Histone Acetyltransferase GCN5 Affects Auxin Transport during Root Growth by Modulating Histone Acetylation and Gene Expression of PINs
Abstract
1. Introduction
2. Results
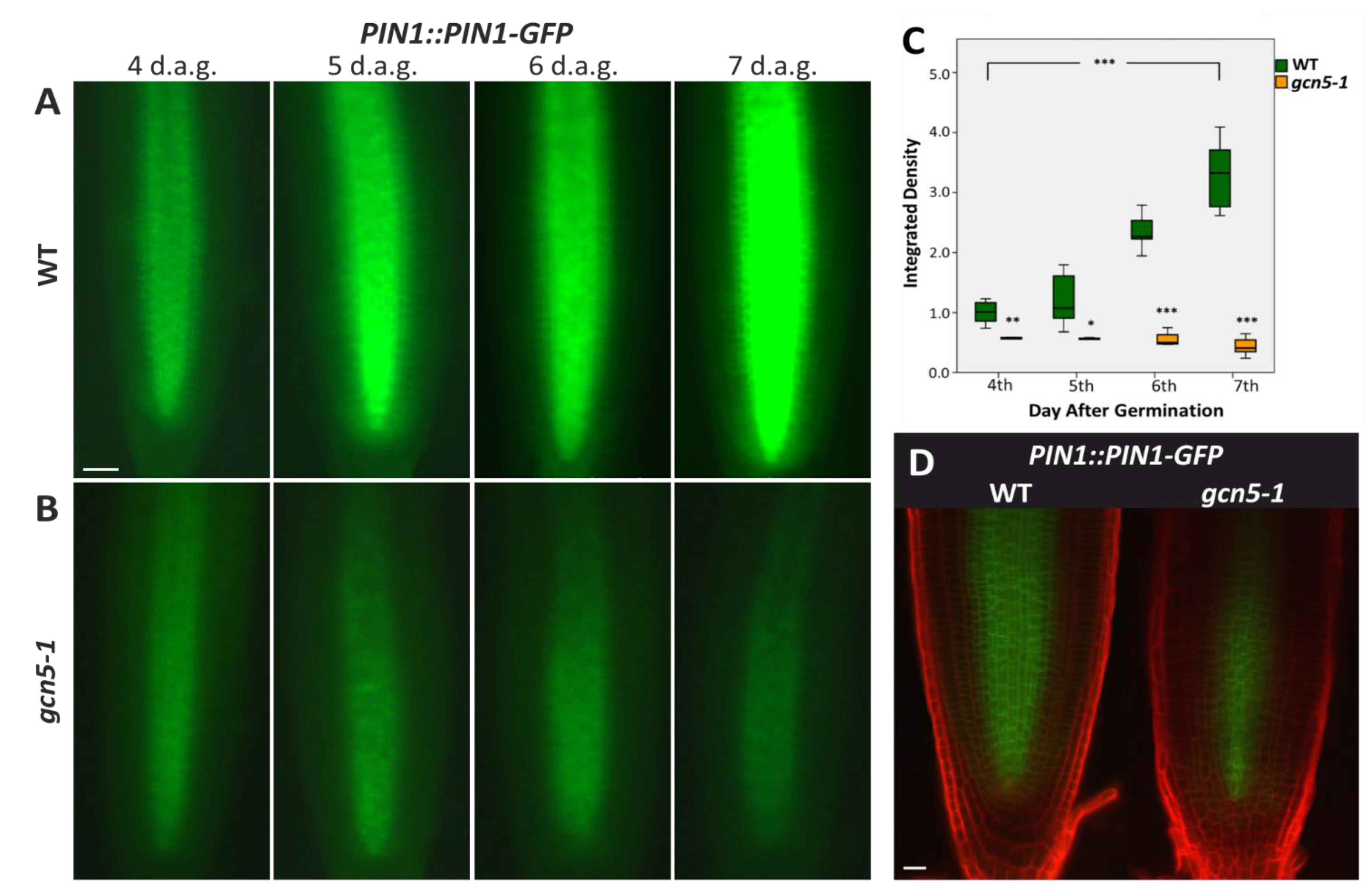
2.1. The Histone Acetyltransferase GCN5 Affects Auxin Transport during Early Root Growth
2.2. ADA2b Affects Auxin Transport during Early Root Growth
2.3. The Histone Acetyltransferase GCN5 and the Transcriptional Adaptor ADA2b Promote the Expression of PINs in the Root of Arabidopsis
2.4. GCN5 and ADA2b Affect Auxin Distribution at the Root Tip during Early Root Growth
2.5. GCN5 and ADA2b Affect the Expression of Genes Involved in Auxin Biosynthesis and Signaling
2.6. GCN5 and ADA2b Affect Histone H3K14 Acetylation on PIN Genomic Loci
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials and Growth
5.2. Genetic Analysis and Genotyping
5.3. Gene Expression Analysis
5.4. Microscopy
5.5. Chromatin Immunoprecipitation (ChIP)
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Michniewicz, M.; Brewer, P.B.; Friml, J.Í. Polar auxin transport and asymmetric auxin distribution. Arab. Book 2007, 5, e0108. [Google Scholar] [CrossRef]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Petrásek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertova, D.; Wisniewska, J.; Tadele, Z.; Kubes, M.; Covanova, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Wisniewska, J.; Xu, J.; Seifertová, D.; Brewer, P.B.; Ruzicka, K.; Billou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef]
- Zhou, J.J.; Luo, J. The PIN-FORMED Auxin Efflux Carriers in Plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Křeček, P.; Skůpa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazimalova, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef]
- Simon, S.; Skůpa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klima, P.; Carna, M.; Rolcik, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Vieten, A.; Vanneste, S.; Wisniewska, J.; Benková, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef] [PubMed]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin: A master regulator in plant root development. Plant Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Mao, Y.; Regier, M.K.; Triezenberg, S.J.; Thomashow, M.F. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001, 29, 1524–1533. [Google Scholar] [CrossRef]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef]
- Vlachonasios, K.; Poulios, S.; Mougiou, N. The Histone Acetyltransferase GCN5 and the Associated Coactivators ADA2: From Evolution of the SAGA Complex to the Biological Roles in Plants. Plants 2021, 10, 308. [Google Scholar] [CrossRef]
- Grasser, K.D.; Rubio, V.; Barneche, F. Multifaceted activities of the plant SAGA complex. Biochim. Biophys. Acta—Gene Regul. Mech. 2021, 1864, 194613. [Google Scholar] [CrossRef]
- Vlachonasios, K.E.; Thomashow, M.F.; Triezenberg, S.J. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 2003, 15, 626–638. [Google Scholar] [CrossRef]
- Bertrand, C.; Bergounioux, C.; Domenichini, S.; Delarue, M.; Zhou, D.-X. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 2003, 278, 28246–28251. [Google Scholar] [CrossRef]
- Kornet, N.; Scheres, B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 2009, 21, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Tsilimigka, F.; Poulios, S.; Mallioura, A.; Vlachonasios, K. ADA2b and GCN5 Affect Cytokinin Signaling by Modulating Histone Acetylation and Gene Expression during Root Growth of Arabidopsis thaliana. Plants 2022, 11, 1335. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Liu, Z.Z.; Wei, L.; Li, L.; Chen, S.; He, X.J. Three functionally redundant plant-specific paralogs are core subunits of the SAGA histone acetyltransferase complex in Arabidopsis. Mol. Plant 2021, 14, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Pavangadkar, K.A.; Thomashow, M.F.; Triezenberg, S.J. Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta 2006, 1759, 69–79. [Google Scholar] [CrossRef]
- Poulios, S.; Dadarou, D.; Gavriilidis, M.; Mougiou, N.; Kargios, N.; Maliori, V.; Hark, A.T.; Doonan, J.H.; Vlachonasios, K.E. The Transcriptional Adaptor Protein ADA3a Modulates Flowering of Arabidopsis thaliana. Cells 2021, 10, 904. [Google Scholar] [CrossRef]
- Hark, A.T.; Vlachonasios, K.E.; Pavangadkar, K.A.; Rao, S.; Gordon, H.; Adamakis, I.-D.; Kaldis, A.; Thomashow, M.F.; Triezenberg, S.J. Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim. Biophys. Acta—Gene Regul. Mech. 2009, 1789, 117–124. [Google Scholar] [CrossRef]
- Sieberer, T.; Hauser, M.T.; Seifert, G.J.; Luschnig, C. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr. Biol. 2003, 13, 837–842. [Google Scholar] [CrossRef]
- Gonin, M.; Bergougnoux, V.; Nguyen, T.D.; Gantet, P.; Champion, A. What Makes Adventitious Roots? Plants 2019, 8, 240. [Google Scholar] [CrossRef]
- Díaz-Sala, C. A Perspective on Adventitious Root Formation in Tree Species. Plants 2020, 9, 1789. [Google Scholar] [CrossRef]
- Poulios, S.; Vlachonasios, K.E. Synergistic action of GCN5 and CLAVATA1 in the regulation of gynoecium development in Arabidopsis thaliana. New Phytol. 2018, 220, 593–608. [Google Scholar] [CrossRef] [PubMed]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Omelyanchuk, N.A.; Kovrizhnykh, V.V.; Oshchepkova, E.A.; Pasternak, T.; Palme, K.; Mironova, V.V. A detailed expression map of the PIN1 auxin transporter in Arabidopsis thaliana root. BMC Plant Biol. 2016, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dai, X.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jurgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Kawashima, M.; Ichikawa, T.; Takahashi, N.; Shimada, H.; Manabe, K.; Matsui, M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004, 37, 471–483. [Google Scholar] [CrossRef]
- Dreher, K.A.; Brown, J.; Saw, R.E.; Callis, J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 2006, 18, 699–714. [Google Scholar] [CrossRef]
- Benhamed, M.; Bertrand, C.; Servet, C.; Zhou, D.-X. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 2006, 18, 2893–2903. [Google Scholar] [CrossRef]
- Earley, K.W.; Shook, M.S.; Brower-Toland, B.; Hicks, L.; Pikaard, C.S. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J. 2007, 52, 615–626. [Google Scholar] [CrossRef]
- Yamoune, A.; Cuyacot, A.R.; Zdarska, M.; Hejatko, J. Hormonal orchestration of root apical meristem formation and maintenance in Arabidopsis. J. Exp. Bot. 2021, 72, 6768–6788. [Google Scholar] [CrossRef] [PubMed]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Ugartechea-Chirino, Y.; Swarup, R.; Swarup, K.; Péret, B.; Whitworth, M.; Bennett, M.; Bougourd, S. The AUX1 LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana. Ann. Bot. 2010, 105, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, J.J.; Bandyopadhyay, A.; Lee, O.R.; Mravec, J.; Titapiwatanakun, B.; Sauer, M.; Makam, S.N.; Cheng, Y.; Bouchard, R.; Adamec, J.; et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 2007, 19, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Heidstra, R.; Sabatini, S. Plant and animal stem cells: Similar yet different. Nat. Rev. Mol. Cell Biol. 2014, 15, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Di Mambro, R.; Svolacchia, N.; Ioio, R.D.; Pierdonati, E.; Salvi, E.; Pedrazzini, E.; Vitale, A.; Perilli, S.; Sozzani, R.; Benfey, P.N.; et al. The lateral root cap acts as an auxin sink that controls meristem size. Curr. Biol. 2019, 29, 1199–1205. [Google Scholar] [CrossRef]
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 2018, 47, 306–318. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Benfey, P.N. Plant stem cell niches: Standing the test of time. Cell 2008, 132, 553–557. [Google Scholar] [CrossRef]
- Salvi, E.; Di Mambro, R.; Sabatini, S. Dissecting mechanisms in root growth from the transition zone perspective. J. Exp. Bot. 2020, 71, 2390–2396. [Google Scholar] [CrossRef]
- Weiste, C.; Dröge-Laser, W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 2014, 5, 3883. [Google Scholar] [CrossRef]
- Anzola, J.M.; Sieberer, T.; Ortbauer, M.; Butt, H.; Korbei, B.; Weinhofer, I.; Müllner, A.E.; Luschnig, C. Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. USA 2010, 107, 10308–10313. [Google Scholar] [CrossRef] [PubMed]
- Pernisová, M.; Klíma, P.; Horák, J.; Válková, M.; Malbeck, J.; Souček, P.; Reichman, P.; Hoyerová, K.; Dubová, J.; Friml, J.; et al. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. USA 2009, 106, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, K.; Simásková, M.; Duclercq, J.; Petrásek, J.; Zazímalová, E.; Simon, S.; Friml, J.; Van Montagu, M.C.; Benková, E. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 2009, 106, 4284–4289. [Google Scholar] [CrossRef] [PubMed]
- Simaskova, M.; O’Brien, J.A.; Khan, M.; Van Noorden, G.; Otvos, K.; Vieten, A.; De Clercq, I.; Van Haperen, J.M.A.; Cuesta, C.; Hoyerova, K.; et al. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat. Commun. 2015, 6, 8717. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–43. [Google Scholar]
- Poulios, S.; Vlachonasios, K.E. Synergistic action of histone acetyltransferase GCN5 and receptor CLAVATA1 negatively affects ethylene responses in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 905–918. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poulios, S.; Tsilimigka, F.; Mallioura, A.; Pappas, D.; Seira, E.; Vlachonasios, K. Histone Acetyltransferase GCN5 Affects Auxin Transport during Root Growth by Modulating Histone Acetylation and Gene Expression of PINs. Plants 2022, 11, 3572. https://doi.org/10.3390/plants11243572
Poulios S, Tsilimigka F, Mallioura A, Pappas D, Seira E, Vlachonasios K. Histone Acetyltransferase GCN5 Affects Auxin Transport during Root Growth by Modulating Histone Acetylation and Gene Expression of PINs. Plants. 2022; 11(24):3572. https://doi.org/10.3390/plants11243572
Chicago/Turabian StylePoulios, Stylianos, Foteini Tsilimigka, Areti Mallioura, Dimitris Pappas, Eleftheria Seira, and Konstantinos Vlachonasios. 2022. "Histone Acetyltransferase GCN5 Affects Auxin Transport during Root Growth by Modulating Histone Acetylation and Gene Expression of PINs" Plants 11, no. 24: 3572. https://doi.org/10.3390/plants11243572
APA StylePoulios, S., Tsilimigka, F., Mallioura, A., Pappas, D., Seira, E., & Vlachonasios, K. (2022). Histone Acetyltransferase GCN5 Affects Auxin Transport during Root Growth by Modulating Histone Acetylation and Gene Expression of PINs. Plants, 11(24), 3572. https://doi.org/10.3390/plants11243572







