Characterization of an Arabidopsis Defensin-like Gene Conferring Resistance against Nematodes
Abstract
1. Introduction
2. Results
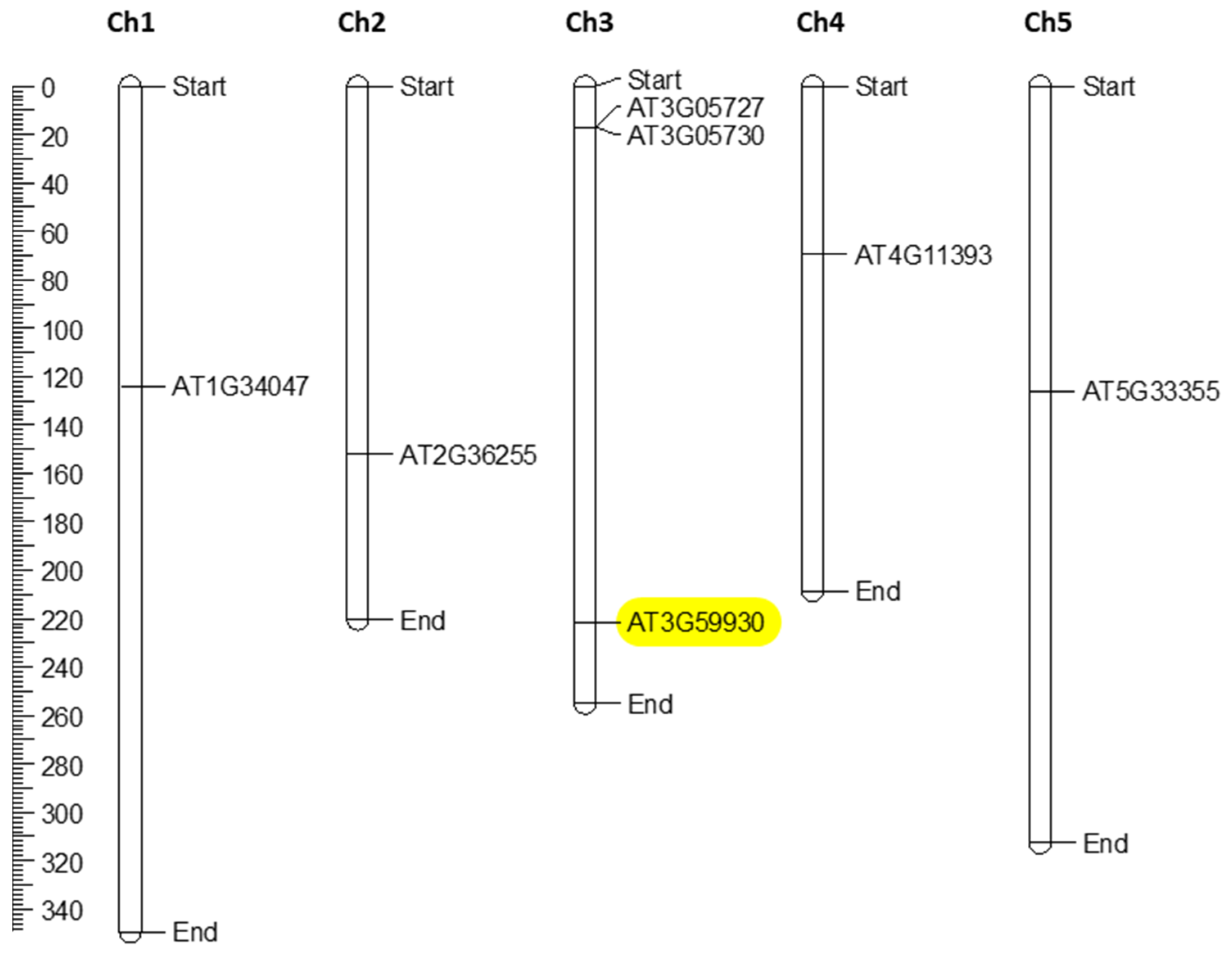
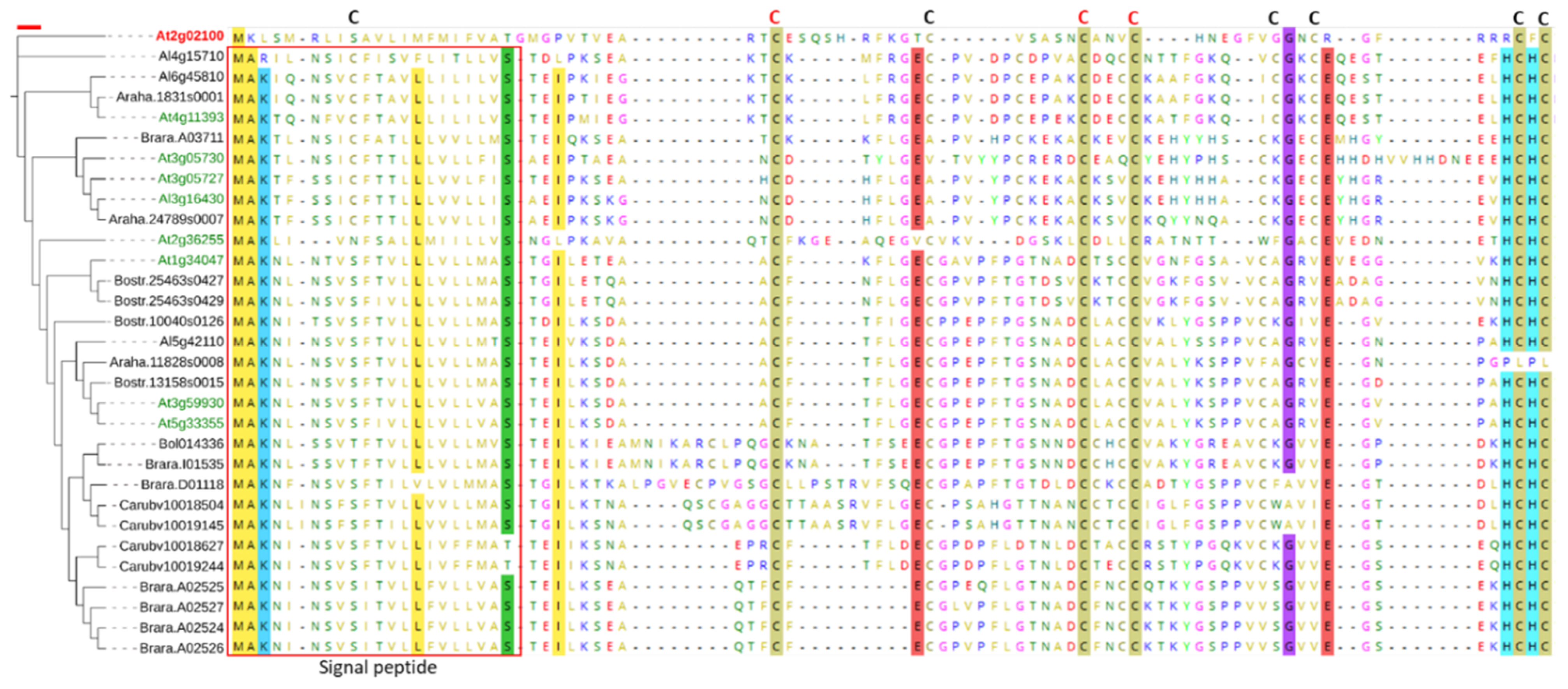
2.1. In Silico Characterization of CRP0770 Genes
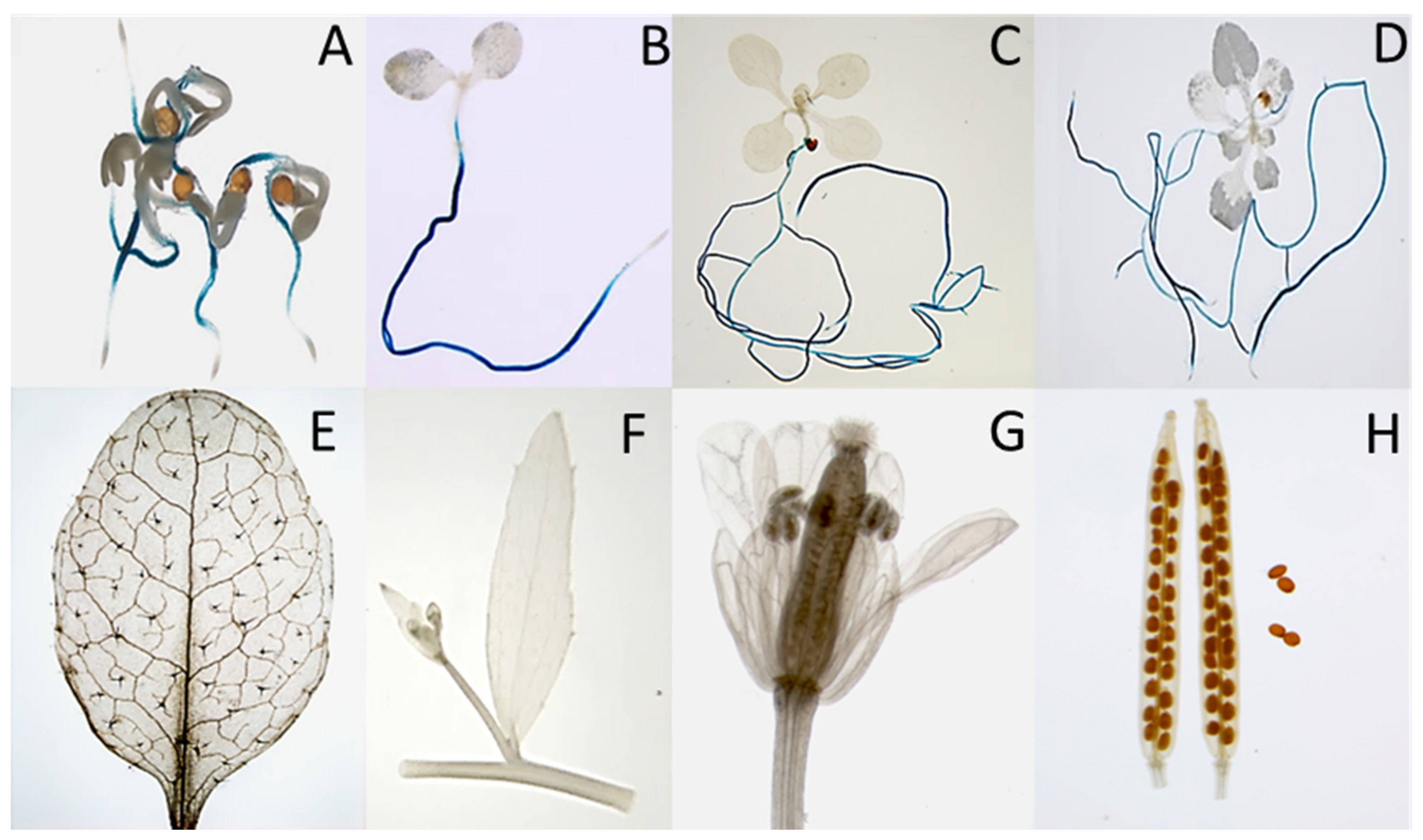
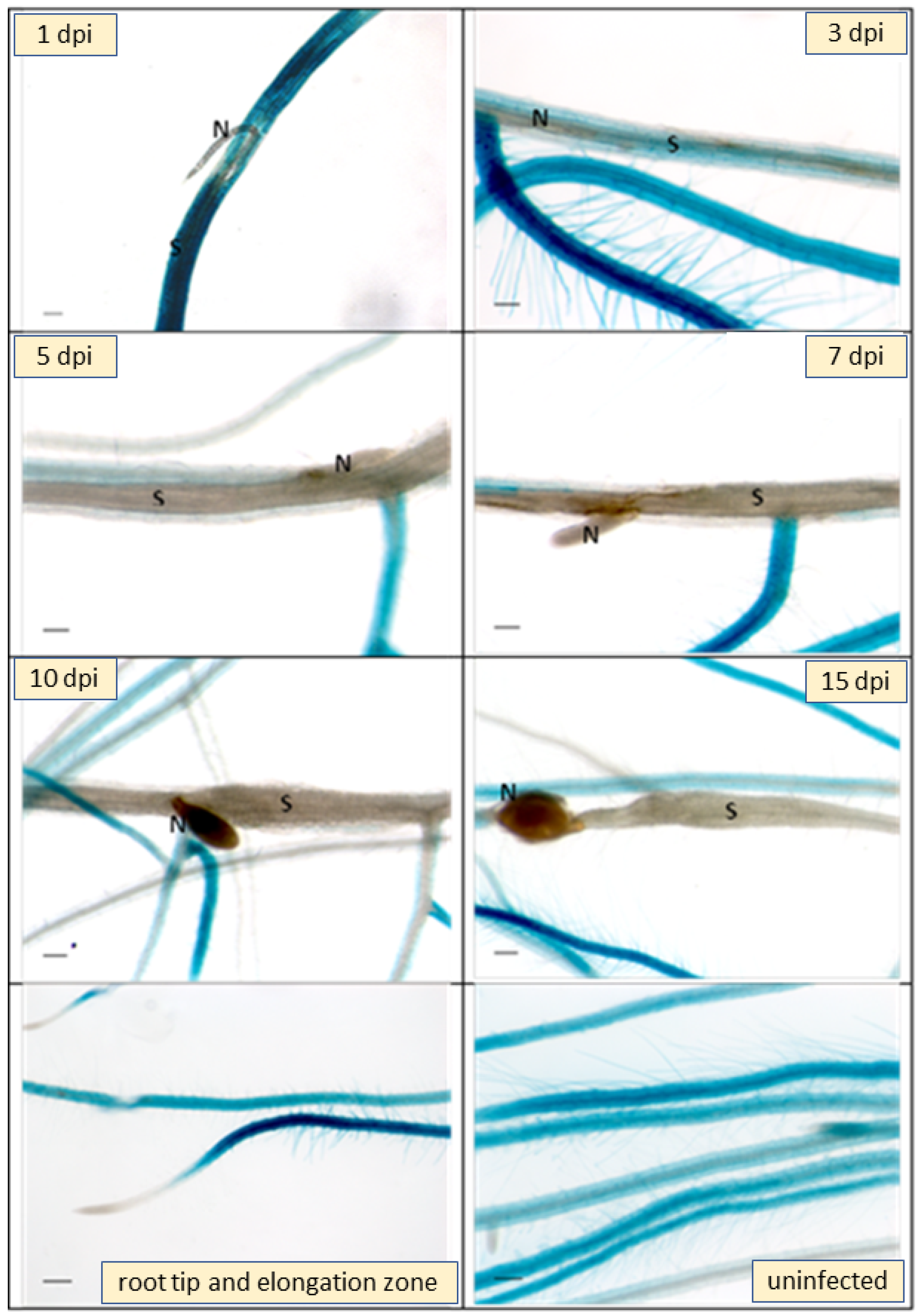
2.2. Expression Analysis of At3g59930
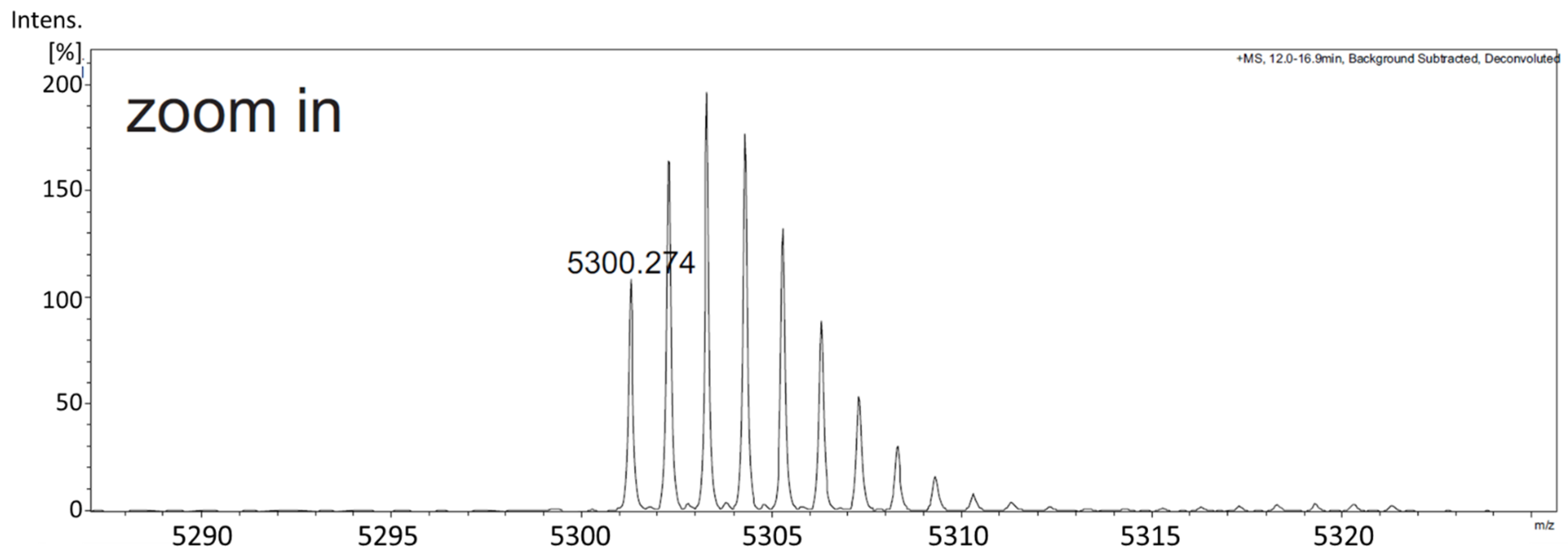
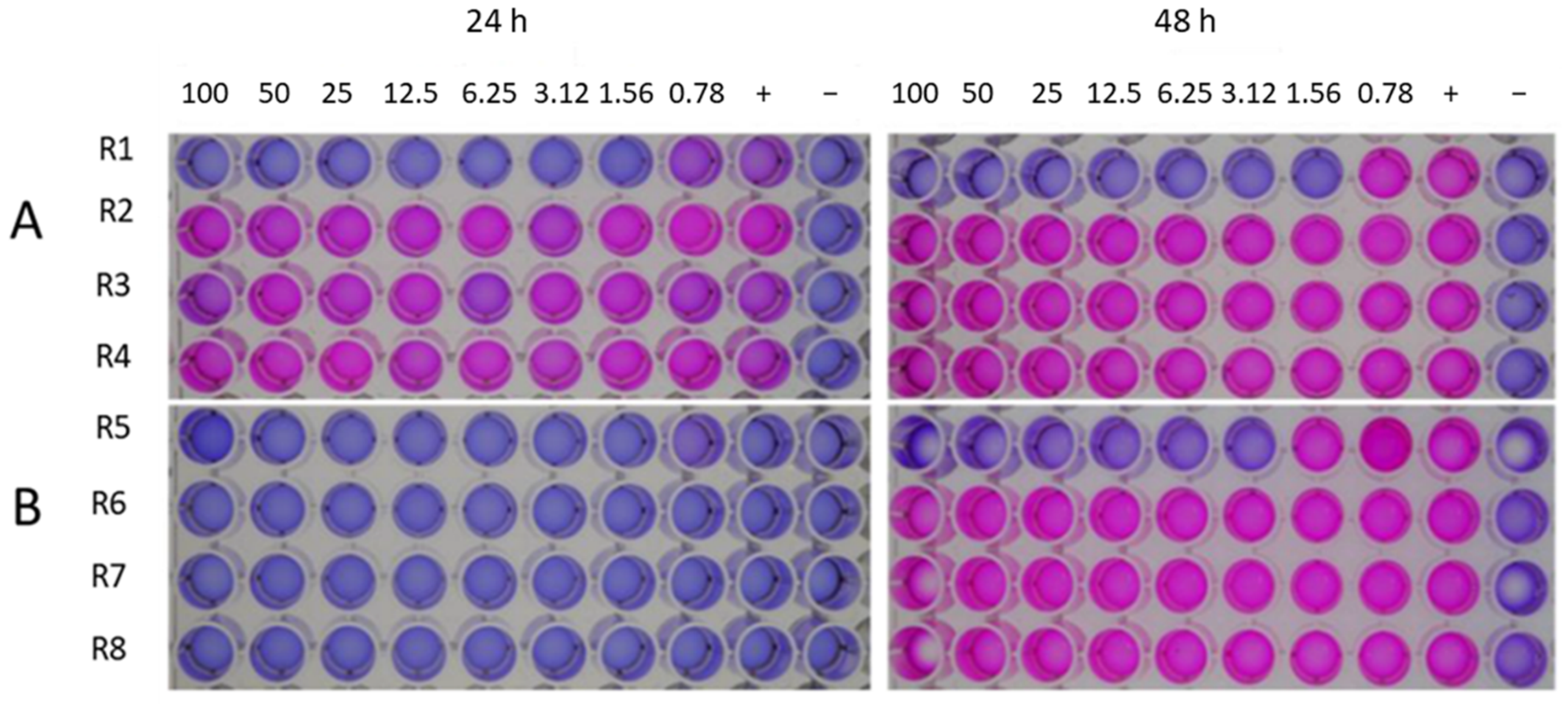
2.3. In Vitro Antimicrobial Assays
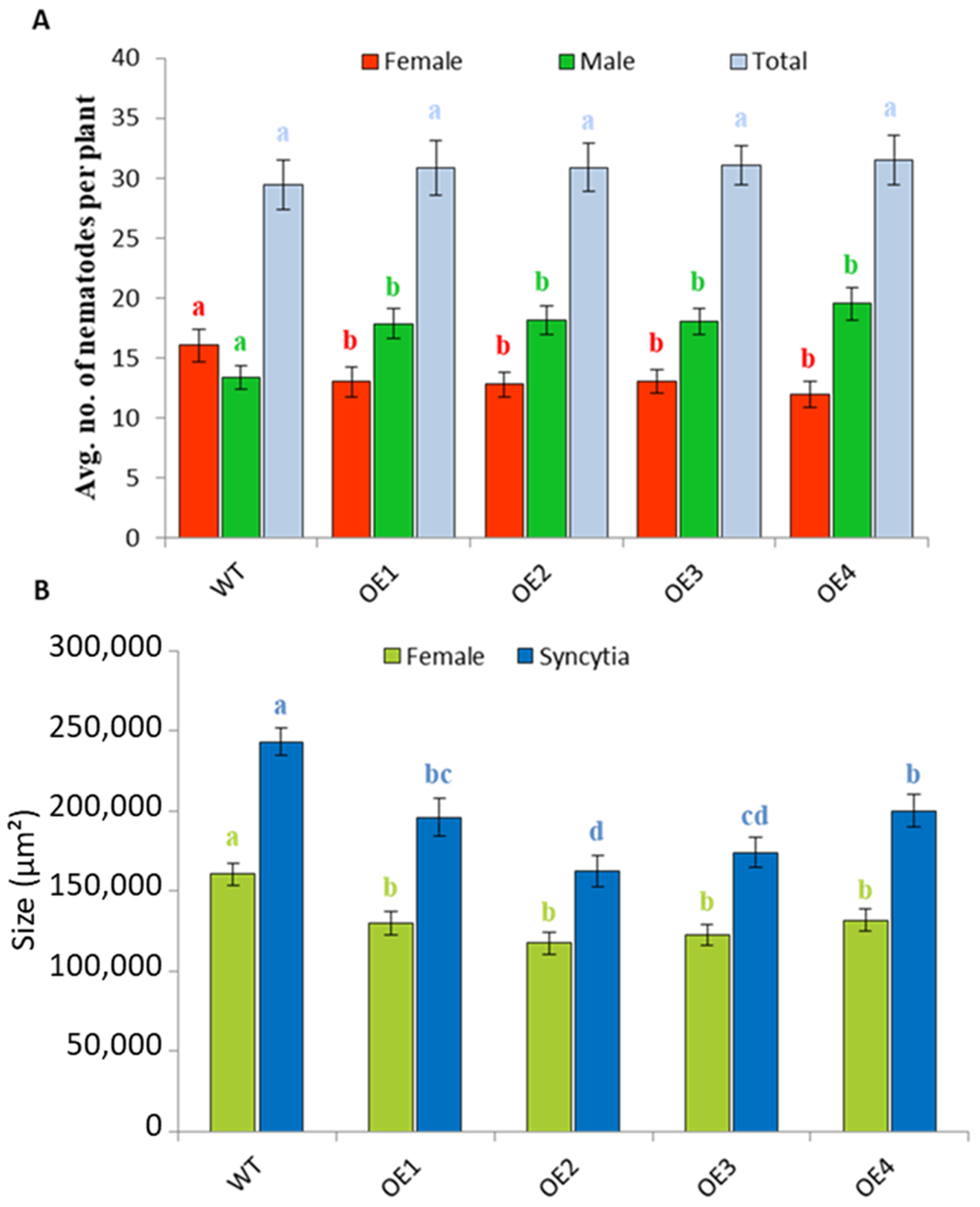
2.4. Overexpression of At3g59930
3. Discussion
3.1. In Silico Characterization of CRP0770 Genes
3.2. Expression
3.3. Antimicrobial Activity and Effect of Overexpression
4. Materials and Methods
4.1. Cloning of Vectors
4.2. Plant Material and Growth Conditions
4.3. Arabidopsis Transformation
4.4. RNA Isolation
4.5. Reverse Transcriptase PCR (RT-PCR)
4.6. RNAseq
4.7. GUS Reporter Analysis
4.8. Nematode Resistance Tests
4.9. Fungal Infection Assays
4.10. Expression and Purification of At3g59930 Peptide
4.11. Antimicrobial Assays
4.12. In Silico Analysis of CRP0770 Genes
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMP | antimicrobial peptide |
| CRP | cysteine-rich peptide |
| DEFL | defensin-like peptide |
| dpi | days post inoculation |
| IPTG | Isopropyl-β-D-thiogalactopyranosid |
| CV | column volume |
| TEV | tobacco etch virus |
| PAGE | polyacrylamide gel electrophoresis |
References
- Bohlmann, H. Introductory chapter on the basic biology of cyst nematodes. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2015; pp. 33–59. ISBN 9780124171619. [Google Scholar]
- Jones, M.G.K.; Northcote, D.H. Nematode-induced syncytium-A multinucleate transfer cell. J. Cell Sci. 1972, 10, 789–809. [Google Scholar] [CrossRef]
- Sobczak, M.; Golinowski, W. The Structure of Syncytia. In Genomics and Molecular Genetics of Plant-Nematode Interaction; Springer: Dordrecht, The Netherlands, 2011; pp. 61–82. ISBN 9789401155960. [Google Scholar]
- Ali, M.A.; Azeem, F.; Li, H.; Bohlmann, H. Smart parasitic nematodes use multifaceted strategies to parasitize plants. Front. Plant Sci. 2017, 8, 1699. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, K.; Golecki, B.; Gerdes, L.; Heinen, P.; Szakasits, D.; Durachko, D.M.; Cosgrove, D.J.; Kreil, D.P.; Puzio, P.S.; Bohlmann, H.; et al. Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J. 2006, 48, 98–112. [Google Scholar] [CrossRef]
- Bohlmann, H.; Sobczak, M. The plant cell wall in the feeding sites of cyst nematodes. Front. Plant Sci. 2014, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Siddique, S.; Sobczak, M.; Tenhaken, R.; Grundler, F.M.W.; Bohlmann, H. Cell wall ingrowths in nematode induced syncytia require UGD2 and UGD3. PLoS ONE 2012, 7, e41515. [Google Scholar] [CrossRef] [PubMed]
- Sobczak, M.; Golinowski, W.; Grundler, F.M. Changes in the structure of Arabidopsis thaliana roots induced during development of males of the plant parasitic nematode Heterodera schachtii. Eur. J. Plant Pathol. 1997, 97, 131–141. [Google Scholar] [CrossRef]
- Niebel, A.; de Almeida Engler, J.; Hemerly, A.; Ferreira, P.; Inzé, D.; Van Montagu, M.; Gheysen, G. Induction of cdc2a and cyc1At expression in Arabidopsis thaliana during early phases of nematode-induced feeding cell formation. Plant J. 1996, 10, 1037–1043. [Google Scholar] [CrossRef]
- Jaubert, S.; Ledger, T.N.; Laffaire, J.B.; Piotte, C.; Abad, P.; Rosso, M.N. Direct identification of stylet secreted proteins from root-knot nematodes by a proteomic approach. Mol. Biochem. Parasitol. 2002, 121, 205–211. [Google Scholar] [CrossRef]
- Hewezi, T.; Baum, T.J. Manipulation of plant cells by cyst and root-knot nematode effectors. Mol. Plant-Microbe Interact. 2013, 26, 9–16. [Google Scholar] [CrossRef]
- Gilchrist, D.G. Programmed cell death in plant disease: The purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 1998, 36, 393–414. [Google Scholar] [CrossRef]
- Piffanelli, P.; Devoto, A.; Schulze-Lefert, P. Defence signalling pathways in cereals. Curr. Opin. Plant Biol. 1999, 2, 295–300. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, H.; Apel, K. Thionins. Annu. Rev. Plant Biol. 1991, 42, 227–240. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Terras, F.; Cammue, B.; Osborn, R.W. Plant Defensins: Novel Antimicrobial Peptides as Components of the Host Defense System. Plant Physiol. 1995, 108, 1353–1358. [Google Scholar] [CrossRef]
- Garcia-Olmedo, F.; Molina, A.; Segura, A.; Moreno, M. The defensive role of nonspecific lipid-transfer proteins in plants. Trends Microbiol. 1995, 3, 72–74. [Google Scholar] [CrossRef]
- Cammue, B.P.; De Bolle, M.F.; Terras, F.R.; Proost, P.; Van Damme, J.; Rees, S.B.; Vanderleyden, J.; Broekaert, W.F. Isolation and Characterization of a Novel Class of Plant Antimicrobial Peptides from Mirabilis jalapa L. seeds. J. Biol. Chem. 1992, 267, 2228–2233. [Google Scholar] [CrossRef]
- Nielsen, K.K.; Nielsen, J.E.; Madrid, S.M.; Mikkelsen, J.D. Characterization of a new antifungal chitin-binding peptide from sugar beet leaves. Plant Physiol. 1997, 113, 83–91. [Google Scholar] [CrossRef][Green Version]
- Craik, D.J. Host-defense activities of cyclotides. Toxins 2012, 4, 139–156. [Google Scholar] [CrossRef]
- Segura, A.; Moreno, M.; Madueño, F.; Molina, A.; García-olmedo, F. Snakin-1, a Peptide from Potato That Is Active Against Plant Pathogens. Mol. Plant-Microbe Interact. 1999, 12, 16–23. [Google Scholar] [CrossRef]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Sathoff, A.E.; Velivelli, S.; Shah, D.M.; Samac, D.A. Plant defensin peptides have antifungal and antibacterial activity against human and plant pathogens. Phytopathology 2019, 109, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-C.; Lin, C.-Y.; Kuan, C.-C.; Sung, H.-Y.; Chen, C.-S. A Novel Defensin Encoded by a Mungbean cDNA Exhibits Insecticidal Activity against Bruchid. J. Agric. Food Chem. 2003, 51, 530. [Google Scholar] [CrossRef][Green Version]
- Mirouze, M.; Sels, J.; Richard, O.; Czernic, P.; Loubet, S.; Jacquier, A.; François, I.E.J.A.; Cammue, B.P.A.; Lebrun, M.; Berthomieu, P.; et al. A putative novel role for plant defensins: A defensin from the zinc hyper-accumulating plant, Arabidopsis halleri, confers zinc tolerance. Plant J. 2006, 47, 329–342. [Google Scholar] [CrossRef]
- Bloch, C.; Richardson, M. A new family of small (5 kDa) protein inhibitors of insect α-amylases from seeds or sorghum (Sorghum bicolor (L) Moench) have sequence homologies with wheat γ-purothionins. FEBS Lett. 1991, 279, 101–104. [Google Scholar] [CrossRef]
- Melo, F.R.; Rigden, D.J.; Franco, O.L.; Mello, L.V.; Ary, M.B.; Grossi-De-Sa, M.F.; Bloch, C., Jr. Inhibition of trypsin by cowpea thionin: Characterization, molecular modeling, and docking. Proteins Struct. Funct. Genet. 2002, 48, 311–319. [Google Scholar] [CrossRef]
- Coninck, B.D.E.; Cammue, B.P.A.; Thevissen, K. Modes of antifungal action and in planta functions of plant defensins and defensin-like peptides. Fungal Biol. Rev. 2013, 26, 109–120. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, M.; Zhang, Z.; Ren, L.; Du, L.; Zhang, B.; Xu, H.; Xin, Z. Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct. Integr. Genom. 2011, 11, 63–70. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J.; Cammue, B.P.A.; Thevissen, K. Plant defensins. Planta 2002, 216, 193–202. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef]
- Penninckx, I.A.M.A.; Thomma, B.P.H.J.; Buchala, A.; Métraux, J.P.; Broekaert, W.F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 1998, 10, 2103–2113. [Google Scholar] [CrossRef]
- De Coninck, B.M.A.; Sels, J.; Venmans, E.; Thys, W.; Goderis, I.J.W.M.; Carron, D.; Delauré, S.L.; Cammue, B.P.A.; De Bolle, M.F.C.; Mathys, J. Arabidopsis thaliana plant defensin AtPDF1.1 is involved in the plant response to biotic stress. New Phytol. 2010, 187, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, P.Y.; Cheng, C.P.; Koh, K.W.; Chan, M.T. The Arabidopsis defensin gene, AtPDF1.1, mediates defence against Pectobacterium carotovorum subsp. carotovorum via an iron-withholding defence system. Sci. Rep. 2017, 7, 9175. [Google Scholar] [CrossRef]
- Luo, J.S.; Yang, Y.; Gu, T.; Wu, Z.; Zhang, Z. The Arabidopsis defensin gene AtPDF2.5 mediates cadmium tolerance and accumulation. Plant Cell Environ. 2019, 42, 2681–2695. [Google Scholar] [CrossRef]
- Luo, J.S.; Gu, T.; Yang, Y.; Zhang, Z. A non-secreted plant defensin AtPDF2.6 conferred cadmium tolerance via its chelation in Arabidopsis. Plant Mol. Biol. 2019, 100, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, K.A.T.; Graham, M.A.; Paape, T.D.; Vandenbosch, K.A. Genome Organization of More Than 300 Defensin-Like Genes in Arabidopsis 1 [w]. Plant Physiol. 2005, 138, 600–610. [Google Scholar] [CrossRef]
- Takeuchi, H.; Higashiyama, T. A Species-Specific Cluster of Defensin-Like Genes Encodes Diffusible Pollen Tube Attractants in Arabidopsis. PLoS Biol. 2012, 10, e1001449. [Google Scholar] [CrossRef]
- Amien, S.; Kliwer, I.; Márton, M.L.; Debener, T.; Geiger, D.; Becker, D.; Dresselhaus, T. Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol. 2010, 8, e1000388. [Google Scholar] [CrossRef]
- Szakasits, D.; Heinen, P.; Wieczorek, K.; Hofmann, J.; Wagner, F.; Kreil, D.P.; Sykacek, P.; Grundler, F.M.W.; Bohlmann, H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 2009, 57, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jin, X.; Zhao, Y.; Dong, Q.; Jiang, H.; Ma, Q. Evolution of the defensin-like gene family in grass genomes. J. Genet. 2016, 95, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Murat, F.; Louis, A.; Maumus, F.; Armero, A.; Cooke, R.; Quesneville, H.; Crollius, H.R.; Salse, J. Understanding Brassicaceae evolution through ancestral genome reconstruction. Genome Biol. 2015, 16, 262. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, M.; Silverstein, K.A.T.; Nallu, S.; Wang, L.; Botanga, C.J.; Gomez, S.K.; Costa, L.M.; Harrison, M.J.; Samac, D.A.; Glazebrook, J.; et al. Spatio-Temporal Expression Patterns of Arabidopsis thaliana and Medicago truncatula Defensin-Like Genes. PLoS ONE 2013, 8, e58992. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, M.; Ijakipour, H.; Stamboulis, A. Defensin-Like Peptides and Their Antimicrobial Activity in Free-Form and Immobilized on Material Surfaces. In Peptide Synthesis; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Parisi, K.; Shafee, T.M.A.; Quimbar, P.; van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. The evolution, function and mechanisms of action for plant defensins. Semin. Cell Dev. Biol. 2019, 88, 107–118. [Google Scholar] [CrossRef]
- Sathoff, A.E.; Samac, D.A. Antibacterial activity of plant defensins. Mol. Plant-Microbe Interact. 2019, 32, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ip, D.C.W.; Ng, T.B.; Chan, Y.S.; Fang, F.; Pan, W.L. A defensin-like peptide from Phaseolus vulgaris cv. “King Pole Bean”. Food Chem 2012, 135, 408–414. [Google Scholar] [CrossRef]
- Wu, X.; Sun, J.; Zhang, G.; Wang, H.; Ng, T.B. An antifungal defensin from Phaseolus vulgaris cv. “Cloud Bean”. Phytomedicine 2011, 18, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Oddepally, R.; Guruprasad, L. Isolation, purification, and characterization of a stable defensin-like antifungal peptide from Trigonella foenum-graecum (fenugreek) seeds. Biochemistry 2015, 80, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ng, T.B. Gymnin, a potent defensin-like antifungal peptide from the Yunnan bean (Gymnocladus chinensis Baill). Peptides 2003, 24, 963–968. [Google Scholar] [CrossRef]
- Li, F.; Gao, Z.; Wang, K.; Zhao, Y.; Wang, H.; Zhao, M.; Zhao, Y.; Bai, L.; Yu, Z.; Yang, X. A novel defensin-like peptide contributing to antimicrobial and antioxidant capacity of the tick Dermacentor silvarum (Acari: Ixodidae). Exp. Appl. Acarol. 2021, 83, 271–283. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, Q.; Wang, Q.; Chen, L.; Liu, Y.; Cong, M.; Wu, H.; Li, F.; Ji, C.; Zhao, J. A defensin-like antimicrobial peptide from the manila clam Ruditapes philippinarum: Investigation of the antibacterial activities and mode of action. Fish Shellfish Immunol. 2018, 80, 274–280. [Google Scholar] [CrossRef]
- Grundler, F.B.M.; Wyss, U. Influence of Changes in the Nurse Cell System (Syncytium) on Sex Determination and Development of the Cyst Nematode Heterodera schachtii: Total Amounts of Proteins and Amino Acids. Phytopathology 1991, 81, 70–74. [Google Scholar] [CrossRef]
- Lay, F.; Anderson, M. Defensins—Components of the Innate Immune System in Plants. Curr. Protein Pept. Sci. 2005, 6, 85–101. [Google Scholar] [CrossRef]
- Stotz, H.U.; Thomson, J.G.; Wang, Y. Plant defensins: Defense, development and application. Plant Signal. Behav. 2009, 4, 1010–1012. [Google Scholar] [CrossRef]
- Allen, A.; Snyder, A.K.; Preuss, M.; Nielsen, E.E.; Shah, D.M.; Smith, T.J. Plant defensins and virally encoded fungal toxin KP4 inhibit plant root growth. Planta 2008, 227, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.J.; Deng, J.S.; Chen, H.J.; Huang, S.S.; Liao, J.C.; Hou, W.C.; Lin, Y.H. Defensin protein from sweet potato (Ipomoea batatas [L.] Lam ’Tainong 57’) storage roots exhibits antioxidant activities in vitro and ex vivo. Food Chem. 2012, 135, 861–867. [Google Scholar] [CrossRef]
- Van De Mortel, J.E.; Villanueva, L.A.; Schat, H.; Kwekkeboom, J.; Coughlan, S.; Moerland, P.D.; Van Themaat, E.V.L.; Koornneef, M.; Aarts, M.G.M. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2006, 142, 1127–1147. [Google Scholar] [CrossRef]
- Ali, M.A.; Shah, K.H.; Bohlmann, H. pMAA-Red: A new pPZP-derived vector for fast visual screening of transgenic Arabidopsis plants at the seed stage. BMC Biotechnol. 2012, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Bogomolovas, J.; Simon, B.; Sattler, M.; Stier, G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 2009, 64, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Sijmons, P.C.; Grundler, F.M.; von Mende, N.; Burrows, P.R.; Wyss, U. Arabidopsis thaliana as a new model host for plant-parasitic nematodes. Plant J. 1991, 1, 245–254. [Google Scholar] [CrossRef]
- Holsters, M.; de Waele, D.; Depicker, A.; Messens, E.; van Montagu, M.; Schell, J. Transfection and transformation of Agrobacterium tumefaciens. MGG Mol. Gen. Genet. 1978, 163, 181–187. [Google Scholar] [CrossRef]
- Logemann, E.; Birkenbihl, R.P.; Ülker, B.; Somssich, I.E. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2006, 2, 16. [Google Scholar] [CrossRef]
- Jefferson, R.A. The GUS reporter gene system. Nature 1989, 342, 837–838. [Google Scholar] [CrossRef] [PubMed]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2016, 11, 753. [Google Scholar] [CrossRef]
- Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Gharahdaghi, F.; Weinberg, C.R.; Meagher, D.A.; Imai, B.S.; Mische, S.M. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 1999, 20, 601–605. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Terras, F.R.G.; Cammue, B.P.A.; Vanderleyden, J. An automated quantitative assay for fungal growth inhibition. FEMS Microbiol. Lett. 1990, 69, 55–59. [Google Scholar] [CrossRef]
- Sarker, S.D.; Nahar, L.; Kumarasamy, Y. Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods 2007, 42, 321–324. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef]
- Voorrips, R.E. Computer Note MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J. Genome analysis GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hawamda, A.I.M.; Reichert, S.; Ali, M.A.; Nawaz, M.A.; Austerlitz, T.; Schekahn, P.; Abbas, A.; Tenhaken, R.; Bohlmann, H. Characterization of an Arabidopsis Defensin-like Gene Conferring Resistance against Nematodes. Plants 2022, 11, 280. https://doi.org/10.3390/plants11030280
Hawamda AIM, Reichert S, Ali MA, Nawaz MA, Austerlitz T, Schekahn P, Abbas A, Tenhaken R, Bohlmann H. Characterization of an Arabidopsis Defensin-like Gene Conferring Resistance against Nematodes. Plants. 2022; 11(3):280. https://doi.org/10.3390/plants11030280
Chicago/Turabian StyleHawamda, Abdalmenem I. M., Susanne Reichert, Muhammad Amjad Ali, Muhammad Amjad Nawaz, Tina Austerlitz, Patricia Schekahn, Amjad Abbas, Raimund Tenhaken, and Holger Bohlmann. 2022. "Characterization of an Arabidopsis Defensin-like Gene Conferring Resistance against Nematodes" Plants 11, no. 3: 280. https://doi.org/10.3390/plants11030280
APA StyleHawamda, A. I. M., Reichert, S., Ali, M. A., Nawaz, M. A., Austerlitz, T., Schekahn, P., Abbas, A., Tenhaken, R., & Bohlmann, H. (2022). Characterization of an Arabidopsis Defensin-like Gene Conferring Resistance against Nematodes. Plants, 11(3), 280. https://doi.org/10.3390/plants11030280






