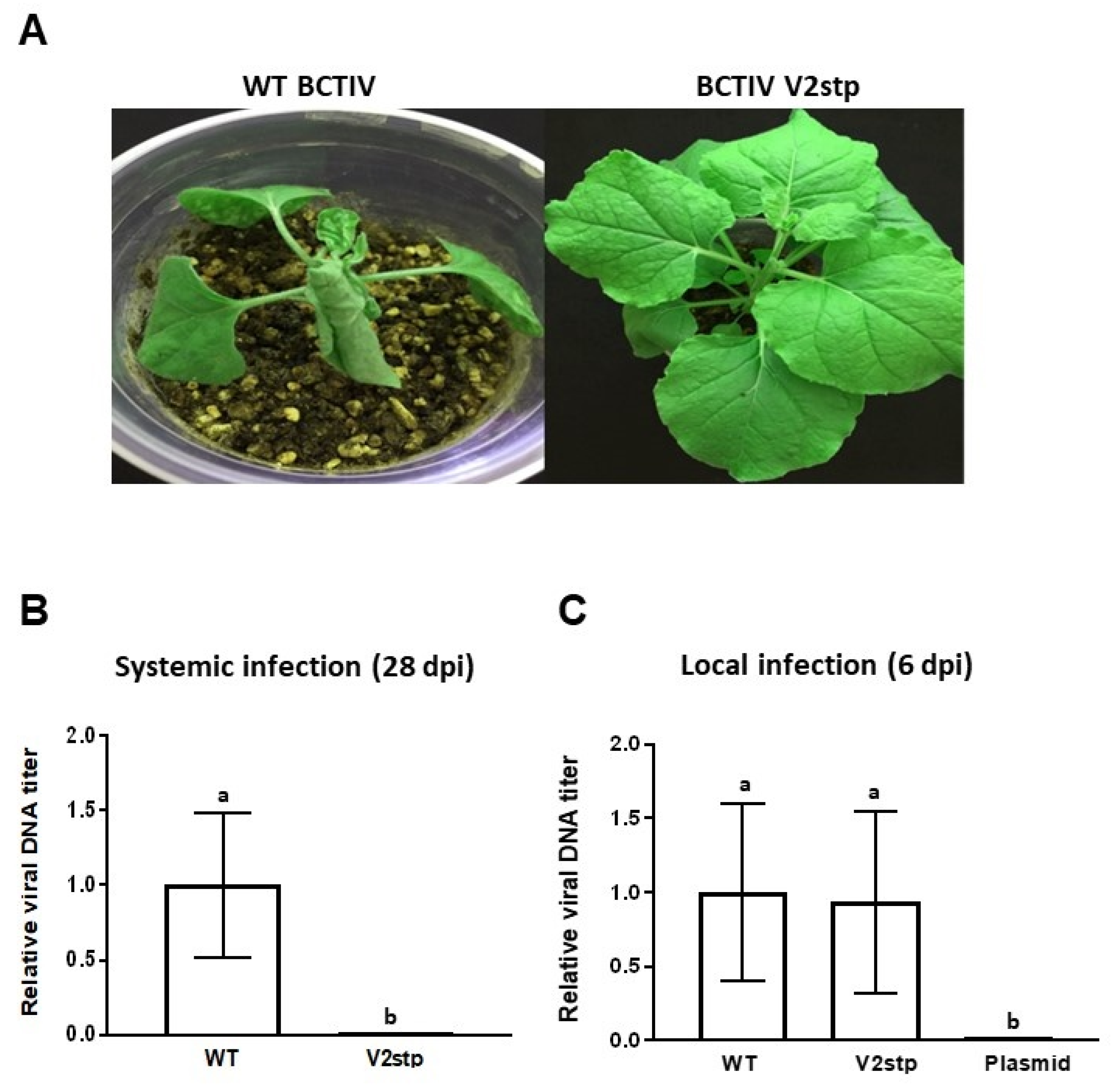
2.7. BCTIV V2 Is Essential for Systemic, but Not for Local Infection of BCTIV
3. Materials and Methods
3.1. Microorganisms, Plant Material, and Growth Conditions
3.2. Sequence Analysis
3.3. Plasmids and Cloning
3.4. Agroinfiltration and Infection Assays
3.5. Subcellular Localization
3.6. Analysis of Nucleic Acids and Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L. Burgyán, viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [PubMed]
- García-Arenal, F.; Zerbini, F.M. Life the Edge: Geminiviruses at the interface between crops and wild plant hosts. Annu. Rev. Virol 2019, 6, 411–433. [Google Scholar] [CrossRef]
- Roumagnac, P.; Lett, J.-M.; Fiallo-Olivé, E.; Navas-Castillo, J.; Zerbini, F.M.; Martin, D.P.; Varsani, A. Establishment of five new genera in the family Geminiviridae: Citlodavirus, Maldovirus, Mulcrilevirus, Opunvirus, and Topilevirus. Arch. Virol. 2022, 2022. 167, 695–710. [Google Scholar] [CrossRef]
- Rojas, M.R.; Jiang, H.; Salati, R.; Xoconostle-Cázares, B.; Sudarshana, M.; Lucas, W.J.; Gilbertson, R.L. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 2001, 291, 110–125. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Pooggin, M.M. How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? Int. J. Mol. Sci. 2013, 14, 15233–15259. [Google Scholar] [CrossRef]
- Zrachya, A.; Glick, E.; Levy, Y.; Arazi, T.; Citovsky, V.; Gafni, Y. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology 2007, 358, 159–165. [Google Scholar] [CrossRef]
- Sharma, P.; Ikegami, M. Tomato leaf curl Java virus V2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology 2010, 396, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant Microbe Interact 2011, 24, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, J.; Xu, Y.; Wu, J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012, 163, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ren, Y.; Sun, S.; Wang, D.; Zhang, F.; Li, D.; Li, S.; Zhou, X. Identification of the potential virulence factors and RNA silencing suppressors of mulberry mosaic dwarf-associated geminivirus. Viruses 2018, 10, 472. [Google Scholar] [CrossRef]
- Mubin, M.; Amin, I.; Amrao, L.; Briddon, R.W.; Mansoor, S. The hypersensitive response induced by the V2 protein of a monopartite begomovirus is countered by the C2 protein. Mol. Plant Pathol. 2010, 11, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.P.; Rodríguez-Negrete, E.A.; Morilla, G.; Wang, L.; Lozano-Durán, R.; Castillo, A.G.; Bejarano, E.R. V2 from a curtovirus is a suppressor of post-transcriptional gene silencing. J. Gen. Virol. 2017, 98, 2607–2614. [Google Scholar] [CrossRef]
- Wang, B.; Li, F.; Huang, C.; Yang, X.; Qian, Y.; Xie, Y.; Zhou, X. V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J. Gen. Virol. 2014, 95, 225–230. [Google Scholar] [CrossRef]
- Li, M.; Li, C.; Jiang, K.; Li, K.; Zhang, J.; Sun, M.; Wu, G.; Qing, L. Characterization of pathogenicity-associated V2 protein of tobacco curly shoot virus. Int. J. Mol. Sci. 2021, 22, 923. [Google Scholar] [CrossRef]
- Zhai, Y.; Roy, A.; Peng, H.; Mullendore, D.L.; Kaur, G.; Mandal, B.; Mukherjee, S.K.; Pappu, H.R. Identification and functional analysis of four RNA silencing suppressors in begomovirus croton yellow vein mosaic virus. Front. Plant Sci. 2022, 12. [Google Scholar] [CrossRef]
- Padidam, M.; Beachy, R.N.; Fauquet, C.M. The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology 1996, 224, 390–404. [Google Scholar] [CrossRef]
- Rothenstein, D.; Krenz, B.; Selchow, O.; Jeske, H. Tissue and cell tropism of Indian cassava mosaic virus (ICMV) and its AV2 (precoat) gene product. Virology 2007, 359, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Moshe, A.; Belausov, E.; Niehl, A.; Heinlein, M.; Czosnek, H.; Gorovits, R. The Tomato yellow leaf curl virus V2 protein forms aggregates depending on the cytoskeleton integrity and binds viral genomic DNA. Sci. Rep. 2015, 5, 9967. [Google Scholar] [CrossRef] [PubMed]
- Bar-Ziv, A.; Levy, Y.; Hak, H.; Mett, A.; Belausov, E.; Citovsky, V.; Gafni, Y. The tomato yellow leaf curl virus (TYLCV) V2 protein interacts with the host papain-like cysteine protease CYP1. Plant Signal. Behav. 2012, 7, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Roshan, P.; Kulshreshtha, A.; Kumar, S.; Purohit, R.; Hallan, V. AV2 protein of tomato leaf curl Palampur virus promotes systemic necrosis in Nicotiana benthamiana and interacts with host Catalase2. Sci. Rep. 2018, 8, 1273. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.P.; Romero-Rodríguez, B.; Rosas-Díaz, T.; Cerero, L.; Rodríguez-Negrete, E.A.; Castillo, A.G.; Bejarano, E.R. Characterization of Curtovirus V2 protein, a functional homolog of begomovirus V2. Front. Plant Sci 2020, 11, 835. [Google Scholar] [CrossRef]
- Heydarnejad, J.; Hosseini Abhari, E.; Bolok Yazdi, H.; Massumi, H. Curly top of cultivated plants and weeds and report of a unique curtovirus from Iran. J. Phytopathol. 2007, 155, 321–325. [Google Scholar] [CrossRef]
- Heydarnejad, J.; Keyvani, N.; Razavinejad, S.; Massumi, H.; Varsani, A. Fulfilling Koch’s postulates for beet curly top Iran virus and proposal for consideration of new genus in the family Geminiviridae. Arch. Virol. 2013, 158, 435–443. [Google Scholar] [CrossRef]
- Yazdi, H.B.; Heydarnejad, J.; Massumi, H. Genome characterization and genetic diversity of beet curly top Iran virus: A geminivirus with a novel nonanucleotide. Virus Genes 2008, 36, 539–545. [Google Scholar] [CrossRef]
- Bozorgi, N.; Heydarnejad, J.; Kamali, M.; Massumi, H. Splicing features in the expression of the complementary-sense genes of Beet curly top Iran virus. Virus Genes 2017, 53, 323–327. [Google Scholar] [CrossRef]
- Chowda-Reddy, R.V.; Achenjang, F.; Felton, C.; Etarock, M.T.; Anangfac, M.T.; Nugent, P.; Fondong, V.N. Role of a geminivirus AV2 protein putative protein kinase C motif on subcellular localization and pathogenicity. Virus Res. 2008, 135, 115–124. [Google Scholar] [CrossRef]
- Dingwall, C.; Laskey, R.A. Nuclear targeting sequences a consensus? Trends Biochem. Sci. 1991, 16, 478–481. [Google Scholar] [CrossRef]
- Robbins, J.; Dilwortht, S.M.; Laskey, R.A.; Dingwall, C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 1991, 64, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, D.; Molnár, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyán, J. A viral protein suppresses RNA silencing and binds silencing-generated, 21-to 25-nucleotide double-stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef]
- Luna, A.P.; Morilla, G.; Voinnet, O.; Bejarano, E.R. Functional analysis of gene-silencing suppressors from tomato yellow leaf curl disease viruses. Mol. Plant-Mic. Interact. 2012, 25, 1294–1306. [Google Scholar] [CrossRef] [PubMed]
- Himber, C.; Dunoyer, P.; Moissiard, G.; Ritzenthaler, C.; Voinnet, O. Transitivity-dependent and-independent cell-to-cell movement of RNA silencing. EMBO J. 2003, 22, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Bucher, E.; Sijen, T.; De Haan, P.; Goldbach, R.; Prins, M. Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol. 2003, 77, 1329–1336. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Sugiyama, K.; Nagano, H.; Mori, M.; Kaido, M.; Mise, K.; Tsuda, S.; Okuno, T. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 2002, 532, 75–79. [Google Scholar] [CrossRef]
- Kotlizky, G.; Boulton, M.I.; Pitaksutheepong, C.; Davies, J.W.; Epel, B.L. Intracellular and intercellular movement of maize streak geminivirus V1 and V2 proteins transiently expressed as green fluorescent protein fusions. Virology 2000, 274, 32–38. [Google Scholar] [CrossRef]
- Zhan, B.; Zhao, W.; Li, S.; Yang, X.; Zhou, X. Functional scanning of apple geminivirus proteins as symptom determinants and suppressors of posttranscriptional gene silencing. Viruses 2018, 10, 488. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Gong, Q.; Ismayil, A.; Yuan, Y.; Lian, B.; Jia, Q.; Han, M.; Deng, H.; Hong, Y.; et al. Geminiviral V2 Protein Suppresses Transcriptional Gene Silencing through Interaction with AGO4. J. Virol. 2019, 93, e01675-18. [Google Scholar] [CrossRef]
- Guo, T.W.; Vimalesvaran, D.; Thompson, J.R.; Perry, K.L.; Krenz, B. Subcellular localization of grapevine red blotch-associated virus ORFs V2 and V3. Virus Genes 2015, 51, 156–158. [Google Scholar] [CrossRef]
- Ruiz, M.T.; Voinnet, O.; Baulcombe, D.C. Initiation and maintenance of virus-induced gene silencing. Plant Cell 1998, 10, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J. Molecular Cloning: A Laboratory Manual; Sambrook, J., Russell, D.W., Eds.; Cold Spring Harbor Laboratory: Long Island, NY, USA, 2001. [Google Scholar]
- Jones, L.; Hamilton, A.J.; Voinnet, O.; Thomas, C.L.; Maule, A.J.; Baulcombe, D.C. RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999, 11, 2291–2301. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Ishiguro, S.; Kimura, T. Gateway vectors for plant transformation. J. Plant Biotechnol. 2009, 26, 275–284. [Google Scholar] [CrossRef]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 2008. 8, 1–10. [Google Scholar] [CrossRef]
- Elmer, J.S.; Brand, L.; Sunter, G.; Gardiner, W.E.; Bisaro, D.M.; Rogers, S.G. Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 1988, 16, 7043–7060. [Google Scholar] [CrossRef]
- Rotenberg, D.; Thompson, T.S.; German, T.L.; Willis, D.K. Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods. 2006, 138, 49–59. [Google Scholar] [CrossRef]
- Tomassi, A.H.; Gagliardi, D.; Cambiagno, D.A.; Manavella, P.A. Nonradioactive detection of small RNAs using digoxigenin-labeled probes. Methods Mol. Biol. 2017, 1640, 199–210. [Google Scholar]
- López-Márquez, D.; Del-Espino, Á.; López-Pagán, N.; Rodríguez-Negrete, E.A.; Rubio-Somoza, I.; Ruiz-Albert, J.; Bejarano, E.R.; Beuzón, C.R. MiR825-5p targets the TIR-NBS-LRR gene MIST1 and down-regulates basal immunity against Pseudomonas syringae in Arabidopsis. J. Exp Bot. 2021, 72, 7316–7334. [Google Scholar] [CrossRef]
- Astaraki, S.; Safaie, N.; Shams-bakhsh, M. Reaction of sugar beet, pepper and bean plants to co-infection with Cucumber mosaic virus and beet curly top viruses. Iran. J. Plant Pathol. 2020, 56, 221–236. [Google Scholar] [CrossRef]
- Aguilar, E.; Almendral, D.; Allende, L.; Pacheco, R.; Chung, B.N.; Canto, T.; Tenllado, F. The P25 protein of Potato virus X (PVX) is the main pathogenicity determinant responsible for systemic necrosis in PVX-associated synergisms. J. Virol. 2015, 89, 2090–2103. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Lozano-Durán, R. The cajal body in Plant-Virus interactions. Viruses 2020, 12, 250. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Z.; Teng, K.; Lai, J.; Zhang, Y.; Huang, Y.; Li, Y.; Liang, L.; Wang, Y.; Chu, C. Up-regulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. Plant J. 2010, 62, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ding, Y.; He, L.; Zhang, G.; Zhu, J.-K.; Lozano-Duran, R. A virus-encoded protein suppresses methylation of the viral genome in the Cajal body through its interaction with AGO4. eLife 2020, 9, e55542. [Google Scholar] [CrossRef]
- Zhao, W.; Wu, S.; Barton, E.; Fan, Y.; Ji, Y.; Wang, X.; Zhou, Y. Tomato yellow leaf curl virus V2 protein plays a critical role in the nuclear export of V1 protein and viral systemic infection. Front. Microbiol. 2020, 11, 1243. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahari, A.; Castillo, A.G.; Safaie, N.; Bejarano, E.R.; Luna, A.P.; Shams-Bakhsh, M. Functional Analysis of V2 Protein of Beet Curly Top Iran Virus. Plants 2022, 11, 3351. https://doi.org/10.3390/plants11233351
Bahari A, Castillo AG, Safaie N, Bejarano ER, Luna AP, Shams-Bakhsh M. Functional Analysis of V2 Protein of Beet Curly Top Iran Virus. Plants. 2022; 11(23):3351. https://doi.org/10.3390/plants11233351
Chicago/Turabian StyleBahari, Atiyeh, Araceli G. Castillo, Naser Safaie, Eduardo R. Bejarano, Ana P. Luna, and Masoud Shams-Bakhsh. 2022. "Functional Analysis of V2 Protein of Beet Curly Top Iran Virus" Plants 11, no. 23: 3351. https://doi.org/10.3390/plants11233351
APA StyleBahari, A., Castillo, A. G., Safaie, N., Bejarano, E. R., Luna, A. P., & Shams-Bakhsh, M. (2022). Functional Analysis of V2 Protein of Beet Curly Top Iran Virus. Plants, 11(23), 3351. https://doi.org/10.3390/plants11233351







