Microbial Intervention: An Approach to Combat the Postharvest Pathogens of Fruits
Abstract
:1. Introduction
2. The Fruit Microbiome: Composition and Community Structure
Microbiome Change in Response to Environmental Factors
3. Action Mechanism of Microbial Antagonist in Postharvest Pathogen Management
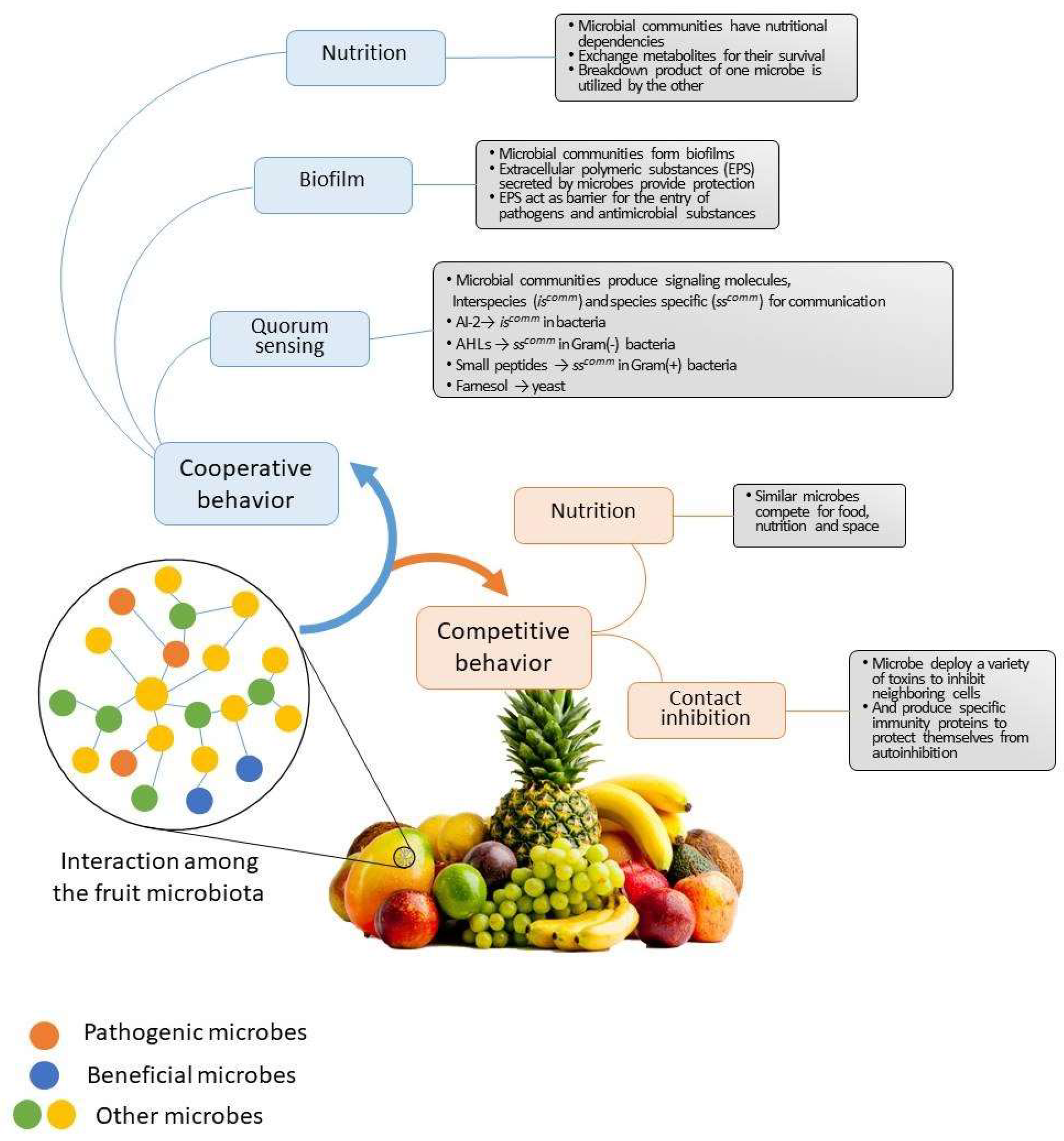
4. Collaborative Interactions among Plant Microbiota
5. Competitive and Co-Exclusion Relationships among Plant Microbiota
6. Microbial Intervention in Postharvest Management
7. Future Perspective
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumari, M.; Qureshi, K.A.; Jaremko, M.; White, J.F.; Singh, S.K.; Sharma, V.K.; Singh, K.K.; Santoyo, G.; Puopolo, G.; Kumar, A. Deciphering the role of endophytic microbiome in postharvest diseases management of fruits: Opportunity areas in commercial up-scale production. Front. Plant Sci. 2022, 13, 1026575. [Google Scholar] [CrossRef] [PubMed]
- Boeing, H.; Bechthold, A.; Bub, A.; Ellinger, S.; Haller, D.; Kroke, A.; Leschik-Bonnet, E.; Müller, M.J.; Oberritter, H.; Schulze, M.; et al. Critical review: Vegetables and fruit in the prevention of chronic diseases. Eur. J. Nutr. 2012, 51, 637–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, G. Fruit and Vegetables: Prevention and Cure? In A Prescription for Healthy Living; Short, E., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 243–253. [Google Scholar] [CrossRef]
- Singh, R.; Rastogi, S.; Dwivedi, U.N. Phenylpropanoid metabolism in ripening fruits. Compr. Rev. Food Sci. Food Saf. 2010, 9, 398–416. [Google Scholar] [CrossRef]
- Maduwanthi, S.D.T.; Marapana, R.A.U.J. Induced ripening agents and their effect on fruit quality of banana. Int. J. Food Sci. 2019, 2019, 2520179. [Google Scholar] [CrossRef] [PubMed]
- Obulesu, M.; Bhattacharya, S. Color changes of tamarind (Tamarindus indica L.) pulp during fruit development, ripening, and storage. Int. J. Food Prop. 2011, 14, 538–549. [Google Scholar] [CrossRef] [Green Version]
- Morales, H.; Marín, S.; Ramos, A.J.; Sanchis, V. Influence of postharvest technologies applied during cold storage of apples in Penicillium expansum growth and patulin accumulation: A review. Food Cont. 2010, 21, 953–962. [Google Scholar] [CrossRef]
- Opara, I.K.; Fawole, O.A.; Opara, U.L. Postharvest Losses of Pomegranate Fruit at the Packhouse and Implications for Sustainability Indicators. Sustainability 2021, 13, 5187. [Google Scholar] [CrossRef]
- Mari, M.; Di Francesco, A.; Bertolini, P. Control of fruit postharvest diseases: Old issues and innovative approaches. Stewart Postharvest Rev. 2014, 10, 1–4. [Google Scholar]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. R. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Pathak, P.; Rai, V.K.; Can, H.; Singh, S.K.; Kumar, D.; Bhardwaj, N.; Roychowdhury, R.; de Azevedo, L.C.B.; Kaushalendra; Verma, H.; et al. Plant-Endophyte Interaction during Biotic Stress Management. Plants 2022, 11, 2203. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Freilich, S.; Bartuv, R.; Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Burchard, E.; Dardick, C.; et al. Global analysis of the apple fruit microbiome: Are all apples the same? Environ. Microbiol. 2021, 23, 6038–6055. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R.; et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sangiorgio, D.; Cellini, A.; Donati, I.; Ferrari, E.; Tanunchai, B.; Wahdan, S.F.M.; Sadubsarn, D.; Farneti, B.; Checcucci, A.; Buscot, F.; et al. Taxonomical and functional composition of strawberry microbiome is genotype-dependent. J. Adv. Res. 2022; in press. [Google Scholar] [CrossRef]
- Zhimo, V.Y.; Kumar, A.; Biasi, A.; Abdelfattah, A.; Sharma, V.K.; Salim, S.; Feygenberg, O.; Bartuv, R.; Freilich, S.; Whitehead, S.R.; et al. Assembly and dynamics of the apple carposphere microbiome during fruit development and storage. Front. Microbiol. 2022, 13, 928888. [Google Scholar] [CrossRef]
- Kecskeméti, E.; Berkelmann-Löhnertz, B.; Reineke, A. Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes? PLoS ONE 2016, 11, e0160852. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, C.R.; Salas-González, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Teixeira, P.J.P.L.; Dangl, J.L. The plant microbiome: From ecology to reductionism and beyond. Annu. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Compant, S.; Van Der Heijden, M.G.; Sessitsch, A. Climate change effects on beneficial plant–microorganism interactions. FEMS Microbiol. Ecol. 2010, 73, 197–214. [Google Scholar] [CrossRef]
- Ho, Y.N.; Mathew, D.C.; Huang, C.C. Plant-microbe ecology: Interactions of plants and symbiotic microbial communities. In Traditional Approaches to Recent Trends; IntechOpen: London, UK, 2017; pp. 93–119. [Google Scholar] [CrossRef]
- Allard, S.M.; Ottesen, A.R.; Micallef, S.A. Rain induces temporary shifts in epiphytic bacterial communities of cucumber and tomato fruit. Sci. Rep. 2020, 10, 1765. [Google Scholar] [CrossRef] [Green Version]
- Louzada Pereira, L.; Carvalho Guarçoni, R.; Soares Cardoso, W.; Côrrea Taques, R.; Rizzo Moreira, T.; da Silva, S.F.; Schwengber ten Caten, C. Influence of solar radiation and wet processing on the final quality of arabica coffee. J. Food Quality 2018, 2018, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Serwah Boateng, N.A.; Ngolong Ngea, G.L.; Shi, Y.; Lin, H.; Yang, Q.; Wang, K.; Zhang, X.; Zhao, L.; Droby, S. Unravelling the fruit microbiome: The key for developing effective biological control strategies for postharvest diseases. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4906–4930. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Droby, S.; Singh, V.K.; Singh, S.K.; White, J.F. Entry, colonization, and distribution of endophytic microorganisms in plants. In Microbial Endophytes; Woodhead Publishing: Sawston, UK, 2020; pp. 1–33. [Google Scholar] [CrossRef]
- Kumar, A.; Zhimo, Y.; Biasi, A.; Salim, S.; Feygenberg, O.; Wisniewski, M.; Droby, S. Endophytic Microbiome in the Carposphere and Its Importance in Fruit Physiology and Pathology. In Postharvest Pathology; Springer: Cham, Switzerland, 2021; pp. 73–88. [Google Scholar] [CrossRef]
- Wisniewski, M.; Droby, S. The postharvest microbiome: The other half of sustainability. Biol. Control 2019, 137, 104025. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duke, A.S.; Pal, S.; Nambi, V.E.; Gupta, R.K.; Singh, R.; Sharma, K.; Vishwakarma, R.K. Exploitation of microbial antagonists for the control of postharvest diseases of fruits: A review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1498–1513. [Google Scholar]
- Janisiewicz, W.J.; Tworkoski, T.J.; Sharer, C.Y.N.T.H.I.A. Characterizing the mechanism of biological control of postharvest diseases on fruits with a simple method to study competition for nutrients. Phytopathology 2000, 90, 1196–1200. [Google Scholar] [CrossRef] [Green Version]
- Janisiewicz, W.J.; Korsten, L. Biological control of postharvest diseases of fruits. Annu. Rev. Phytopathol. 2002, 40, 411–441. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Zhao, L.; Zhang, X.; Foku, J.M.; Li, J.; Hu, W.; Zhang, H. Efficacy of Yarrowia lipolytica in the biocontrol of green mold and blue mold in Citrus reticulata and the mechanisms involved. Biol. Control 2019, 139, 104096. [Google Scholar] [CrossRef]
- Wang, S.; Ruan, C.; Yi, L.; Deng, L.; Yao, S.; Zeng, K. Biocontrol ability and action mechanism of Metschnikowia citriensis against Geotrichum citri-aurantii causing sour rot of postharvest citrus fruit. Food Microbiol. 2020, 87, 103375. [Google Scholar] [CrossRef]
- Abdel-Rahim, I.R.; Abo-Elyousr, K.A. Talaromyces pinophilus strain AUN-1 as a novel mycoparasite of Botrytis cinerea, the pathogen of onion scape and umbel blights. Microbiol. Res. 2018, 212, 1–9. [Google Scholar] [CrossRef]
- Cao, R.; Liu, X.; Gao, K.; Mendgen, K.; Kang, Z.; Gao, J.; Dai, Y.; Wang, X. Mycoparasitism of endophytic fungi isolated from reed on soilborne phytopathogenic fungi and production of cell wall-degrading enzymes in vitro. Curr. Microbiol. 2009, 59, 584–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Ádám, A.L.; Nagy, Z.Á.; Kátay, G.; Mergenthaler, E.; Viczián, O. Signals of systemic immunity in plants: Progress and open questions. Int. J. Mol. Sci. 2018, 19, 1146. [Google Scholar] [CrossRef] [Green Version]
- Van Loon, L.C.; Bakker, P.A.H.M.; Pieterse, C.M.J. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathma, J.; Kennedy, R.K.; Sakthivel, N. Mechanisms of fluorescent pseudomonads that mediate biological control of phytopathogens and plant growth promotion of crop plants. In Bacteria in Agrobiology: Plant Growth Responses; Springer: Berlin/Heidelberg, Germany, 2011; pp. 77–105. [Google Scholar]
- Audenaert, K.; Pattery, T.; Cornelis, P.; Höfte, M. Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: Role of salicylic acid, pyochelin, and pyocyanin. Mol. Plant Microbe Inter. 2002, 15, 1147–1156. [Google Scholar] [CrossRef] [Green Version]
- Maffei, M.E.; Gertsch, J.; Appendino, G. Plant volatiles: Production, function and pharmacology. Nat. Prod. Rep. 2011, 28, 1359–1380. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M.E. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 2015, 6, 151. [Google Scholar] [CrossRef]
- Chen, H.; Xiao, X.; Wang, J.; Wu, L.; Zheng, Z.; Yu, Z. Antagonistic effects of volatiles generated by Bacillus subtilis on spore germination and hyphal growth of the plant pathogen, Botrytis cinerea. Biotechnol. Lett. 2008, 30, 19–923. [Google Scholar] [CrossRef]
- Schink, B. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 2002, 81, 257–261. [Google Scholar] [CrossRef] [Green Version]
- Mee, M.T.; Collins, J.J.; Church, G.M.; Wang, H.H. Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. USA 2014, 111, E2149–E2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gralka, M.; Szabo, R.; Stocker, R.; Cordero, O.X. Trophic interactions and the drivers of microbial community assembly. Curr. Biol. 2020, 30, R1176–R1188. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Acker, H.; Van Dijck, P.; Coenye, T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014, 22, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Mousa, W.K.; Shearer, C.; Limay-Rios, V.; Ettinger, C.; Eisen, J.A.; Raizada, M.N. Root hair-endophyte stacking (RHESt) in an ancient Afro-Indian crop creates an unusual physicochemical barrier to trap pathogen (s). bioRxiv 2016, 071548. [Google Scholar] [CrossRef] [Green Version]
- Klein, M.N.; Kupper, K.C. Biofilm production by Aureobasidium pullulans improves biocontrol against sour rot in citrus. Food Microbiol. 2018, 69, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Madsen, J.S.; Røder, H.L.; Russel, J.; Sørensen, H.; Burmølle, M.; Sørensen, S.J. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Environ. Microbiol. 2016, 18, 2565–2574. [Google Scholar] [CrossRef]
- Tan, C.H.; Koh, K.S.; Xie, C.; Zhang, J.; Tan, X.H.; Lee, G.P.; Zhou, Y.; Ng, W.J.; Rice, S.A.; Kjelleberg, S. Community quorum sensing signalling and quenching: Microbial granular biofilm assembly. npj Biofilms Microbiomes 2015, 1, 1–9. [Google Scholar] [CrossRef]
- March, J.C.; Bentley, W.E. Quorum sensing and bacterial crosstalk in biotechnology. Curr. Opin. Biotechnol. 2004, 15, 495–502. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Jarosz, L.M.; Ovchinnikova, E.S.; Meijler, M.M.; Krom, B.P. Microbial spy games and host response: Roles of a Pseudomonas aeruginosa small molecule in communication with other species. PLoS Pathog. 2011, 7, e1002312. [Google Scholar] [CrossRef] [Green Version]
- Worrich, A.; König, S.; Miltner, A.; Banitz, T.; Centler, F.; Frank, K.; Thullner, M.; Harms, H.; Kästner, M.; Wicka, Y.W. Mycelium-like networks increase bacterial dispersal, growth, and biodegradation in a model ecosystem at various water potentials. Appl. Environ. Microbiol. 2016, 82, 2902–2908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, A.; Bindschedler, S.; Job, D.; Wick, L.Y.; Filippidou, S.; Kooli, W.M.; Verrecchia, E.P.; Junier, P. Exploiting the fungal highway: Development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol. Ecol. 2015, 91, fiv116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koeppel, A.F.; Wu, M. Species matter: The role of competition in the assembly of congeneric bacteria. ISME J. 2014, 8, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, M.M.; Levine, J.M. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 2010, 13, 1085–1093. [Google Scholar] [CrossRef]
- Wei, Z.; Yang, T.; Friman, V.P.; Xu, Y.; Shen, Q.; Jousset, A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 2015, 6, 8413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015, 6, 299. [Google Scholar] [CrossRef] [PubMed]
- Tata, A.; Perez, C.; Campos, M.L.; Bayfield, M.A.; Eberlin, M.N.; Ifa, D.R. Imprint desorption electrospray ionization mass spectrometry imaging for monitoring secondary metabolites production during antagonistic interaction of fungi. Anal. Chem. 2015, 87, 12298–12305. [Google Scholar] [CrossRef]
- Kumar, A. Microbial Biocontrol: Sustainable Agriculture and Phytopathogen Management; Springer Nature Chem: Cham, Switzerland, 2022; Volume 1, pp. 1–369. [Google Scholar]
- Eng, A.; Borenstein, E. Microbial community design: Methods, applications, and opportunities. Curr. Opin. Biotechnol. 2019, 58, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Mitter, B.; Pfaffenbichler, N.; Flavell, R.; Compant, S.; Antonielli, L.; Petric, A.; Berninger, T.; Naveed, M.; Sheibani-Tezerji, R.; von Maltzahn, G.; et al. A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 2017, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Tian, C.; Xiao, J.; Wei, L.; Tian, Y.; Liang, Z. Soil inoculation of Trichoderma asperellum M45a regulates rhizosphere microbes and triggers watermelon resistance to Fusarium wilt. AMB Express 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Alvindia, D.G.; Natsuaki, K.T. Evaluation of fungal epiphytes isolated from banana fruit surfaces for biocontrol of banana crown rot disease. Crop Prot. 2008, 27, 1200–1207. [Google Scholar] [CrossRef]
- Alvindia, D.G.; Natsuaki, K.T. Biocontrol activities of Bacillus amyloliquefaciens DGA14 isolated from banana fruit surface against banana crown rot-causing pathogens. Crop Prot. 2009, 28, 236–242. [Google Scholar] [CrossRef]
- Taqarort, N.; Echairi, A.; Chaussod, R.; Nouaim, R.; Boubaker, H.; Benaoumar, A.A.; Boudyach, E. Screening and identification of epiphytic yeasts with potential for biological control of green mold of citrus fruits. World J. Microbiol. Biotechnol. 2008, 24, 3031–3038. [Google Scholar] [CrossRef]
- Hammami, R.; Oueslati, M.; Smiri, M.; Nefzi, S.; Ruissi, M.; Comitini, F.; Romanazzi, G.; Cacciola, S.O.; Sadfi Zouaoui, N. Epiphytic Yeasts and Bacteria as Candidate Biocontrol Agents of Green and Blue Molds of Citrus Fruits. J. Fungi 2022, 8, 818. [Google Scholar] [CrossRef]
- Pereyra, M.M.; Díaz, M.A.; Soliz-Santander, F.F.; Poehlein, A.; Meinhardt, F.; Daniel, R.; Dib, J.R. Screening Methods for Isolation of Biocontrol Epiphytic Yeasts against Penicillium digitatum in Lemons. J. Fungi 2021, 7, 166. [Google Scholar] [CrossRef]
- Lorenzini, M.; Zapparoli, G. Epiphytic bacteria from withered grapes and their antagonistic effects on grape-rotting fungi. Int. J. Food Microbiol. 2020, 319, 108505. [Google Scholar] [CrossRef]
- Bleve, G.; Grieco, F.; Cozzi, G.; Logrieco, A.; Visconti, A. Isolation of epiphytic yeasts with potential for biocontrol of Aspergillus carbonarius and A. niger on grape. Int. J. Food Microbiol. 2006, 108, 204–209. [Google Scholar] [CrossRef]
- Pusey, P.L.; Stockwell, V.O.; Mazzola, M. Epiphytic bacteria and yeasts on apple blossoms and their potential as antagonists of Erwinia amylovora. Phytopathology 2009, 99, 571–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Niu, C.; Liu, B.; Yuan, Y.; Yue, T. Identification and characterization of epiphytic yeasts on apples in China. Rsc Adv. 2017, 7, 44766–44772. [Google Scholar] [CrossRef] [Green Version]
- Bösch, Y.; Britt, E.; Perren, S.; Naef, A.; Frey, J.E.; Bühlmann, A. Dynamics of the Apple Fruit Microbiome after Harvest and Implications for Fruit Quality. Microorganisms 2021, 9, 272. [Google Scholar] [CrossRef]
- Madbouly, A.K.; Elyousr, K.A.A.; Ismail, I.M. Biocontrol of Monilinia fructigena, causal agent of brown rot of apple fruit, by using endophytic yeasts. Biol. Control 2020, 144, 104239. [Google Scholar] [CrossRef]
- Hipol, R.M.; Magtoto, L.M.; Tamang, S.M.A.; Damatac, A.M.; Amor, M. Antioxidant activities of fungal endophytes isolated from strawberry Fragaria× ananassa fruit. Electron. J. Biol. 2014, 10, 107–112. [Google Scholar]
- Chen, C.; Cao, Z.; Li, J.; Tao, C.; Feng, Y.; Han, Y. A novel endophytic strain of Lactobacillus plantarum CM-3 with antagonistic activity against Botrytis cinerea on strawberry fruit. Biol. Control 2020, 148, 104306. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; Magalhães, K.T.; Lorenzetii, E.R.; Souza, T.P.; Schwan, R.F. A multiphasic approach for the identification of endophytic bacterial in strawberry fruit and their potential for plant growth promotion. Micro. Ecol. 2012, 63, 405–417. [Google Scholar] [CrossRef]
- Abdel-Rahim, I.R.; Abo-Elyousr, K.A. Using of endophytic Saccharomycopsis fibuligera and thyme oil for management of gray mold rot of guava fruits. Biol. Control 2017, 110, 124–131. [Google Scholar] [CrossRef]
- Krishnan, P.; Bhat, R.; Kush, A.; Ravikumar, P. Isolation and functional characterization of bacterial endophytes from Carica papaya fruits. J. Appl. Microbiol. 2012, 113, 308–317. [Google Scholar] [CrossRef]
- Compant, S.; Mitter, B.; Colli-Mull, J.G.; Gangl, H.; Sessitsch, A. Endophytes of grapevine flowers, berries, and seeds: Identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Micro. Ecol. 2011, 62, 188–197. [Google Scholar] [CrossRef]
- Hanin, N.A.; Fitriasari, P.D. Identification of Endophytic Fungi from Fruits and Seeds of Jambolana (Syzygium cumini L.) Skeels. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2019; Volume 276, p. 012060. [Google Scholar]
- Diskin, S.; Feygenberg, O.; Maurer, D.; Droby, S.; Prusky, D.; Alkan, N. Microbiome alterations are correlated with the occurrence of postharvest stem-end rot in mango fruit. Phytobiomes 2017, 1, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Ardanov, P.; Sessitsch, A.; Häggman, H.; Kozyrovska, N.; Pirttilä, A.M. Methylobacterium-induced endophyte community changes correspond with Protection of plants against pathogen attack. PLoS ONE 2012, 7, e46802. [Google Scholar] [CrossRef] [PubMed]
- Preto, G.; Martins, F.; Pereira, J.A.; Baptista, P. Fungal community in olive fruits of cultivars with different susceptibilities to anthracnose and selection of isolates to be used as biocontrol agents. Biol. Control 2017, 110, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Cruz, A.F.; Barka, G.D.; Sylla, J.; Reineke, A. Biocontrol of strawberry fruit infected by Botrytis cinerea: Effects on the microbial communities on fruit assessed by next-generation sequencing. J. Phytopathol. 2018, 166, 403–411. [Google Scholar] [CrossRef]
- Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Toamy, M.A.; Abdelfattaah, A.; Medina, S.; Freilich, S.; Wisniewski, M.; et al. Compositional shifts in the strawberry fruit microbiome in response to near-harvest application of Metschnikowia fructicola, a yeast biocontrol agent. Postharvest Biol. Technol. 2021, 175, 111469. [Google Scholar] [CrossRef]
- Biasi, A.; Zhimo, V.Y.; Kumar, A.; Abdelfattah, A.; Salim, S.; Feygenberg, O.; Wisniewski, M.; Droby, S. Changes in the Fungal Community Assembly of Apple Fruit Following Postharvest Application of the Yeast Biocontrol Agent Metschnikowia fructicola. Horticulturae 2021, 7, 360. [Google Scholar] [CrossRef]
| Fruits | Genera/Strains | Function | References |
|---|---|---|---|
| Epiphytic strains | |||
| Banana | Clonostachys byssicola, C. pallescens, Penicillium oxalicum, and Trichoderma harzianum | Biocontrol agent | [70] |
| Banana | Bacillus amyloliquefaciens | Biocontrol activity against crown-rot-causing pathogens | [71] |
| Citrus | Pichia anomala, Debaryomyces hansenii, Hanseniaspora guilliermondii | Biocontrol activity against P. digitatum | [72] |
| Citrus | Candida oleophila and Debaryomyces hansenii, Bacillus amyloliquefaciens, B. pumilus and B. subtilis | Antagonistic activity against Penicillium digitatum and P. italicum | [73] |
| Lemon | Clavispora lusitaniae | Antagonistic activity against Penicillium digitatum | [74] |
| Withered grapes | Bacillus, Brevibacillus, Curtobacterium, Micrococcus, Pseudomonas, Staphylococcus | Antagonistic effects on grape-rotting fungi | [75] |
| Grape berries | Issatchenkia orientalis, Metschnikowia pulcherrima, Kluyveromyces thermotolerans, Issatchenkia terricola and Candida incommunis, | Killer activity against Aspergillus carbonarius and A. niger | [76] |
| Apple blossoms | Pantoea agglomerans and Pseudomonas spp. Cryptococcus spp. | Biocontrol activity against Erwinia amylovora | [77] |
| Apple | Aureobasidium pullulans and Hanseniaspora uvarum | Not mentioned | [78] |
| Apple | Aureobasidium, Metschnikowia, and Rhodotorula | Not mentioned | [79] |
| Endophytic strains | |||
| Apple | Schwanniomyces vanrijiae, Galactomyces geotrichum, Pichia kudriavzevii, Debaryomyces hansenii, and Rhodotorula glutini | Biocontrol activity against Monilinia fructigena | [80] |
| Strawberry | Sporidiobolus sp., Rhodotorula sp., Pilidium concavum, Corynespora cassiicola, Neodeightonia subglobosa, Aspergillus awamori, and Aspergillus sp. | Antioxidant activity | [81] |
| Strawberry | Lactobacillus plantarum | Antagonistic activity against Botrytis cinerea | [82] |
| Strawberry | B. subtilis, Enterobacter sp., Pseudomonas sp. | Plant growth promotion | [83] |
| Guava | Saccharomycopsis fibuligera | Management of gray mold rot of guava | [84] |
| papaya | Kocuria, Acinetobacter, Enterobacter, Bacillus Staphylococcus | Not mentioned | [85] |
| Grapes | Bacillus cereus | Not mentioned | [86] |
| Jambolana | Neofusicoccum parvum, Pestalotiopsis | Not mentioned | [87] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, S.; Azevedo, L.C.B.; Pandey, J.; Khusharia, S.; Kumari, M.; Kumar, D.; Kaushalendra; Bhardwaj, N.; Teotia, P.; Kumar, A. Microbial Intervention: An Approach to Combat the Postharvest Pathogens of Fruits. Plants 2022, 11, 3452. https://doi.org/10.3390/plants11243452
Verma S, Azevedo LCB, Pandey J, Khusharia S, Kumari M, Kumar D, Kaushalendra, Bhardwaj N, Teotia P, Kumar A. Microbial Intervention: An Approach to Combat the Postharvest Pathogens of Fruits. Plants. 2022; 11(24):3452. https://doi.org/10.3390/plants11243452
Chicago/Turabian StyleVerma, Sargam, Lucas Carvalho Basilio Azevedo, Jyoti Pandey, Saksham Khusharia, Madhuree Kumari, Dharmendra Kumar, Kaushalendra, Nikunj Bhardwaj, Pratibha Teotia, and Ajay Kumar. 2022. "Microbial Intervention: An Approach to Combat the Postharvest Pathogens of Fruits" Plants 11, no. 24: 3452. https://doi.org/10.3390/plants11243452








