Investigation and Expression Analysis of R2R3-MYBs and Anthocyanin Biosynthesis-Related Genes during Seed Color Development of Common Bean (Phaseolus vulgaris)
Abstract
1. Introduction
2. Results
2.1. Identification, Classification, and Constructional Analysis of MYB Gene Family Members
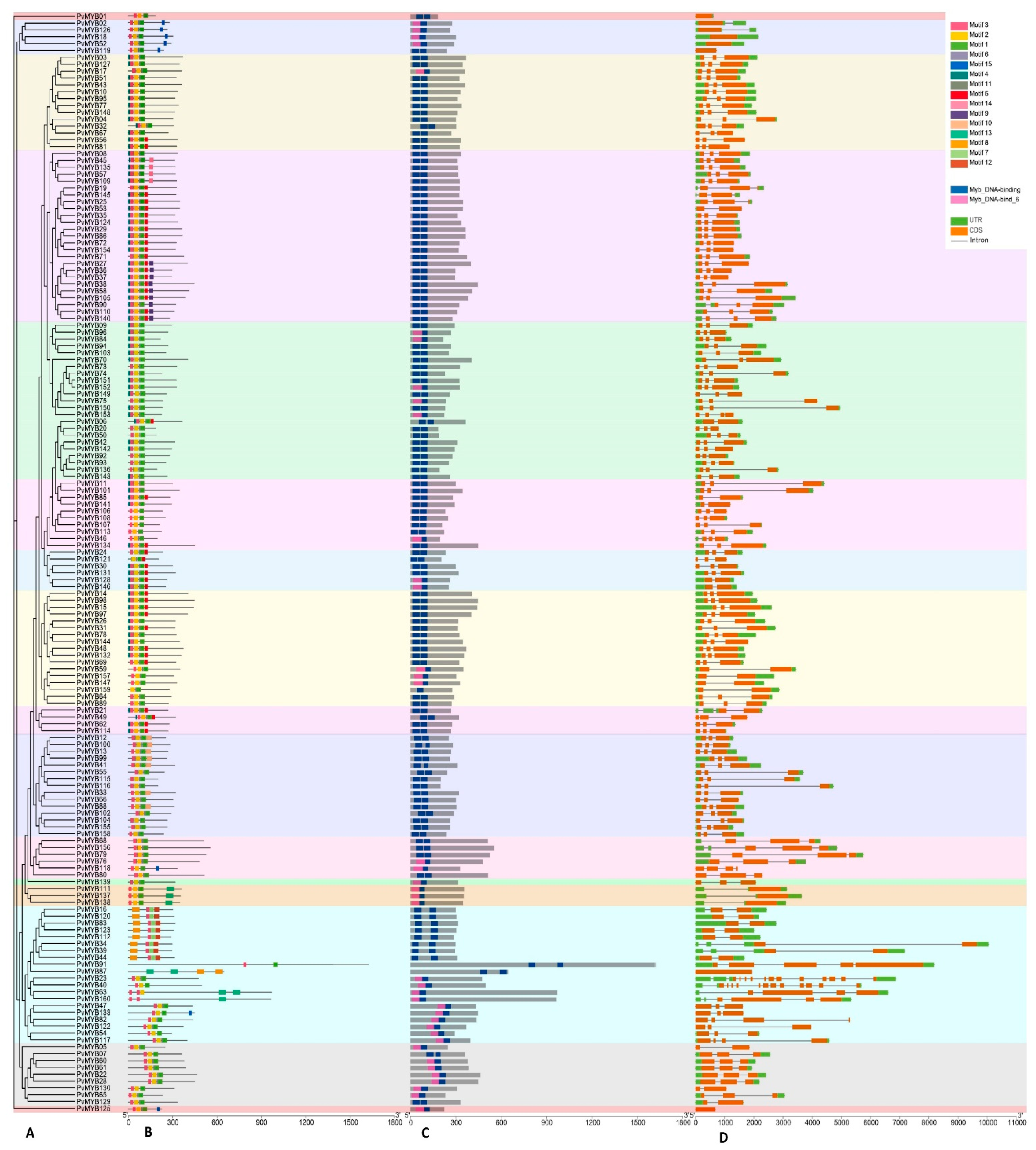
2.2. Phylogenetic Relationship, Conserved Motifs, and Gene Structures of the PvMYB Gene Family Members
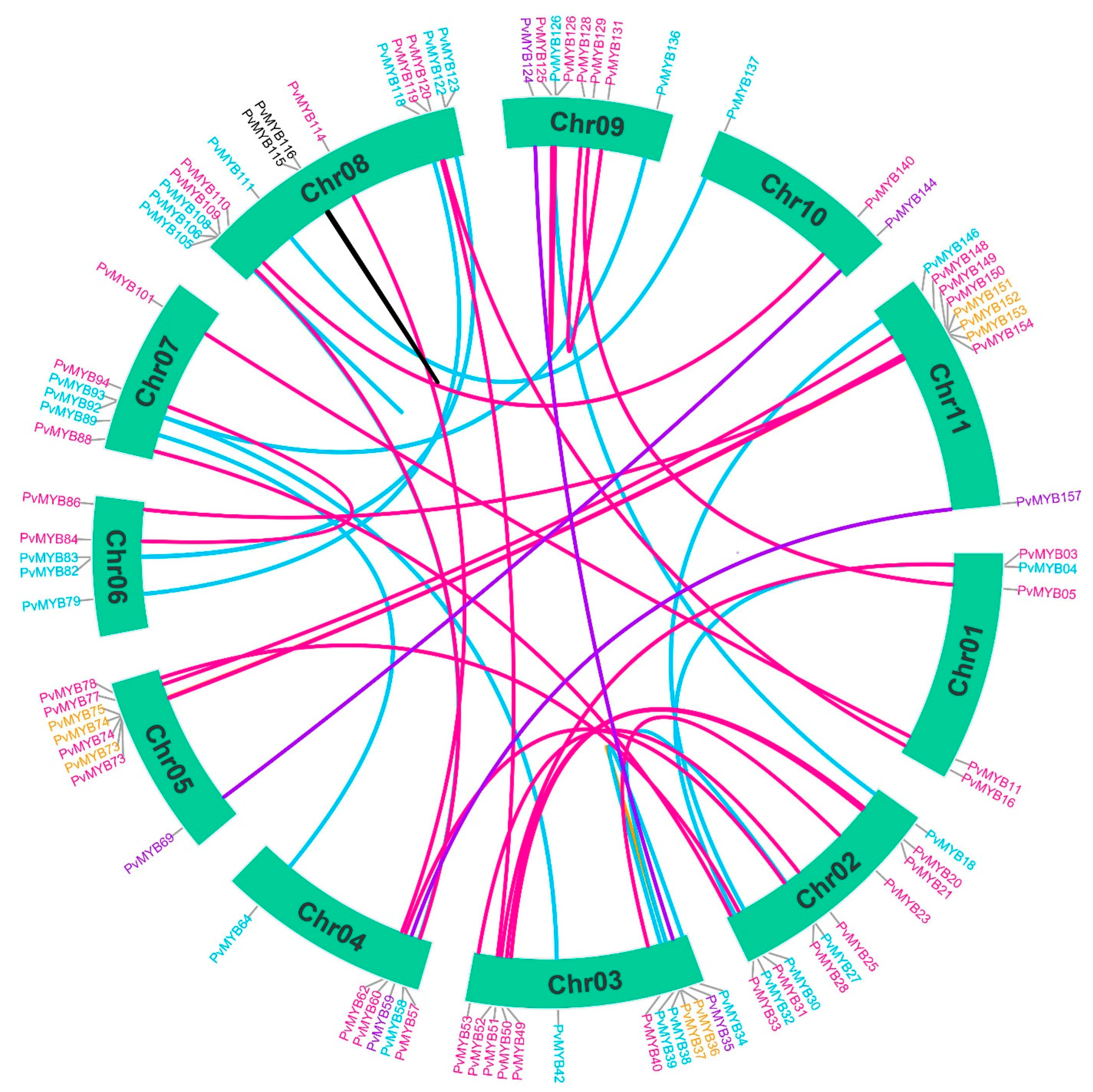
2.3. Analysis of Evolutionary Divergence in PvMYB Transcription Factors
2.4. Prediction of Cis-Elements and miRNAs Targeting PvMYB Genes
2.5. Analysis of the Core Gene Set in Common Bean (P. vulgaris) Seed Color
3. Discussion
3.1. R2R3-MYB Proteins Associated with Seed Coat Color in P. vulgaris
3.2. Prediction of Putative Suppressors among PvMYB Genes
3.3. miRNAs Targeting Expression of PvMYB Genes
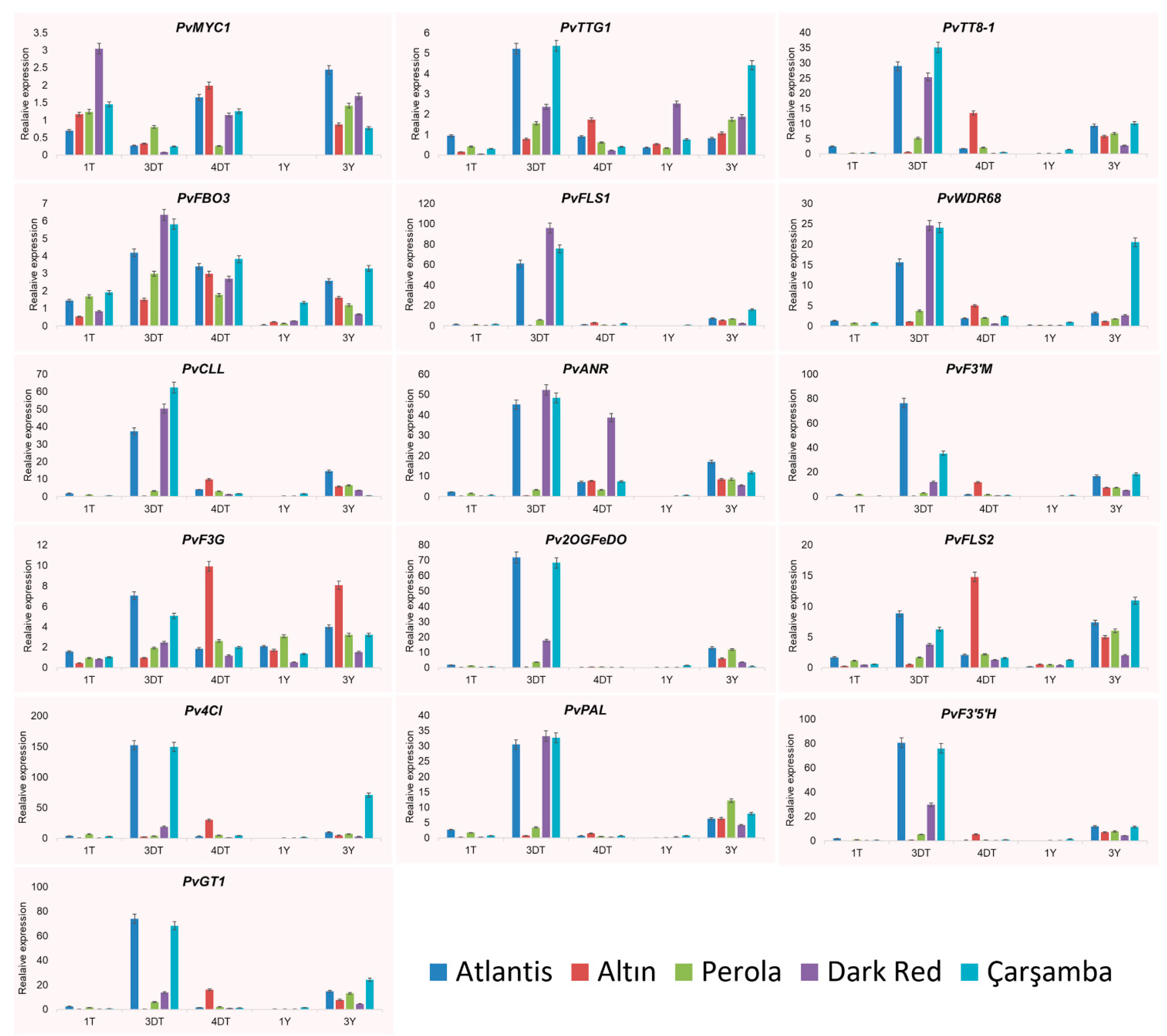
3.4. Expression of Candidate PvMYB Genes and Anthocyanin Regulatory Genes Using RT-qPCR
4. Materials and Methods
4.1. Plant Material
4.2. Expression Analysis Using Quantitative RT-PCR
4.3. Determination of MYB Members in the Common Bean Genome
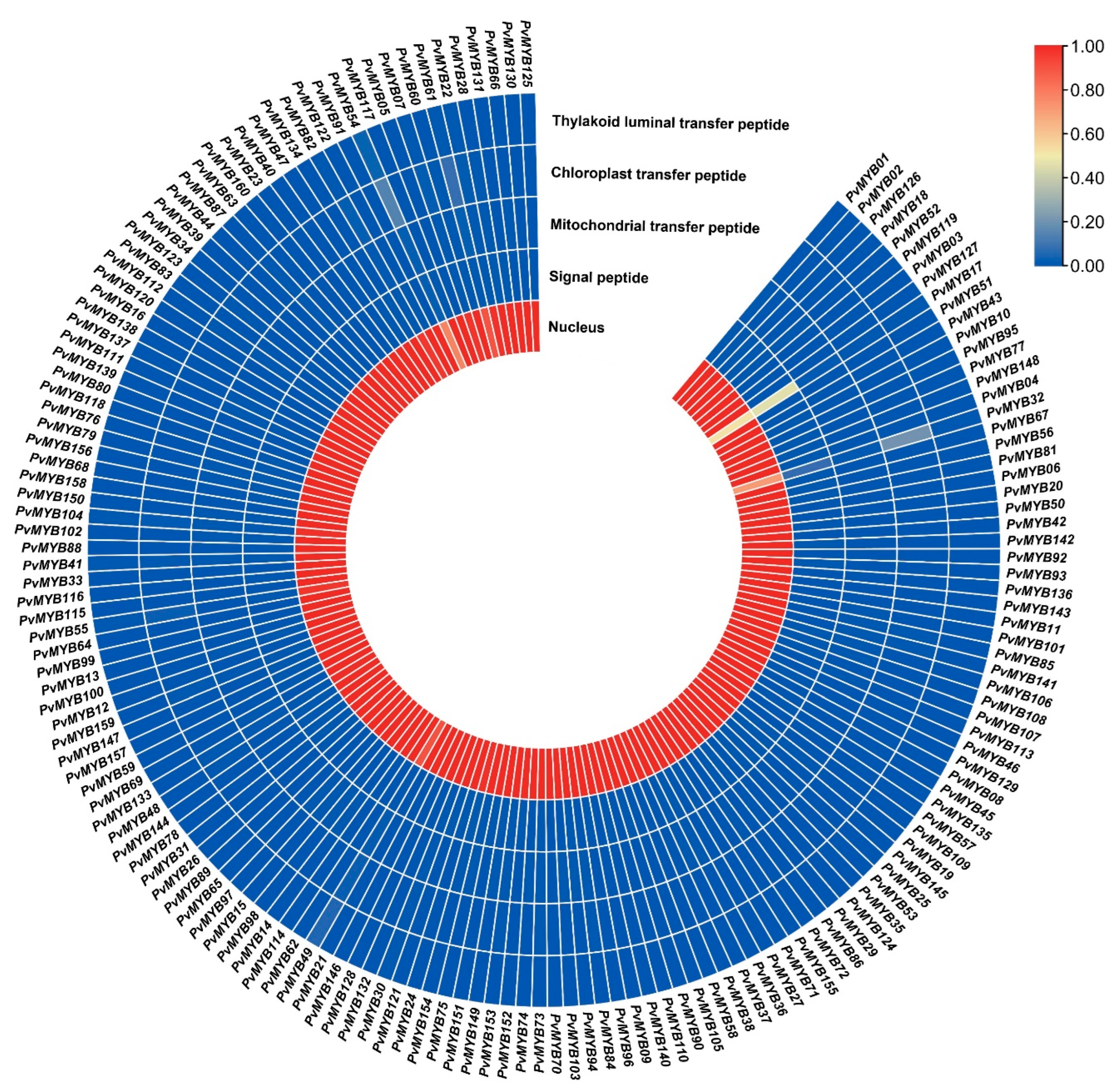
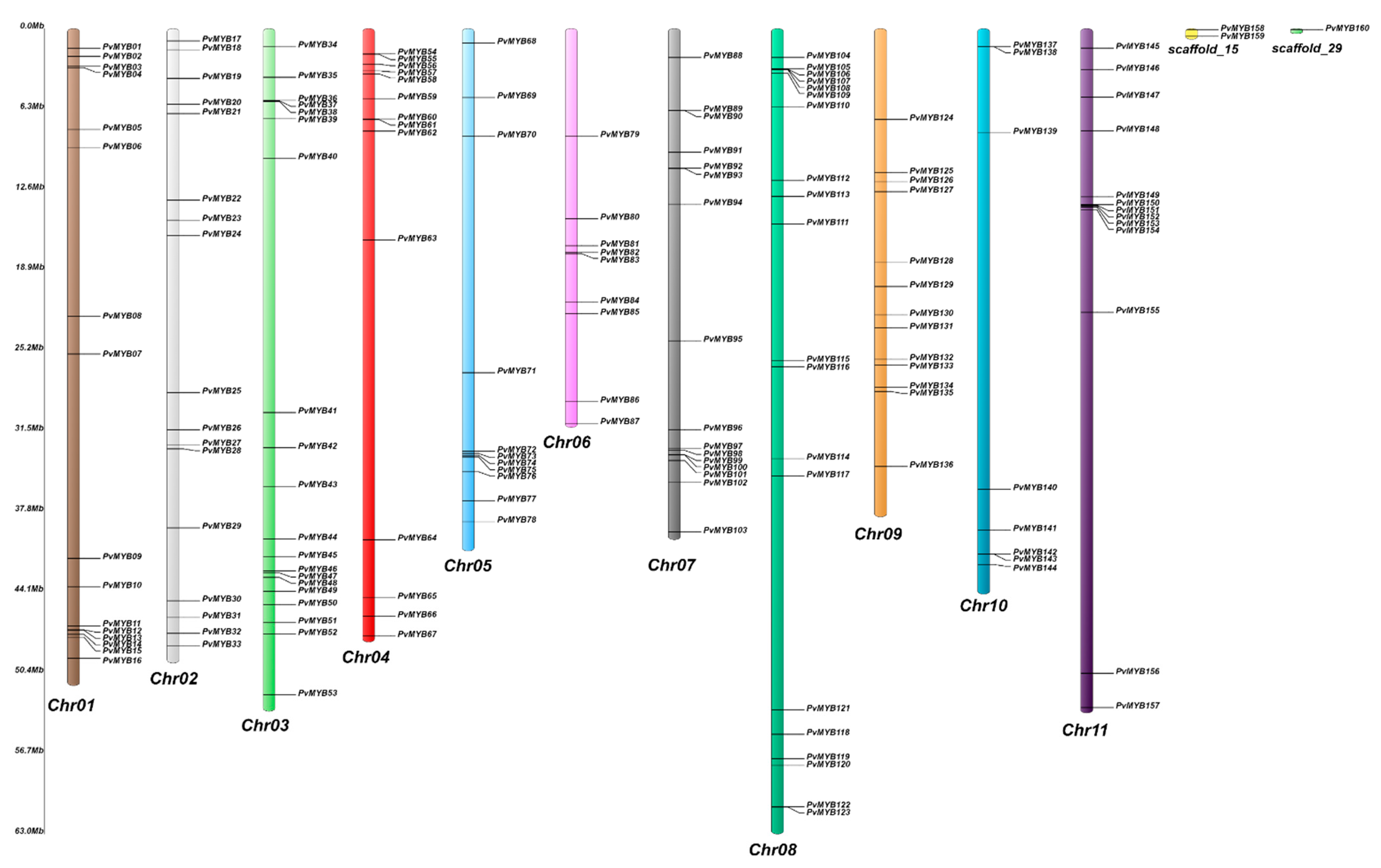
4.4. Chromosomal Position, GO Annotation, and Subcellular Localization in Common Bean
4.5. Identification of the Cis-Acting Elements on the Putative Promoter Region of Analyzed Genes
4.6. Prediction of miRNA Targeting PvMYB Genes
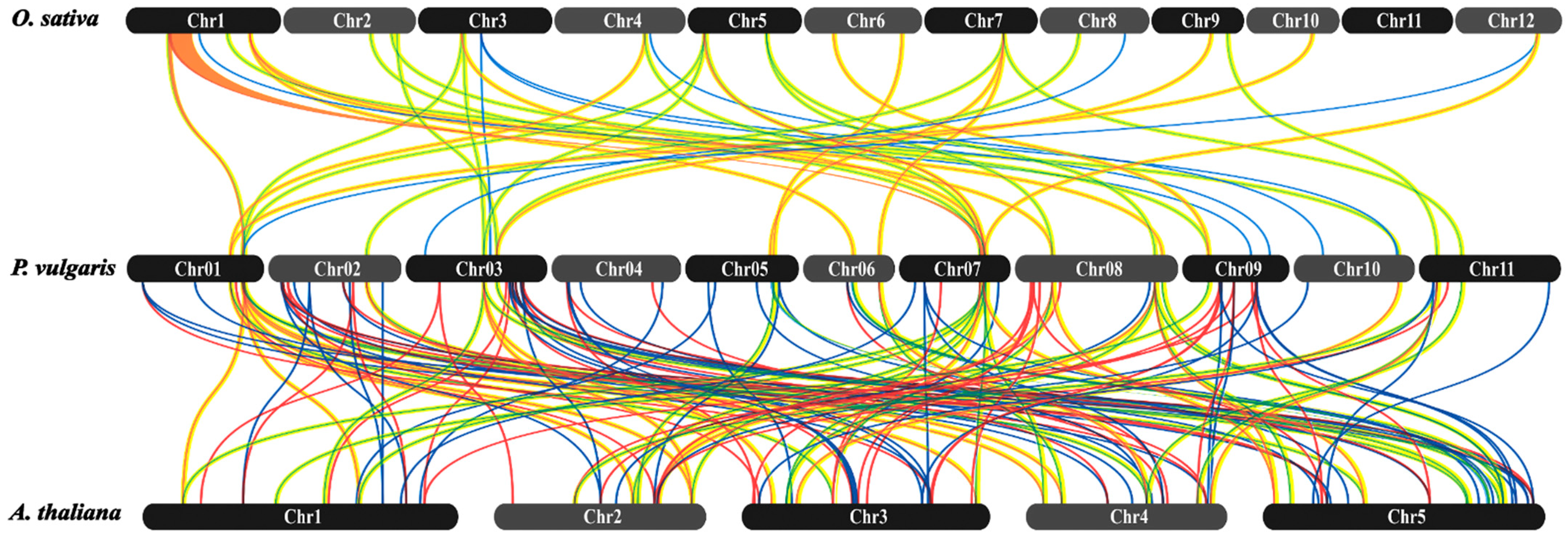
4.7. Evolutionary Investigation of PvMYB Using the Phylogenetic Tree, Gene Duplication, and Synteny Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Díaz-Batalla, L.; Widholm, J.M.; Fahey, G.C.; Castaño-Tostado, E.; Paredes-López, O. Chemical components with health implications in wild and cultivated mexican common bean seeds (phaseolus vulgaris l.). J. Agric. Food Chem. 2006, 54, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Lomas-Soria, C.; Pérez-Ramírez, I.F.; Caballero-Pérez, J.; Guevara-Gonzalez, R.G.; Guevara-Olvera, L.; Loarca-Piña, G.; Guzman-Maldonado, H.S.; Reynoso-Camacho, R. Cooked common beans (phaseolus vulgaris l.) modulate renal genes in streptozotocin-induced diabetic rats. J. Nutr. Biochem. 2015, 26, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Loko, L.E.Y.; Orobiyi, A.; Adjatin, A.; Akpo, J.; Toffa, J.; Djedatin, G.; Dansi, A. Morphological characterization of common bean (Phaseolus vulgaris L.) landraces of central region of benin republic. J. Plant Breed. Crop Sci. 2018, 10, 304–318. [Google Scholar] [CrossRef]
- de la Rosa, L.A.; Moreno-Escamilla, J.O.; Rodrigo-García, J.; Alvarez-Parrilla, E. Chapter 12—phenolic compounds. In Postharvest Physiology and Biochemistry of Fruits and Vegetables; Yahia, E.M., Ed.; Woodhead Publishing: Sawston, UK, 2019; pp. 253–271. [Google Scholar]
- Koornneef, M. Mutations affecting the testa color in arabidopsis. Arab. Inf. Serv. 1990, 28, 1–4. Available online: https://ci.nii.ac.jp/naid/10003761038/en/ (accessed on 6 November 2022).
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, R.; Xu, J.; Yan, J.; Guo, L.; Song, J.; Feng, R.; Yu, M. Genome-wide identification and classification of myb superfamily genes in peach. PLoS ONE 2018, 13, e0199192. [Google Scholar] [CrossRef]
- Rosinski, J.A.; Atchley, W.R. Molecular evolution of the myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The r2r3-myb gene family in arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Du, H.; Yang, S.-S.; Liang, Z.; Feng, B.-R.; Liu, L.; Huang, Y.-B.; Tang, Y.-X. Genome-wide analysis of the myb transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant myb-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Gu, J.; Chopra, S.; Gu, X.; Peterson, T. Ordered origin of the typical two- and three-repeat myb genes. Gene 2004, 326, 13–22. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. Myb transcription factors in arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Schwinn, K.; Venail, J.; Shang, Y.; Mackay, S.; Alm, V.; Butelli, E.; Oyama, R.; Bailey, P.; Davies, K.; Martin, C. A small family of myb-regulatory genes controls floral pigmentation intensity and patterning in the genus antirrhinum. Plant Cell 2006, 18, 831–851. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. Myb transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the ttg1/bhlh/myb transcriptional complex in arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Sabir, I.A.; Manzoor, M.A.; Shah, I.H.; Liu, X.; Zahid, M.S.; Jiu, S.; Wang, J.; Abdullah, M.; Zhang, C. Myb transcription factor family in sweet cherry (prunus avium l.): Genome-wide investigation, evolution, structure, characterization and expression patterns. BMC Plant Biol. 2022, 22, 2. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of myb transcription factor families in rice and arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. Myb repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two r2r3-myb proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. Ear motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Feng, B.-R.; Yang, S.-S.; Huang, Y.-B.; Tang, Y.-X. The r2r3-myb transcription factor gene family in maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef]
- Wang, N.; Ma, Q.; Ma, J.; Pei, W.; Liu, G.; Cui, Y.; Wu, M.; Zang, X.; Zhang, J.; Yu, S.; et al. A comparative genome-wide analysis of the r2r3-myb gene family among four gossypium species and their sequence variation and association with fiber quality traits in an interspecific g. Hirsutum × g. Barbadense population. Front. Genet. 2019, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the populus r2r3-myb family of transcription factors. Plant Physiol. 2008, 149, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape myb r2r3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across vitis and arabidopsis genomes. BMC Plant Biol. 2008, 8, 83. [Google Scholar] [CrossRef]
- Lavin, M.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary rates analysis of leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol 2005, 54, 575–594. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nat 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Nakai, K. Prediction of subcellular locations of proteins: Where to proceed? Proteomics 2010, 10, 3970–3983. [Google Scholar] [CrossRef]
- Bhandari, B.; Roesler, W.J.; DeLisio, K.D.; Klemm, D.J.; Ross, N.S.; Miller, R.E. A functional promoter flanks an intronless glutamine synthetase gene. J. Biol. Chem. 1991, 266, 7784–7792. [Google Scholar] [CrossRef]
- Gatermann, K.B.; Hoffmann, A.; Rosenberg, G.H.; Käufer, N.F. Introduction of functional artificial introns into the naturally intronless ura4 gene of schizosaccharomyces pombe. Mol. Cell Biol. 1989, 9, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Guo, B.; He, S. The roles and evolutionary patterns of intronless genes in deuterostomes. Comp. Funct Genom 2011, 2011, 680673. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.-M.; Zhang, S.; Yang, Q.-S.; Gao, H.-J.; Sheng, O.; Bi, F.-C.; Li, C.-Y.; Dong, T.; Yi, G.-J.; He, W.-D.; et al. Mamyb4, an r2r3-myb repressor transcription factor, negatively regulates the biosynthesis of anthocyanin in banana. Front. Plant Sci. 2021, 11. [Google Scholar] [CrossRef]
- Chorostecki, U.; Moro, B.; Rojas, A.M.L.; Debernardi, J.M.; Schapire, A.L.; Notredame, C.; Palatnik, J.F. Evolutionary footprints reveal insights into plant microrna biogenesis. Plant Cell 2017, 29, 1248–1261. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. Micrornas and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Biswas, S.; Hazra, S.; Chattopadhyay, S. Identification of conserved mirnas and their putative target genes in podophyllum hexandrum (himalayan mayapple). Plant Gene 2016, 6, 82–89. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Stellwag, E.J. Identification of soybean microRNAs and their targets. Planta 2008, 229, 161–182. [Google Scholar] [CrossRef]
- He, J.; Xu, M.; Willmann, M.R.; McCormick, K.; Hu, T.; Yang, L.; Starker, C.G.; Voytas, D.F.; Meyers, B.C.; Poethig, R.S. Threshold-dependent repression of spl gene expression by mir156/mir157 controls vegetative phase change in arabidopsis thaliana. PLoS Genet 2018, 14, e1007337. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; MacLean, D.; Jikumaru, Y.; Hill, L.; Yamaguchi, S.; Kamiya, Y.; Jones, J.D.G. The microrna mir393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 2011, 67, 218–231. [Google Scholar] [CrossRef]
- Gupta, O.P.; Dahuja, A.; Sachdev, A.; Kumari, S.; Jain, P.K.; Vinutha, T.; Praveen, S. Conserved mirnas modulate the expression of potential transcription factors of isoflavonoid biosynthetic pathway in soybean seeds. Mol. Biol. Rep. 2019, 46, 3713–3730. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in arabidopsis thalianaby mir156 and its target spl3. Dev 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Czech, B.; Weigel, D. Mir156-regulated spl transcription factors define an endogenous flowering pathway in arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Coruh, C.; Axtell, M.J. Mir156 and mir390 regulate tasirna accumulation and developmental timing in physcomitrella patens. Plant Cell 2012, 24, 4837–4849. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Paterson, A.H. Genome and gene duplications and gene expression divergence: A view from plants. Ann. N. Y. Acad. Sci. 2012, 1256, 1–14. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Chen, X.; Kong, Q. Characterization and divergence analysis of duplicated r2r3-myb genes in watermelon. J. Am. Soc. Hortic. Sci. 2020, 145, 281–288. [Google Scholar] [CrossRef]
- Xiao, C.; Jia, Y.; Watt, C.; Chen, W.; Zhang, Y.; Li, C. Genome-wide identification and transcriptional analyses of the r2r3-myb gene family in wheat. Biorxiv 2020. [Google Scholar] [CrossRef]
- Yamagishi, M.; Shimoyamada, Y.; Nakatsuka, T.; Masuda, K. Two r2r3-myb genes, homologs of petunia an2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol. 2010, 51, 463–474. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Tong, L.; Wei, C.; Lu, K.; Li, S.; Kimani, S.; Wang, S.; Wang, L.; Gao, X. The conserved and particular roles of the r2r3-myb regulator fhpap1 from freesia hybrida in flower anthocyanin biosynthesis. Plant Cell Physiol. 2020, 61, 1365–1380. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an r2r3-myb transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef]
- Lin, R.-C.; Rausher, M.D. R2r3-myb genes control petal pigmentation patterning in clarkia gracilis ssp. Sonomensis (onagraceae). New Phytol. 2021, 229, 1147–1162. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved myb regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.-i.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of arabidopsis plants over-expressing an myb transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; de Vos, C.H.R.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.M.; O’Connell, A.P. The strawberry famyb1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef]
- Chen, X.; Li, M.; Ni, J.; Hou, J.; Shu, X.; Zhao, W.; Su, P.; Wang, D.; Shah, F.A.; Huang, S.; et al. The r2r3-myb transcription factor ssmyb1 positively regulates anthocyanin biosynthesis and determines leaf color in chinese tallow (sapium sebiferum roxb.). Ind. Crops Prod. 2021, 164, 113335. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.-J.; Wang, Y.; Geng, Z.; Ding, B.; Jiang, J.; Chen, S.; Chen, F. An r2r3-myb transcription factor cmmyb21 represses anthocyanin biosynthesis in color fading petals of chrysanthemum. Sci. Hortic. 2022, 293, 110674. [Google Scholar] [CrossRef]
- Hu, J.; Chen, G.; Zhang, Y.; Cui, B.; Yin, W.; Yu, X.; Zhu, Z.; Hu, Z. Anthocyanin composition and expression analysis of anthocyanin biosynthetic genes in kidney bean pod. Plant Physiol. Biochem. 2015, 97, 304–312. [Google Scholar] [CrossRef]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.; et al. Tomato r2r3-myb proteins slant1 and slan2: Same protein activity, different roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The r2r3 myb transcription factor pavmyb10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (prunus avium l.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef]
- Gonzalez, A.; Mendenhall, J.; Huo, Y.; Lloyd, A. Ttg1 complex mybs, myb5 and tt2, control outer seed coat differentiation. Dev. Biol. 2009, 325, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. Tt2, tt8, and ttg1 synergistically specify the expression of banyuls and proanthocyanidin biosynthesis in arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; Molthoff, J.; de Vos, R.; Hekkert, B.; Orzaez, D.; Fernández-Moreno, J.P.; Tripodi, P.; Grandillo, S.; Martin, C.; Heldens, J.; et al. Biochemical and molecular analysis of pink tomatoes: Deregulated expression of the gene encoding transcription factor slmyb12 leads to pink tomato fruit color. Plant Physiol. 2010, 152, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Kavas, M.; Yıldırım, K.; Seçgin, Z.; Abdulla, M.F.; Gökdemir, G. Genome-wide identification of the burp domain-containing genes in phaseolus vulgaris. Physiol. Mol. Biol. Plants 2021, 27, 1885–1902. [Google Scholar] [CrossRef] [PubMed]
- Kavas, M.; Mostafa, K.; Seçgin, Z.; Yerlikaya, B.A.; Yıldırım, K.; Gökdemir, G. Genome-wide analysis of duf221 domain-containing gene family in common bean and identification of its role on abiotic and phytohormone stress response. Genet. Res. Crop Evol. 2022. [Google Scholar] [CrossRef]
- Kavas, M.; Baloğlu, M.C.; Atabay, E.S.; Ziplar, U.T.; Daşgan, H.Y.; Ünver, T. Genome-wide characterization and expression analysis of common bean bhlh transcription factors in response to excess salt concentration. Mol. Genet. Genom. 2016, 291, 129–143. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. Plantregmap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. Cdd/sparcle: The conserved domain database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouzé, P.; Rombauts, S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L.; et al. Pmiren2.0: From data annotation to functional exploration of plant micrornas. Nucleic Acids Res. 2021, 50, D1475–D1482. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. Psrnatarget: A plant small rna target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-iq-tree: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. Modelfinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. Ufboot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (itol) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]



| Gene Name | Phytozome Identifier | Chromosome | Length (bp) | CDS (pb) | Protein Length (A.A) | NCBI Accession Number |
|---|---|---|---|---|---|---|
| PvMYB01 | Phvul.001G019200.1.v2.1 | chr01 | 610 | 555 | 185 | XP_007160819.1 |
| PvMYB02 | Phvul.001G025200.1.v2.1 | chr01 | 1727 | 831 | 277 | XP_007160886.1 |
| PvMYB03 | Phvul.001G032100.1.v2.1 | chr01 | 2118 | 1104 | 368 | XP_007160964.1 |
| PvMYB04 | Phvul.001G032600.1.v2.1 | chr01 | 2793 | 903 | 301 | XP_007160971.1 |
| PvMYB05 | Phvul.001G064600.1.v2.1 | chr01 | 1847 | 741 | 247 | XP_007161388.1 |
| PvMYB06 | Phvul.001G071000.1.v2.1 | chr01 | 1613 | 1092 | 364 | XP_007161463.1 |
| PvMYB07 | Phvul.001G106800.1.v2.1 | chr01 | 2610 | 1083 | 361 | XP_007161897.1 |
| PvMYB08 | Phvul.001G107600.1.v2.1 | chr01 | 1861 | 1002 | 334 | XP_007161907.1 |
| PvMYB09 | Phvul.001G161000.1.v2.1 | chr01 | 1965 | 879 | 293 | XP_007162544.1 |
| PvMYB10 | Phvul.001G179400.1.v2.1 | chr01 | 2079 | 996 | 332 | XP_007162775.1 |
| PvMYB11 | Phvul.001G211700.1.v2.1 | chr01 | 4406 | 900 | 300 | XP_007163163.1 |
| PvMYB12 | Phvul.001G215100.1.v2.1 | chr01 | 1290 | 762 | 254 | XP_007163206.1 |
| PvMYB13 | Phvul.001G215200.1.v2.1 | chr01 | 1416 | 804 | 268 | XP_007163207.1 |
| PvMYB14 | Phvul.001G219000.1.v2.1 | chr01 | 1964 | 1215 | 405 | XP_007163255.1 |
| PvMYB15 | Phvul.001G221500.1.v2.1 | chr01 | 2611 | 1326 | 442 | XP_007163283.1 |
| PvMYB16 | Phvul.001G240300.1.v2.1 | chr01 | 2438 | 900 | 300 | XP_007163513.1 |
| PvMYB17 | Phvul.002G008800.1.v2.1 | chr02 | 1717 | 1038 | 346 | XP_007156687.1 |
| PvMYB18 | Phvul.002G015100.1.v2.1 | chr02 | 2145 | 936 | 312 | XP_007156759.1 |
| PvMYB19 | Phvul.002G040900.1.v2.1 | chr02 | 2335 | 975 | 325 | XP_007157076.1 |
| PvMYB20 | Phvul.002G056900.1.v2.1 | chr02 | 796 | 555 | 185 | XP_007157272.1 |
| PvMYB21 | Phvul.002G060500.1.v2.1 | chr02 | 2293 | 804 | 268 | XP_007157323.1 |
| PvMYB22 | Phvul.002G083800.1.v2.1 | chr02 | 2419 | 1389 | 463 | XP_007157609.1 |
| PvMYB23 | Phvul.002G088900.1.v2.1 | chr02 | 6929 | 1422 | 474 | XP_007157671.1 |
| PvMYB24 | Phvul.002G092100.1.v2.1 | chr02 | 1612 | 696 | 232 | XP_007157714.1 |
| PvMYB25 | Phvul.002G139500.1.v2.1 | chr02 | 1942 | 1044 | 348 | XP_007158283.1 |
| PvMYB26 | Phvul.002G159700.1.v2.1 | chr02 | 2386 | 951 | 317 | XP_007158529.1 |
| PvMYB27 | Phvul.002G170000.1.v2.1 | chr02 | 1828 | 1200 | 400 | XP_007158642.1 |
| PvMYB28 | Phvul.002G170500.1.v2.1 | chr02 | 2190 | 1344 | 448 | XP_007158647.1 |
| PvMYB29 | Phvul.002G221000.1.v2.1 | chr02 | 1525 | 1089 | 363 | XP_007159239.1 |
| PvMYB30 | Phvul.002G279000.1.v2.1 | chr02 | 1460 | 897 | 299 | XP_007159923.1 |
| PvMYB31 | Phvul.002G292600.1.v2.1 | chr02 | 2737 | 942 | 314 | XP_007160103.1 |
| PvMYB32 | Phvul.002G306000.1.v2.1 | chr02 | 1652 | 912 | 304 | XP_007160258.1 |
| PvMYB33 | Phvul.002G317000.1.v2.1 | chr02 | 1623 | 960 | 320 | XP_007160381.1 |
| PvMYB34 | Phvul.003G013600.1.v2.1 | chr03 | 10049 | 891 | 297 | XP_007153178.1 |
| PvMYB35 | Phvul.003G036400.1.v2.1 | chr03 | 1452 | 939 | 313 | XP_007153451.1 |
| PvMYB36 | Phvul.003G046200.1.v2.1 | chr03 | 1235 | 888 | 296 | XP_007153564.1 |
| PvMYB37 | Phvul.003G046300.1.v2.1 | chr03 | 1124 | 882 | 294 | XP_007153565.1 |
| PvMYB38 | Phvul.003G046400.1.v2.1 | chr03 | 3148 | 1335 | 445 | XP_007153567.1 |
| PvMYB39 | Phvul.003G054100.1.v2.1 | chr03 | 7172 | 885 | 295 | XP_007153659.1 |
| PvMYB40 | Phvul.003G067800.1.v2.1 | chr03 | 5691 | 1491 | 497 | XP_007153822.1 |
| PvMYB41 | Phvul.003G118450.1.v2.1 | Chr03 | 2246 | 939 | 313 | XP_007154150.1 |
| PvMYB42 | Phvul.003G132100.1.v2.1 | chr03 | 1755 | 936 | 312 | XP_007154596.1 |
| PvMYB43 | Phvul.003G148200.1.v2.1 | chr03 | 2020 | 1086 | 362 | XP_007154791.1 |
| PvMYB44 | Phvul.003G176800.1.v2.1 | chr03 | 1677 | 930 | 310 | XP_007155139.1 |
| PvMYB45 | Phvul.003G190400.1.v2.1 | chr03 | 1520 | 933 | 311 | XP_007155313.1 |
| PvMYB46 | Phvul.003G200100.1.v2.1 | chr03 | 1107 | 588 | 196 | XP_007155422.1 |
| PvMYB47 | Phvul.003G201300.1.v2.1 | chr03 | 1623 | 1302 | 434 | XP_007155438.1 |
| PvMYB48 | Phvul.003G203900.1.v2.1 | chr03 | 1668 | 1110 | 370 | XP_007155468.1 |
| PvMYB49 | Phvul.003G214300.1.v2.1 | chr03 | 1771 | 960 | 320 | XP_007155582.1 |
| PvMYB50 | Phvul.003G222400.1.v2.1 | chr03 | 1548 | 567 | 189 | XP_007155685.1 |
| PvMYB51 | Phvul.003G232300.1.v2.1 | chr03 | 1545 | 972 | 324 | XP_007155794.1 |
| PvMYB52 | Phvul.003G240200.1.v2.1 | chr03 | 1671 | 870 | 290 | XP_007155887.1 |
| PvMYB53 | Phvul.003G284000.1.v2.1 | chr03 | 1582 | 1041 | 347 | XP_007156413.1 |
| PvMYB54 | Phvul.004G011400.1.v2.1 | chr04 | 2184 | 879 | 293 | XP_007151013.1 |
| PvMYB55 | Phvul.004G012000.1.v2.1 | chr04 | 3691 | 660 | 220 | XP_007151019.1 |
| PvMYB56 | Phvul.004G024200.1.v2.1 | chr04 | 1690 | 996 | 332 | XP_007151175.1 |
| PvMYB57 | Phvul.004G028500.1.v2.1 | chr04 | 1890 | 954 | 318 | XP_007151227.1 |
| PvMYB58 | Phvul.004G029800.1.v2.1 | chr04 | 3046 | 1227 | 409 | XP_007151240.1 |
| PvMYB59 | Phvul.004G046000.1.v2.1 | chr04 | 3445 | 1050 | 350 | XP_007151435.1 |
| PvMYB60 | Phvul.004G053500.1.v2.1 | chr04 | 2056 | 1131 | 377 | XP_007151515.1 |
| PvMYB61 | Phvul.004G053600.1.v2.1 | chr04 | 2104 | 1158 | 386 | XP_007151516.1 |
| PvMYB62 | Phvul.004G057800.1.v2.1 | chr04 | 1357 | 828 | 276 | XP_007151568.1 |
| PvMYB63 | Phvul.004G090900.1.v2.1 | chr04 | 6608 | 2910 | 970 | XP_007151972.1 |
| PvMYB64 | Phvul.004G116500.1.v2.1 | chr04 | 2635 | 870 | 290 | XP_007158818.1 |
| PvMYB65 | Phvul.004G144900.1.v2.1 | chr04 | 3058 | 693 | 231 | XP_007152283.1 |
| PvMYB66 | Phvul.004G156154.1.v2.1 | Chr04 | 1483 | 903 | 301 | XP_007152615.1 |
| PvMYB67 | Phvul.004G173500.1.v2.1 | chr04 | 1282 | 810 | 270 | XP_007152944.1 |
| PvMYB68 | Phvul.005G012900.1.v2.1 | chr05 | 4278 | 1536 | 512 | XP_007148771.1 |
| PvMYB69 | Phvul.005G047400.1.v2.1 | chr05 | 1644 | 969 | 323 | XP_007149171.1 |
| PvMYB70 | Phvul.005G060000.1.v2.1 | chr05 | 2932 | 1212 | 404 | XP_007149313.1 |
| PvMYB71 | Phvul.005G087400.1.v2.1 | chr05 | 1861 | 1125 | 375 | XP_007149649.1 |
| PvMYB72 | Phvul.005G109100.1.v2.1 | chr05 | 1316 | 972 | 324 | XP_007149908.1 |
| PvMYB73 | Phvul.005G109700.1.v2.1 | chr05 | 1454 | 981 | 327 | XP_007149915.1 |
| PvMYB74 | Phvul.005G109800.1.v2.1 | chr05 | 3191 | 684 | 228 | XP_007149916.1 |
| PvMYB75 | Phvul.005G109900.1.v2.1 | chr05 | 4174 | 699 | 233 | XP_007149917.1 |
| PvMYB76 | Phvul.005G115500.1.v2.1 | chr05 | 3778 | 1437 | 479 | XP_007149977.1 |
| PvMYB77 | Phvul.005G131300.1.v2.1 | chr05 | 1935 | 1014 | 338 | XP_007150152.1 |
| PvMYB78 | Phvul.005G157600.1.v2.1 | chr05 | 2073 | 786 | 262 | XP_007150497.1 |
| PvMYB79 | Phvul.006G020200.1.v2.1 | chr06 | 5751 | 1578 | 526 | XP_007146191.1 |
| PvMYB80 | Phvul.006G045300.1.v2.1 | chr06 | 2290 | 1539 | 513 | XP_007146493.1 |
| PvMYB81 | Phvul.006G061600.1.v2.1 | chr06 | 1169 | 978 | 326 | XP_007146695.1 |
| PvMYB82 | Phvul.006G064600.1.v2.1 | chr06 | 5300 | 1311 | 437 | XP_007146726.1 |
| PvMYB83 | Phvul.006G065700.1.v2.1 | chr06 | 2767 | 945 | 315 | XP_007146740.1 |
| PvMYB84 | Phvul.006G105200.1.v2.1 | chr06 | 1229 | 648 | 216 | XP_007147213.1 |
| PvMYB85 | Phvul.006G114800.1.v2.1 | chr06 | 1622 | 846 | 282 | XP_007147330.1 |
| PvMYB86 | Phvul.006G192900.1.v2.1 | chr06 | 1577 | 1092 | 364 | XP_007148253.1 |
| PvMYB87 | Phvul.006G217200.1.v2.1 | chr06 | 1941 | 1941 | 647 | XP_007148540.1 |
| PvMYB88 | Phvul.007G028700.1.v2.1 | chr07 | 1672 | 918 | 306 | XP_007142925.1 |
| PvMYB89 | Phvul.007G069200.1.v2.1 | chr07 | 2444 | 813 | 271 | XP_007143397.1 |
| PvMYB90 | Phvul.007G069400.1.v2.1 | chr07 | 3042 | 966 | 322 | XP_007143400.1 |
| PvMYB91 | Phvul.007G093100.1.v2.1 | chr07 | 8179 | 4878 | 1626 | XP_007143686.1 |
| PvMYB92 | Phvul.007G099900.1.v2.1 | chr07 | 1116 | 837 | 279 | XP_007143768.1 |
| PvMYB93 | Phvul.007G100100.1.v2.1 | chr07 | 1333 | 765 | 255 | XP_007143770.1 |
| PvMYB94 | Phvul.007G108500.1.v2.1 | chr07 | 2434 | 804 | 268 | XP_007143866.1 |
| PvMYB95 | Phvul.007G147600.1.v2.1 | chr07 | 2077 | 939 | 313 | XP_007144336.1 |
| PvMYB96 | Phvul.007G192900.1.v2.1 | chr07 | 1061 | 804 | 268 | XP_007144899.1 |
| PvMYB97 | Phvul.007G206200.1.v2.1 | chr07 | 2040 | 1206 | 402 | XP_007145057.1 |
| PvMYB98 | Phvul.007G208400.1.v2.1 | chr07 | 2113 | 1338 | 446 | XP_007145085.1 |
| PvMYB99 | Phvul.007G211800.1.v2.1 | chr07 | 1765 | 777 | 259 | XP_007145121.1 |
| PvMYB100 | Phvul.007G211900.1.v2.1 | chr07 | 1202 | 846 | 282 | XP_007145122.1 |
| PvMYB101 | Phvul.007G215800.1.v2.1 | chr07 | 4034 | 1035 | 345 | XP_007145164.1 |
| PvMYB102 | Phvul.007G231800.1.v2.1 | chr07 | 1407 | 864 | 288 | XP_007145351.1 |
| PvMYB103 | Phvul.007G273400.1.v2.1 | chr07 | 2248 | 765 | 255 | XP_007145850.1 |
| PvMYB104 | Phvul.008G028000.1.v2.1 | chr08 | 1673 | 786 | 262 | XP_007139421.1 |
| PvMYB105 | Phvul.008G038000.1.v2.1 | chr08 | 3435 | 1146 | 382 | XP_007139534.1 |
| PvMYB106 | Phvul.008G038200.1.v2.1 | chr08 | 1070 | 693 | 231 | XP_007139536.1 |
| PvMYB107 | Phvul.008G038400.1.v2.1 | chr08 | 2275 | 642 | 214 | XP_007139538.1 |
| PvMYB108 | Phvul.008G038600.1.v2.1 | chr08 | 1081 | 753 | 251 | XP_007139540.1 |
| PvMYB109 | Phvul.008G041500.1.v2.1 | chr08 | 1506 | 975 | 325 | XP_007139579.1 |
| PvMYB110 | Phvul.008G067300.1.v2.1 | chr08 | 2643 | 927 | 309 | XP_007139892.1 |
| PvMYB111 | Phvul.008G102300.1.v2.1 | chr08 | 3132 | 1071 | 357 | XP_007140319.1 |
| PvMYB112 | Phvul.008G107000.1.v2.1 | chr08 | 2224 | 861 | 287 | XP_007140381.1 |
| PvMYB113 | Phvul.008G113300.1.v2.1 | chr08 | 1965 | 672 | 224 | XP_007140451.1 |
| PvMYB114 | Phvul.008G146900.1.v2.1 | chr08 | 1056 | 807 | 269 | XP_007140849.1 |
| PvMYB115 | Phvul.008G155700.1.v2.1 | chr08 | 3582 | 603 | 201 | XP_007140962.1 |
| PvMYB116 | Phvul.008G155900.1.v2.1 | chr08 | 4724 | 600 | 200 | XP_007140964.1 |
| PvMYB117 | Phvul.008G156100.1.v2.1 | chr08 | 4583 | 1191 | 397 | XP_007140966.1 |
| PvMYB118 | Phvul.008G205000.1.v2.1 | chr08 | 1443 | 990 | 330 | XP_007141541.1 |
| PvMYB119 | Phvul.008G222600.1.v2.1 | chr08 | 720 | 720 | 240 | XP_007141751.1 |
| PvMYB120 | Phvul.008G226600.1.v2.1 | chr08 | 2172 | 918 | 306 | XP_007141798.1 |
| PvMYB121 | Phvul.008G233800.1.v2.1 | chr08 | 1071 | 612 | 204 | XP_007141884.1 |
| PvMYB122 | Phvul.008G262700.1.v2.1 | chr08 | 3968 | 1110 | 370 | XP_007142225.1 |
| PvMYB123 | Phvul.008G262900.1.v2.1 | chr08 | 2006 | 915 | 305 | XP_007142227.1 |
| PvMYB124 | Phvul.009G031200.1.v2.1 | chr09 | 1512 | 1008 | 336 | XP_007136252.1 |
| PvMYB125 | Phvul.009G062700.1.v2.1 | chr09 | 675 | 675 | 225 | XP_007136655.1 |
| PvMYB126 | Phvul.009G068000.1.v2.1 | chr09 | 2081 | 789 | 263 | XP_007136715.1 |
| PvMYB127 | Phvul.009G075000.1.v2.1 | chr09 | 1808 | 1035 | 345 | XP_007136795. |
| PvMYB128 | Phvul.009G119900.1.v2.1 | chr09 | 1315 | 780 | 260 | XP_007137351.1 |
| PvMYB129 | Phvul.009G133700.1.v2.1 | chr09 | 1643 | 996 | 332 | XP_007134825.1 |
| PvMYB130 | Phvul.009G151000.1.v2.1 | chr09 | 1061 | 924 | 308 | XP_007137516.1 |
| PvMYB131 | Phvul.009G158200.1.v2.1 | chr09 | 1660 | 957 | 319 | XP_007137729.1 |
| PvMYB132 | Phvul.009G174900.1.v2.1 | chr09 | 1707 | 1068 | 356 | XP_007137818.1 |
| PvMYB133 | Phvul.009G177100.1.v2.1 | chr09 | 1640 | 1338 | 446 | XP_007138025.1 |
| PvMYB134 | Phvul.009G185950.1.v2.1 | Chr09 | 2431 | 1347 | 449 | XP_007138059.1 |
| PvMYB135 | Phvul.009G187700.1.v2.1 | chr09 | 1713 | 948 | 316 | XP_007138187.1 |
| PvMYB136 | Phvul.009G228200.1.v2.1 | chr09 | 2842 | 576 | 192 | XP_007138671.1 |
| PvMYB137 | Phvul.010G009800.1.v2.1 | chr10 | 3638 | 1059 | 353 | XP_007133991.1 |
| PvMYB138 | Phvul.010G009900.1.v2.1 | chr10 | 3098 | 1047 | 349 | XP_007133992.1 |
| PvMYB139 | Phvul.010G053200.1.v2.1 | chr10 | 2065 | 948 | 316 | XP_007134506.1 |
| PvMYB140 | Phvul.010G096400.1.v2.1 | chr10 | 2768 | 837 | 279 | XP_007135039.1 |
| PvMYB141 | Phvul.010G115500.1.v2.1 | chr10 | 1194 | 879 | 293 | XP_007135274.1 |
| PvMYB142 | Phvul.010G130500.1.v2.1 | chr10 | 1279 | 879 | 293 | XP_007135450.1 |
| PvMYB143 | Phvul.010G130600.1.v2.1 | chr10 | 1506 | 786 | 262 | XP_007135451.1 |
| PvMYB144 | Phvul.010G137500.1.v2.1 | chr10 | 1804 | 1041 | 347 | XP_007135535.1 |
| PvMYB145 | Phvul.011G019200.1.v2.1 | chr11 | 1516 | 969 | 323 | XP_007131513.1 |
| PvMYB146 | Phvul.011G034900.1.v2.1 | chr11 | 1416 | 762 | 254 | XP_007131703.1 |
| PvMYB147 | Phvul.011G059800.1.v2.1 | chr11 | 2344 | 984 | 328 | XP_007132019.1 |
| PvMYB148 | Phvul.011G084500.1.v2.1 | chr11 | 2090 | 936 | 312 | XP_007132317.1 |
| PvMYB149 | Phvul.011G109400.1.v2.1 | chr11 | 1593 | 774 | 258 | XP_007132609.1 |
| PvMYB150 | Phvul.011G109500.1.v2.1 | chr11 | 4958 | 690 | 230 | XP_007144143.1 |
| PvMYB151 | Phvul.011G109600.1.v2.1 | chr11 | 1458 | 972 | 324 | XP_007132610.1 |
| PvMYB152 | Phvul.011G109700.1.v2.1 | chr11 | 1493 | 975 | 325 | XP_007132611.1 |
| PvMYB153 | Phvul.011G109800.1.v2.1 | chr11 | 1301 | 675 | 225 | XP_007132612.1 |
| PvMYB154 | Phvul.011G110500.1.v2.1 | chr11 | 1300 | 957 | 319 | XP_007132613.1 |
| PvMYB155 | Phvul.011G115230.1.v2.1 | Chr11 | 1292 | 789 | 263 | XP_007132622.1 |
| PvMYB156 | Phvul.011G191300.1.v2.1 | chr11 | 4855 | 1665 | 555 | XP_007133583.1 |
| PvMYB157 | Phvul.011G212000.1.v2.1 | chr11 | 2698 | 915 | 305 | XP_007133825.1 |
| PvMYB158 | Phvul.L002743.1.v2.1 | scaffold_15 | 1663 | 717 | 239 | XP_007150291.1 |
| PvMYB159 | Phvul.L009043.1.v2.1 | scaffold_15 | 2870 | 834 | 278 | XP_007150366.1 |
| PvMYB160 | Phvul.L001860.1.v2.1 | scaffold_29 | 5338 | 2892 | 964 | XP_007144020.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavas, M.; Abdulla, M.F.; Mostafa, K.; Seçgin, Z.; Yerlikaya, B.A.; Otur, Ç.; Gökdemir, G.; Kurt Kızıldoğan, A.; Al-Khayri, J.M.; Jain, S.M. Investigation and Expression Analysis of R2R3-MYBs and Anthocyanin Biosynthesis-Related Genes during Seed Color Development of Common Bean (Phaseolus vulgaris). Plants 2022, 11, 3386. https://doi.org/10.3390/plants11233386
Kavas M, Abdulla MF, Mostafa K, Seçgin Z, Yerlikaya BA, Otur Ç, Gökdemir G, Kurt Kızıldoğan A, Al-Khayri JM, Jain SM. Investigation and Expression Analysis of R2R3-MYBs and Anthocyanin Biosynthesis-Related Genes during Seed Color Development of Common Bean (Phaseolus vulgaris). Plants. 2022; 11(23):3386. https://doi.org/10.3390/plants11233386
Chicago/Turabian StyleKavas, Musa, Mohamed Farah Abdulla, Karam Mostafa, Zafer Seçgin, Bayram Ali Yerlikaya, Çiğdem Otur, Gökhan Gökdemir, Aslıhan Kurt Kızıldoğan, Jameel Mohammed Al-Khayri, and Shri Mohan Jain. 2022. "Investigation and Expression Analysis of R2R3-MYBs and Anthocyanin Biosynthesis-Related Genes during Seed Color Development of Common Bean (Phaseolus vulgaris)" Plants 11, no. 23: 3386. https://doi.org/10.3390/plants11233386
APA StyleKavas, M., Abdulla, M. F., Mostafa, K., Seçgin, Z., Yerlikaya, B. A., Otur, Ç., Gökdemir, G., Kurt Kızıldoğan, A., Al-Khayri, J. M., & Jain, S. M. (2022). Investigation and Expression Analysis of R2R3-MYBs and Anthocyanin Biosynthesis-Related Genes during Seed Color Development of Common Bean (Phaseolus vulgaris). Plants, 11(23), 3386. https://doi.org/10.3390/plants11233386












