Identification of Novel Candidate Genes Involved in Apple Cuticle Integrity and Russeting-Associated Triterpene Synthesis Using Metabolomic, Proteomic, and Transcriptomic Data
Abstract
:1. Introduction
2. Results
2.1. Phenotypic and Genetic Characterization
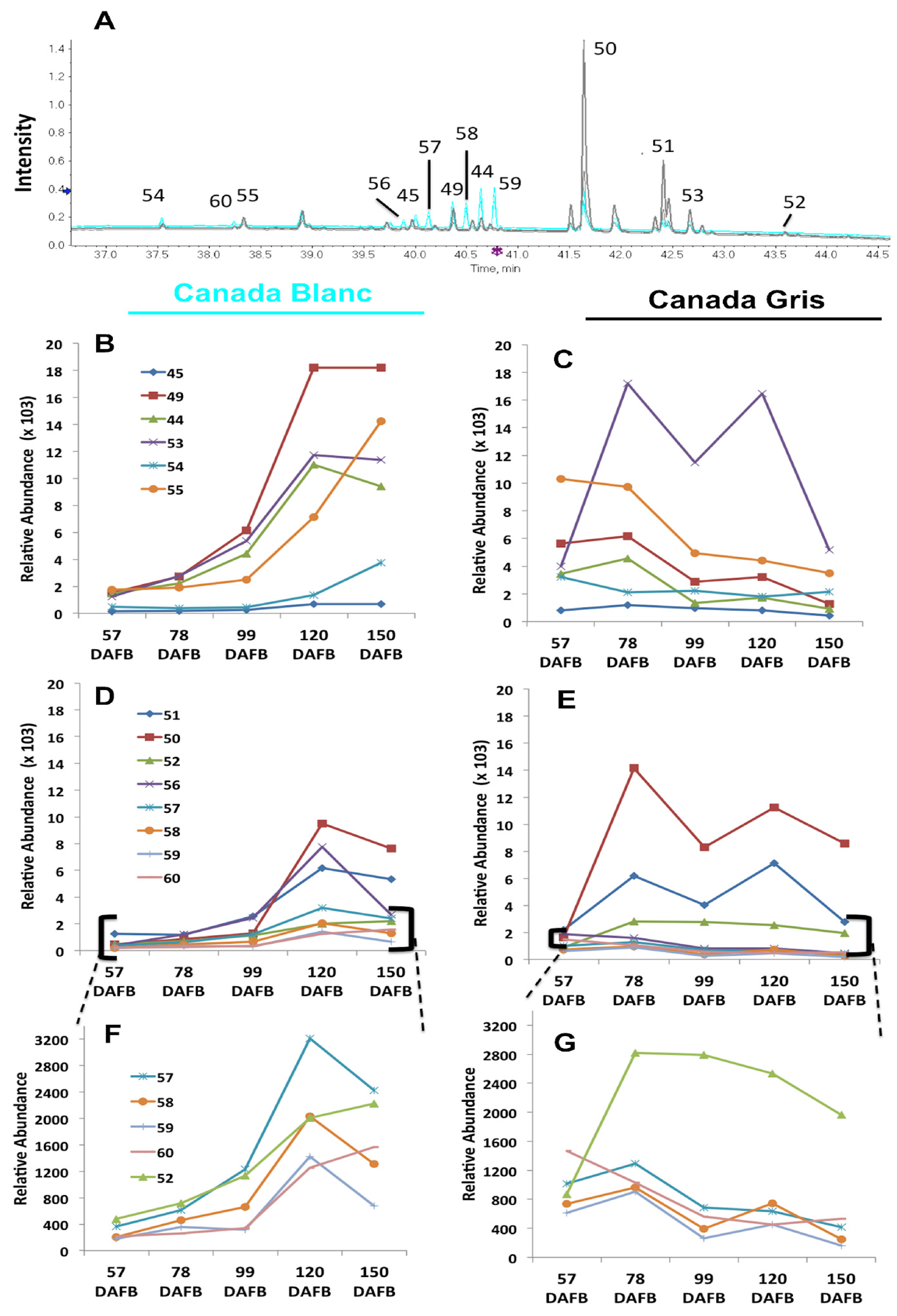
2.2. Untargeted Metabolomic Analysis between Russet CG and Non-Russet CB Skin during Fruit Development
2.3. Targeted Triterpene-Hydroxycinnamate Analysis
2.4. Proteome Analysis Overview
2.5. Differentially Expressed Genes between CB and CG
2.6. Cuticle Deposition Markers Show Increased Expression in the Waxy CB Cultivar Both at the Transcriptional and Protein Level
2.7. Increased Abundance of Suberin-, Cell Wall- and Triterpene-Related Transcript and Proteins in the Russet CG Cultivar
3. Discussion
3.1. The CB/CG Apple Mutational Sports Present Different Skin Phenotype
3.2. Metabolomics Analysis Revealed Key Phenolic and Triterpene Compounds Associated with Skin Phenotype
3.3. Loss of Cuticle Integrity in CG Is Associated with Low Abundance of Proteins and Transcripts Coding for Lipid Transfer Protein 3 (LTP3)
3.4. Identification of Potential BAHD Acyltransferases Involved in the Esterification of Triterpenes with Hydroxycinnamic Acids
4. Materials and Methods
4.1. Plant Material
4.2. Genotypic Characterization
4.3. Metabolomics Analysis
4.4. RNA Extraction and Sequencing
4.5. Proteomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Curry, E. Increase in Epidermal Planar Cell Density Accompanies Decreased Russeting of ‘Golden Delicious’ Apples Treated with Gibberellin A4+7. HortScience 2012, 47, 232–237. [Google Scholar] [CrossRef] [Green Version]
- Knoche, M.; Grimm, E. Surface Moisture Induces Microcracks in the Cuticle of ‘Golden Delicious’ Apple. HortScience 2008, 43, 1929–1931. [Google Scholar] [CrossRef] [Green Version]
- Straube, J.; Chen, Y.-H.; Khanal, B.P.; Shumbusho, A.; Zeisler-Diehl, V.; Suresh, K.; Schreiber, L.; Knoche, M.; Debener, T. Russeting in Apple is Initiated after Exposure to Moisture Ends: Molecular and Biochemical Evidence. Plants 2020, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Khanal, B.P.; Grimm, E.; Knoche, M. Russeting in apple and pear: A plastic periderm replaces a stiff cuticle. AoB Plants 2012, 5, pls048. [Google Scholar] [CrossRef] [Green Version]
- Lashbrooke, J.; Aharoni, A.; Costa, F. Genome investigation suggests MdSHN3, an APETALA2-domain transcription factor gene, to be a positive regulator of apple fruit cuticle formation and an inhibitor of russet development. J. Exp. Bot. 2015, 66, 6579–6589. [Google Scholar] [CrossRef] [Green Version]
- Legay, S.; Guerriero, G.; Deleruelle, A.; Lateur, M.; Evers, D.; André, C.M.; Hausman, J.-F. Apple russeting as seen through the RNA-seq lens: Strong alterations in the exocarp cell wall. Plant Mol. Biol. 2015, 88, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Legay, S.; Cocco, E.; André, C.M.; Guignard, C.; Hausman, J.-F.; Guerriero, G. Differential Lipid Composition and Gene Expression in the Semi-Russeted “Cox Orange Pippin” Apple Variety. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Brendolise, C.; Yauk, Y.-K.; Eberhard, E.D.; Wang, M.; Chagne, D.; Andre, C.; Greenwood, D.R.; Beuning, L.L. An unusual plant triterpene synthase with predominant α-amyrin-producing activity identified by characterizing oxidosqualene cyclases from Malus × domestica. FEBS J. 2011, 278, 2485–2499. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Larsen, L.; Burgess, E.J.; Jensen, D.J.; Cooney, J.M.; Evers, D.; Zhang, J.; Perry, N.B.; Laing, W.A. Unusual immuno-modulatory triterpene-caffeates in the skins of russeted varieties of apples and pears. J. Agric. Food Chem. 2013, 61, 2773–2779. [Google Scholar] [CrossRef]
- Andre, C.M.; Legay, S.; Deleruelle, A.; Nieuwenhuizen, N.; Punter, M.; Brendolise, C.; Cooney, J.M.; Lateur, M.; Hausman, J.-F.; Larondelle, Y.; et al. Multifunctional oxidosqualene cyclases and cytochrome P450 involved in the biosynthesis of apple fruit triterpenic acids. New Phytol. 2016, 211, 1279–1294. [Google Scholar] [CrossRef] [Green Version]
- McGhie, T.K.; Hudault, S.; Lunken, R.C.M.; Christeller, J.T. Apple peels, from seven cultivars, have lipase-inhibitory activity and contain numerous ursenoic acids as identified by LC-ESI-QTOF-HRMS. J. Agric. Food Chem. 2012, 60, 482–491. [Google Scholar] [CrossRef]
- He, X.; Liu, R.H. Triterpenoids isolated from apple peels have potent antiproliferative activity and may be partially responsible for apple’s anticancer activity. J. Agric. Food Chem. 2007, 55, 4366–4370. [Google Scholar] [CrossRef]
- Pan, H.; Lundgren, L.N.; Andersson, R. Triterpene caffeates from bark of Betula pubescens. Phytochemistry 1994, 37, 795–799. [Google Scholar] [CrossRef]
- Jiang, Z.-H.; Tanaka, T.; Kouno, I. A lupane triterpene and two triterpene caffeates from Rhoiptelea chiliantha. Phytochemistry 1995, 40, 1223–1226. [Google Scholar] [CrossRef]
- Falginella, L.; Andre, C.M.; Legay, S.; Lin-Wang, K.; Dare, A.P.; Deng, C.; Rebstock, R.; Plunkett, B.J.; Guo, L.; Cipriani, G.; et al. Differential regulation of triterpene biosynthesis induced by an early failure in cuticle formation in apple. Hortic. Res. 2021, 8, 1–15. [Google Scholar] [CrossRef]
- Kosma, D.K.; Murmu, J.; Razeq, F.M.; Santos, P.; Bourgault, R.; Molina, I.; Rowland, O. AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J. Cell Mol. Biol. 2014, 80, 216–229. [Google Scholar] [CrossRef] [Green Version]
- Lashbrooke, J.; Cohen, H.; Levy-Samocha, D.; Tzfadia, O.; Panizel, I.; Zeisler, V.; Massalha, H.; Stern, A.; Trainotti, L.; Schreiber, L.; et al. MYB107 and MYB9 Homologs Regulate Suberin Deposition in Angiosperms. Plant Cell 2016, 28, 2097–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legay, S.; Guerriero, G.; André, C.; Guignard, C.; Cocco, E.; Charton, S.; Boutry, M.; Rowland, O.; Hausman, J.-F. MdMyb93 is a regulator of suberin deposition in russeted apple fruit skins. New Phytol. 2016, 212, 977–991. [Google Scholar] [CrossRef]
- Andre, C.M.; Greenwood, J.M.; Walker, E.G.; Rassam, M.; Sullivan, M.; Evers, D.; Perry, N.B.; Laing, W.A. Anti-Inflammatory Procyanidins and Triterpenes in 109 Apple Varieties. J. Agric. Food Chem. 2012, 60, 10546–10554. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. Cell Mol. Biol. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Domergue, F.; Kosma, D.K. Occurrence and Biosynthesis of Alkyl Hydroxycinnamates in Plant Lipid Barriers. Plants 2017, 6, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Auria, J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Falginella, L.; Cipriani, G.; Monte, C.; Gregori, R.; Testolin, R.; Velasco, R.; Troggio, M.; Tartarini, S. A major QTL controlling apple skin russeting maps on the linkage group 12 of ‘Renetta Grigia di Torriana’. BMC Plant Biol. 2015, 15, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, B.L.; Zhong, G.-Y.; Brown, S.K. Increased phloridzin content associated with russeting in apple (Malus domestica (Suckow) Borkh.) fruit. Genet. Resour. Crop Evol. 2018, 65, 2135–2149. [Google Scholar] [CrossRef]
- Andre, C.M.; Larondelle, Y.; Evers, D. Dietary Antioxidants and Oxidative Stress from a Human and Plant Perspective: A Review. Curr. Nutr. Food Sci. 2010, 6, 2–12. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.; Gao, J.; Feng, X.; Shi, Z.; Gao, F.; Xu, X.; Yang, L. Inhibitory effect of chlorogenic acid on fruit russeting in ‘Golden Delicious’ apple. Sci. Hortic. 2014, 178, 14–22. [Google Scholar] [CrossRef]
- McFarlane, H.E.; Shin, J.J.H.; Bird, D.A.; Samuels, A.L. Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 2010, 22, 3066–3075. [Google Scholar] [CrossRef] [Green Version]
- Fabre, G.; Garroum, I.; Mazurek, S.; Daraspe, J.; Mucciolo, A.; Sankar, M.; Humbel, B.M.; Nawrath, C. The ABCG transporter PEC1/ABCG32 is required for the formation of the developing leaf cuticle in Arabidopsis. New Phytol. 2016, 209, 192–201. [Google Scholar] [CrossRef]
- Qin, P.; Tu, B.; Wang, Y.; Deng, L.; Quilichini, T.D.; Li, T.; Wang, H.; Ma, B.; Li, S. ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 2013, 54, 138–154. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Yu, K.; Gao, Q.-M.; Wilson, E.V.; Navarre, D.; Kachroo, P.; Kachroo, A. Acyl CoA Binding Proteins are Required for Cuticle Formation and Plant Responses to Microbes. Front. Plant Sci. 2012, 3, 224. [Google Scholar] [CrossRef] [Green Version]
- Arondel, V.; Vergnolle, C.; Cantrel, C.; Kader, J. Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci. Int. J. Exp. Plant Biol. 2000, 157, 1–12. [Google Scholar] [CrossRef]
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 2018, 59, 1374–1382. [Google Scholar] [CrossRef] [Green Version]
- Yadav, V.; Molina, I.; Ranathunge, K.; Castillo, I.Q.; Rothstein, S.J.; Reed, J.W. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 2014, 26, 3569–3588. [Google Scholar] [CrossRef] [Green Version]
- Bird, D.; Beisson, F.; Brigham, A.; Shin, J.; Greer, S.; Jetter, R.; Kunst, L.; Wu, X.; Yephremov, A.; Samuels, L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. Cell Mol. Biol. 2007, 52, 485–498. [Google Scholar] [CrossRef]
- Panikashvili, D.; Shi, J.X.; Bocobza, S.; Franke, R.B.; Schreiber, L.; Aharoni, A. The Arabidopsis DSO/ABCG11 transporter affects cutin metabolism in reproductive organs and suberin in roots. Mol. Plant 2010, 3, 563–575. [Google Scholar] [CrossRef]
- Edstam, M.M.; Blomqvist, K.; Eklöf, A.; Wennergren, U.; Edqvist, J. Coexpression patterns indicate that GPI-anchored non-specific lipid transfer proteins are involved in accumulation of cuticular wax, suberin and sporopollenin. Plant Mol. Biol. 2013, 83, 625–649. [Google Scholar] [CrossRef] [PubMed]
- Molina, I.; Kosma, D. Role of HXXXD-motif/BAHD acyltransferases in the biosynthesis of extracellular lipids. Plant Cell Rep. 2015, 34, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Tsubaki, S.; Sugimura, K.; Teramoto, Y.; Yonemori, K.; Azuma, J. Cuticular Membrane of Fuyu Persimmon Fruit Is Strengthened by Triterpenoid Nano-Fillers. PLoS ONE 2013, 8, e75275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, M.-H.; Chen, C.-M.; Lee, S.-W.; Chen, Z.-T. Cytotoxic triterpenoids from the root bark of Helicteres angustifolia. Chem. Biodivers. 2008, 5, 565–574. [Google Scholar] [CrossRef]
- Guilford, P.; Prakash, S.; Zhu, J.M.; Rikkerink, E.; Gardiner, S.; Bassett, H.; Forster, R. Microsatellites in Malus X domestica (apple): Abundance, polymorphism and cultivar identification. Theor. Appl. Genet. 1997, 94, 249–254. [Google Scholar] [CrossRef]
- Gianfranceschi, L.; Seglias, N.; Tarchini, R.; Komjanc, M.; Gessler, C. Simple sequence repeats for the genetic analysis of apple. Theor. Appl. Genet. 1998, 96, 1069–1076. [Google Scholar] [CrossRef]
- Liebhard, R.; Gianfranceschi, L.; Koller, B.; Ryder, C.D.; Tarchini, R.; Van De Weg, E.; Gessler, C. Development and characterisation of 140 new microsatellites in apple (Malus x domestica Borkh.). Mol. Breed. 2002, 10, 217–241. [Google Scholar] [CrossRef]
- Gasic, K.; Hernandez, A.; Korban, S.S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Report. 2004, 22, 437–438. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaut, J.; Lutts, S.; Hoffmann, L.; Hausman, J.-F. Responses of poplar to chilling temperatures: Proteomic and physiological aspects. Plant Biol. Stuttg. Ger. 2004, 6, 81–90. [Google Scholar] [CrossRef]
- Chabi, M.; Goulas, E.; Leclercq, C.C.; de Waele, I.; Rihouey, C.; Cenci, U.; Day, A.; Blervacq, A.-S.; Neutelings, G.; Duponchel, L.; et al. A Cell Wall Proteome and Targeted Cell Wall Analyses Provide Novel Information on Hemicellulose Metabolism in Flax. Mol. Cell. Proteomics MCP 2017, 16, 1634–1651. [Google Scholar] [CrossRef] [Green Version]
- Bohn, T.; Planchon, S.; Leclercq, C.C.; Renaut, J.; Mihaly, J.; Beke, G.; Rühl, R. Proteomic responses of carotenoid and retinol administration to Mongolian gerbils. Food Funct. 2018, 9, 3835–3844. [Google Scholar] [CrossRef]
- Busatto, N.; Matsumoto, D.; Tadiello, A.; Vrhovsek, U.; Costa, F. Multifaceted analyses disclose the role of fruit size and skin-russeting in the accumulation pattern of phenolic compounds in apple. PLoS ONE 2019, 14, e0219354. [Google Scholar] [CrossRef] [Green Version]
- Yuan, G.; Bian, S.; Han, X.; He, S.; Liu, K.; Zhang, C.; Cong, P. An Integrated Transcriptome and Proteome Analysis Reveals New Insights into Russeting of Bagging and Non-Bagging “Golden Delicious” Apple. Int. J. Mol. Sci. 2019, 20, 4462. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Putative Identity | Compound Number | tr (min) | [M+H]+/[M−H]− | Error (ppm) | Molecular Formula | Average EIC (‘Canada Blanc’) | Average EIC (‘Canada Gris’) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 57 DAFB | 78 DAFB | 99 DAFB | 120 DAFB | 150 DAFB | 57 DAFB | 78 DAFB | 99 DAFB | 120 DAFB | 150 DAFB | ||||||
| Negative Mode | |||||||||||||||
| - Negatively Associated with fruit development | |||||||||||||||
| Quinic Acid | 1 | 0.44 | 191.05611 | 2.6 | C7H12O6 | 74,765 | 60,322 | 49,342 | 43,609 | 31,943 | 78,172 | 68,535 | 58,939 | 37,821 | 24,492 |
| Quinic acid- dihexoside | 5 | 0.5 | 533.1709 | −2.7 | C19H34017 | 20,370 | 23,395 | 25,050 | 20,919 | 12,613 | 27,378 | 22,885 | 22,526 | 11,415 | 6370 |
| Hydroxybenzoic acid—hexoside | 7 | 0.8 | 315.0742 | 0.1 | C13H16O9 | 1830 | 649 | 606 | 475 | 390 | 1648 | 1008 | 913 | 658 | 670 |
| Quinic acid ethyl ester | 8 | 2.43 | 219.0877 | 1.3 | C8H1606 | 774 | 317 | 196 | 111 | 30 | 1538 | 822 | 422 | 95 | 46 |
| Caffeoyl-hexoside | 10 | 7.03 | 341.0873 | −0.51 | C15H1809 | 42,252 | 32,268 | 32,255 | 22,397 | 15,770 | 48,418 | 39,677 | 40,931 | 31,002 | 16,502 |
| Feruloyl-hexoside | 12 | 14.72 | 355.1032 | −0.7 | C16H20O9 | 8596 | 5889 | 4823 | 4063 | 2550 | 8949 | 6785 | 5650 | 4182 | 2703 |
| Flavonol-dimethyl ether-hexoside | 16 | 21.41 | 491.1178 | −1.6 | C23H24O12 | 1194 | 463 | 290 | 232 | 139 | 1575 | 1077 | 789 | 416 | 205 |
| Trihydroxy-dimethoxyflavanone—hexoside | 19 | 24.5 | 493.1355 | 0.7 | C23H26O12 | 5213 | 1808 | 1844 | 1371 | 1241 | 3874 | 2591 | 2050 | 1885 | 1524 |
| Quercetin dimethyl ether—hexoside | 20 | 24.53 | 509.1292 | −1.7 | C23H26013 | 1396 | 483 | 513 | 382 | 351 | 1033 | 689 | 566 | 524 | 1396 |
| Coumaroyl-caffeoyl-hexoside | 23 | 25.28 | 487.1264 | 3.7 | C24H24011 | 2069 | 1607 | 1796 | 1463 | 943 | 2408 | 2339 | 2110 | 1108 | 802 |
| Negative Mode | |||||||||||||||
| - Positively Associated with fruit development | |||||||||||||||
| Methyl-phloracetophenone-hexoside (domesticoside) | 2 | 0.49 | 343.1021 | −4 | C15H20O9 | 777 | 1504 | 3498 | 4891 | 7718 | 653 | 1361 | 2565 | 3458 | 6172 |
| Ethyl ester di-hexoside | 3 | 0.5 | 387.112 | −6.2 | C13H26013 | 162 | 229 | 906 | 1248 | 2301 | 140 | 242 | 630 | 990 | 2164 |
| Di-saccharide | 4 | 0.5 | 341.1073 | −1.9 | C12H22O11 | 19,711 | 35,366 | 84,004 | 112,633 | 166,024 | 18,672 | 33,367 | 65,512 | 87,756 | 147,611 |
| Di-saccharide | 6 | 0.52 | 341.1086 | −3.7 | C12H22O11 | 1503 | 2858 | 7144 | 10,344 | 15,288 | 1430 | 2573 | 5375 | 6963 | 13,122 |
| Vomifoliol-pentosyl-hexoside Isomer 1 | 13 | 18.89 | 517.2282 | −1.6 | C24H38O12 | 50 | 240 | 647 | 870 | 3139 | 229 | 777 | 1098 | 2606 | 3501 |
| Vomifoliol-pentosyl-hexoside Isomer 2 | 14 | 19.67 | 517.2282 | −1.6 | C24H38O12 | 7826 | 12,156 | 21,726 | 22,111 | 41,273 | 7592 | 16,393 | 20,763 | 22,169 | 32,980 |
| Negative Mode | |||||||||||||||
| - Positively Associated with russeting | |||||||||||||||
| Neochlorogenic acid | 9 | 4.19 | 353.0872 | −2 | C16H1809 | 326 | 455 | 426 | 313 | 518 | 991 | 595 | 1166 | 931 | 1046 |
| p-coumaroyl quinic acid Isomer | 11 | 8.38 | 337.0928 | −3.5 | C16H1808 | 3611 | 5511 | 6108 | 6378 | 5238 | 7120 | 5940 | 9170 | 8391 | 8850 |
| Hydroxy-Phloretin-hexoside | 17 | 21.8 | 451.124 | −1.3 | C21H24O11 | 7414 | 10,524 | 16,784 | 14,432 | 16,392 | 25,917 | 15,281 | 41,500 | 32,953 | 43,274 |
| Phloretin-pentoside | 21 | 24.64 | 405.1185 | −1.5 | C20H2209 | 237 | 307 | 399 | 393 | 598 | 1054 | 481 | 1240 | 1301 | 1466 |
| Hydroxy-phloretin | 22 | 24.73 | 289.0718 | 0.1 | C15H1406 | 23 | 28 | 54 | 101 | 105 | 186 | 277 | 1210 | 1707 | 1820 |
| Phlroretin | 25 | 26.89 | 273.0772 | 1.3 | C15H14O5 | 68 | 89 | 129 | 687 | 864 | 1281 | 1449 | 5384 | 10,097 | 8984 |
| Negative Mode | |||||||||||||||
| - Negatively Associated with russeting | |||||||||||||||
| Quercetin-hexoside | 15 | 20.42 | 463.0876 | 1.3 | C21H20O12 | 1825 | 1505 | 3197 | 1841 | 4088 | 1092 | 1529 | 1345 | 961 | 696 |
| Quercitrin | 18 | 22.12 | 447.0931 | −0.4 | C21H20O11 | 72,487 | 71,476 | 123,369 | 81,673 | 122,670 | 37,825 | 52,949 | 43,901 | 32,276 | 28,207 |
| Unknown triterpene | 26 | 31.74 | 489.358 | −1.1 | C30H50O5 | 735 | 756 | 1034 | 1231 | 1315 | 601 | 440 | 268 | 205 | 126 |
| Maslinic Acid | 27 | 34.36 | 471.3482 | 0.5 | C30H4804 | 233 | 460 | 789 | 1021 | 818 | 429 | 515 | 146 | 266 | 206 |
| Corosolic Acid | 28 | 35.34 | 471.3482 | 0.5 | C30H4804 | 295 | 277 | 553 | 569 | 684 | 310 | 282 | 265 | 211 | 185 |
| Positive Mode | |||||||||||||||
| - Negatively Associated with fruit development | |||||||||||||||
| Benzoic acid derivative (sodium adduct) | 29 | 0.5 | 219.0263 | −0.4 | C9H8O5 | 1434 | 1585 | 1143 | 935 | 841 | 1150 | 1143 | 1144 | 953 | 817 |
| Quinic Acid | 1 | 0.57 | 193.0702 | −1.9 | C7H12O6 | 5940 | 3579 | 2159 | 1348 | 531 | 8569 | 5824 | 2468 | 1150 | 398 |
| Hydroxybenzoic acid | 30 | 0.57 | 139.0389 | 1 | C7H6O3 | 1490 | 1049 | 700 | 439 | 177 | 2070 | 1512 | 772 | 368 | 128 |
| Feruloyl-hexoside (sodium adduct) | 12 | 15.59 | 379.0998 | −0.4 | C16H20O9 | 5531 | 4695 | 3952 | 3525 | 1853 | 5187 | 5252 | 4128 | 3211 | 1994 |
| Positive Mode | |||||||||||||||
| - Positively Associated with fruit development | |||||||||||||||
| Tetra-saccharide (sodium adduct) | 31 | 0.58 | 689.2096 | −2.4 | C24H42O21 | 230 | 370 | 748 | 963 | 1352 | 227 | 380 | 794 | 929 | 1380 |
| Tri-saccharide (sodium adduct) | 32 | 0.62 | 527.1574 | −1.6 | C18H32O16 | 812 | 1400 | 3036 | 4386 | 5653 | 1006 | 1670 | 3397 | 4398 | 5730 |
| Phenol-hexoside (sodium adduct) | 35 | 15.17 | 381.1154 | −0.6 | C16H22O9 | 112 | 146 | 338 | 671 | 1277 | 243 | 401 | 829 | 727 | 1227 |
| Vomifoliol-hexoside (sodium adduct) | 36 | 19.12 | 387.2009 | −0.9 | C19H30O8 | 992 | 1474 | 2637 | 2674 | 3953 | 1034 | 2093 | 2439 | 2634 | 3798 |
| Vomifoliol-pentosyl-hexoside | 14 | 19.89 | 519.2435 | −0.2 | C24H38O12 | 579 | 956 | 1848 | 1876 | 2694 | 667 | 1548 | 1954 | 1916 | 2609 |
| Positive Mode | |||||||||||||||
| - Positively Associated with russeting | |||||||||||||||
| Chlorogeno-quinone | 33 | 10.82 | 353.0868 | 0.3 | C16H16O9 | 3826 | 6638 | 5381 | 3701 | 8393 | 30,027 | 12,450 | 26,518 | 23,452 | 15,823 |
| Cryptochlorogeno-quinone | 34 | 13.13 | 353.0868 | 0.3 | C16H16O9 | 236 | 367 | 244 | 179 | 151 | 969 | 663 | 727 | 596 | 335 |
| Coumaroyl-quinic acid isomer | 24 | 17.26 | 339.107 | −1.3 | C16H18O8 | 107 | 231 | 217 | 214 | 244 | 2044 | 334 | 1129 | 1191 | 804 |
| Coumaroyl-hexoside derivative | 38 | 26.34 | 309.0968 | −0.3 | C15H16O7 | 822 | 731 | 789 | 675 | 651 | 2194 | 1522 | 1944 | 1345 | 963 |
| Positive Mode | |||||||||||||||
| - Negatively Associated with russeting | |||||||||||||||
| Quercitrin | 18 | 22.9 | 449.1075 | 2.1 | C21H20O11 | 4186 | 3864 | 6774 | 4526 | 6222 | 1986 | 3110 | 2380 | 1729 | 1658 |
| Quercetin-acetyl-hexoside | 37 | 23.69 | 507.1132 | −0.2 | C23H22O13 | 773 | 832 | 3296 | 1854 | 5250 | 793 | 1586 | 714 | 528 | 719 |
| Linolenic acid | 39 | 39.2 | 279.2318 | −0.2 | C18H30O2 | 380 | 480 | 797 | 707 | 364 | 196 | 188 | 78 | 61 | 78 |
| Ursa/Olea-dien-one | 40 | 39.375 | 423.3617 | −1 | C30H46O | 719 | 960 | 1310 | 1639 | 1508 | 327 | 428 | 207 | 195 | 175 |
| Alpha/Beta-amyrone | 41 | 39.69 | 425.3771 | −1.6 | C30H48O | 972 | 1459 | 2516 | 2704 | 3274 | 932 | 1470 | 1152 | 1107 | 945 |
| Olea/Ursa-dien-one | 42 | 40.07 | 423.3613 | −2 | C30H46O | 386 | 502 | 726 | 661 | 581 | 237 | 345 | 157 | 182 | 210 |
| Ursolic/Oleanolic Aldehyde | 43 | 40.09 | 441.3723 | 1,2 | C30H48O2 | 1883 | 2098 | 3222 | 3211 | 1898 | 927 | 672 | 583 | 750 | 505 |
| Putative Identity | Compound Number | tr (min) | Observed [M+H]+/ [M−H]− | Error (ppm) | Molecular Formula | Average EIC (‘Canada Blanc’) | Average EIC (‘Canada Gris’) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 57 DAFB | 78 DAFB | 99 DAFB | 120 DAFB | 150 DAFB | 57 DAFB | 78 DAFB | 99 DAFB | 120 DAFB | 150 DAFB | ||||||
| Positive Mode | |||||||||||||||
| Chlorogeno-quinone | 33 | 10.82 | 353.0868 | 0.3 | C16H16O9 | 3826 | 6638 | 5381 | 3701 | 8393 | 30,027 | 12,450 | 26,518 | 23,452 | 15,823 |
| Cryptochlorogeno-quinone | 34 | 13.13 | 353.0867 | 0.01 | C16H16O9 | 1496 | 2167 | 1379 | 1148 | 958 | 8100 | 3916 | 4626 | 3993 | 2438 |
| Coumaroyl-quinic acid isomer | 24 | 17.26 | 339.107 | −1.3 | C16H18O8 | 107 | 231 | 217 | 214 | 244 | 2044 | 334 | 1129 | 1191 | 804 |
| Maslinic Acid | 27 | 39.09 | 473.3604 | −4.9 | C30H48O4 | 1298 | 1412 | 4466 | 6990 | 8421 | 5139 | 3872 | 1601 | 3338 | 3219 |
| Corosolic Acid | 28 | 39.24 | 473.3616 | −2 | C30H48O4 | 242 | 467 | 1376 | 1997 | 3415 | 697 | 1037 | 533 | 1148 | 1163 |
| p-coumaroyloxy-hydroxy-urs/olean-12-en-28-oic acid | 44 | 40.68 | 619.3993 | 0 | C39H5406 | 1309 | 1413 | 2318 | 7016 | 6210 | 3720 | 4989 | 956 | 2369 | 1498 |
| Negative Mode | |||||||||||||||
| Quercetin-hexoside | 15 | 20.42 | 463.0876 | 1.3 | C21H20O12 | 1825 | 1505 | 3197 | 1841 | 4088 | 1092 | 1529 | 1345 | 961 | 696 |
| Phlroretin | 25 | 26.89 | 273.0772 | 1.3 | C15H14O5 | 68 | 89 | 129 | 687 | 864 | 1281 | 1449 | 5384 | 10,097 | 8984 |
| p-coumaroyloxy-hydroxy-urs/olean-12-en-28-oic acid | 45 | 39.65 | 617.3848 | 0.1 | C39H5406 | 150 | 178 | 253 | 667 | 688 | 810 | 1175 | 485 | 797 | 430 |
| Caffeoyl-quinic isomer | 46 | 2.1 | 353.0853 | −7.1 | C16H1809 | 3011 | 4441 | 4188 | 2301 | 5588 | 29,751 | 8479 | 19,423 | 20,194 | 14,270 |
| Unknown Triterpene | 47 | 35.91 | 473.3634 | −0.5 | C30H50O4 | 4931 | 5962 | 7696 | 9044 | 8975 | 3040 | 2879 | 1283 | 1041 | 790 |
| 3-oxo-hydroxy-urs-12-en-28-oic acid | 48 | 38.04 | 469.3321 | 1.8 | C30H46O4 | 258 | 245 | 389 | 561 | 672 | 584 | 585 | 866 | 970 | 354 |
| Dataset | Contig Code | Gene Symbol | Annotation Description | ‘Canada Blanc’ | ‘Canada Gris’ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 57 DAFB | 78 DAFB | 99 DAFB | 120 DAFB | 150 DAFB | 57 DAFB | 78 DAFB | 99 DAFB | 120 DAFB | 150 DAFB | ||||
| Features found in the RNA-Seq and proteomic data | |||||||||||||
| Cutin and wax related | |||||||||||||
| LC | MDP0000120594 | HXXXD-type acyl-transferase family protein | 0.25 | ||||||||||
| RNAseq | MDP0000120594 | HXXXD-type acyl-transferase family protein | 270.7 | 252.8 | 332.3 | 207.8 | 455.1 | 138.7 | 98.7 | 75.3 | 64.6 | 115.3 | |
| LC | MDP0000166457 | HXXXD-type acyl-transferase family protein | 0.23 | ||||||||||
| RNAseq | MDP0000166457 | HXXXD-type acyl-transferase family protein | 4.8 | 6.3 | 15.3 | 8.3 | 30.0 | 1.1 | 0.3 | 0.4 | 0.2 | 3.5 | |
| LC | MDP0000184619 | Li-tolerant lipase 1 | 1.98 | 1.76 | 2.42 | 2.38 | 0.68 | ||||||
| RNAseq | MDP0000184619 | ATLTL1, LTL1 | Li-tolerant lipase 1 | 1967.7 | 1372.9 | 2047.1 | 560.5 | 59.0 | 450.2 | 302.3 | 47.2 | 6.0 | 0.3 |
| LC | MDP0000231084 | acetoacetyl-CoA thiolase 2 | 0.47 | 0.18 | 0.23 | 0.25 | 0.96 | 0.28 | |||||
| RNAseq | MDP0000231084 | ACAT2 | acetoacetyl-CoA thiolase 2 | 1195.0 | 848.8 | 1219.7 | 834.9 | 1366.9 | 532.4 | 287.9 | 282.8 | 218.9 | 396.1 |
| LC | MDP0000285074 | lipid transfer protein 3 | 16.35 | 10.52 | 24.23 | 22.81 | 11.37 | 10.84 | 5.03 | 9.44 | 14.46 | 6.51 | |
| RNAseq | MDP0000285074 | LTP3 | lipid transfer protein 3 | 23,540.0 | 29,906.7 | 49,936.6 | 32,077.5 | 64,941.5 | 6517.5 | 5625.1 | 2116.9 | 2379.1 | 19,183.5 |
| LC | MDP0000304369 | lipid transfer protein 3 | 5.98 | 4.45 | 8.75 | 8.04 | 1.25 | 6.90 | |||||
| RNAseq | MDP0000304369 | LTP3 | lipid transfer protein 3 | 543.4 | 284.9 | 245.1 | 150.0 | 778.1 | 194.2 | 62.9 | 25.3 | 26.6 | 539.6 |
| LC | MDP0000319048 | Peroxidase superfamily protein | 3.06 | 2.04 | 2.48 | 1.37 | 0.68 | 2.36 | 1.29 | 1.06 | |||
| RNAseq | MDP0000319048 | PRX52 | Peroxidase superfamily protein | 297.8 | 133.7 | 149.7 | 44.0 | 29.1 | 80.9 | 28.3 | 4.4 | 1.5 | 3.6 |
| Suberin-related | |||||||||||||
| LC | MDP0000197743 | Bifunctional inhibitor/lipid-transfer protein | 0.87 | ||||||||||
| RNAseq | MDP0000197743 | Bifunctional inhibitor/lipid-transfer protein | 0.6 | 1.5 | 4.4 | 8.5 | 13.0 | 11.3 | 18.6 | 23.9 | 16.5 | 10.2 | |
| LC | MDP0000208152 | peroxidase 2 | 0.62 | 0.18 | 0.23 | 0.71 | 0.35 | 0.79 | 1.94 | 0.26 | |||
| RNAseq | MDP0000208152 | ATPA2, PA2 | peroxidase 2 | 0.4 | 2.8 | 3.2 | 13.3 | 10.1 | 1.7 | 8.5 | 20.5 | 32.1 | 16.0 |
| LC | MDP0000306867 | Peroxidase superfamily protein | 0.55 | 0.47 | 1.15 | 0.74 | 0.67 | ||||||
| RNAseq | MDP0000306867 | PRX52 | Peroxidase superfamily protein | 22.6 | 29.3 | 77.2 | 183.4 | 210.9 | 127.6 | 204.2 | 138.5 | 128.5 | 107.5 |
| LC | MDP0000312095 | Peroxidase superfamily protein | 0.21 | ||||||||||
| RNAseq | MDP0000312095 | PRX72 | Peroxidase superfamily protein | 4.2 | 3.0 | 7.5 | 14.0 | 19.7 | 13.1 | 19.8 | 26.0 | 23.1 | 32.0 |
| Features found in the proteomic data only | |||||||||||||
| Cutin and wax related | |||||||||||||
| LC | MDP0000231084 | acetoacetyl-CoA thiolase 2 | 0.47 | 0.18 | 0.23 | 0.25 | 0.96 | 0.28 | |||||
| LC | MDP0000157284 | acetyl co-A carboxylase | 0.73 | ||||||||||
| LC | MDP0000166116 | acyl activating enzyme 5 | 0.81 | ||||||||||
| LC | MDP0000323883 | acyl-CoA oxidase 1 | 0.60 | ||||||||||
| LC | MDP0000129664 | peroxisomal 3-ketoacyl-CoA thiolase 3 | 0.48 | ||||||||||
| LC | MDP0000231304 | peroxisomal 3-ketoacyl-CoA thiolase 3 | 0.62 | ||||||||||
| LC | MDP0000251803 | alpha/beta-Hydrolases superfamily protein | 0.35 | ||||||||||
| LC | MDP0000697378 | alpha/beta-Hydrolases superfamily protein | 0.49 | ||||||||||
| LC | MDP0000122785 | alpha/beta-Hydrolases superfamily protein | 0.83 | ||||||||||
| LC | MDP0000476415 | LTL1, GDSL1 | Li-tolerant lipase 1 | 1.17 | |||||||||
| LC | MDP0000496434 | LTL1, GDSL1 | Li-tolerant lipase 1 | 2.62 | 2.00 | ||||||||
| LC | MDP0000322755 | CCR | cinnamoyl coa reductase 1 | 0.48 | |||||||||
| LC | MDP0000227287 | Terpenoid cyclases family protein | 0.51 | ||||||||||
| LC | MDP0000478473 | CYP716A1 | cytochrome P450, family 716A1 | 0.21 | |||||||||
| LC | MDP0000130449 | CYP716A1 | cytochrome P450, family 716A1 | 0.22 | |||||||||
| LC | MDP0000432497 | EXPA15 | expansin A15 | 0.23 | |||||||||
| LC | MDP0000431696 | EXPA8 | expansin A8 | 0.52 | |||||||||
| LC | MDP0000220665 | PA2, PRX53 | peroxidase 2 | 0.32 | 0.25 | ||||||||
| LC | MDP0000539299 | PA2, PRX53 | peroxidase 2 | 0.72 | |||||||||
| LC | MDP0000319048 | Peroxidase superfamily protein | 3.06 | 2.04 | 2.48 | 1.37 | 0.68 | 2.36 | 1.29 | 1.06 | |||
| LC | MDP0000127521 | MYB17 | myb domain protein 17 | 0.18 | |||||||||
| Suberin-related | |||||||||||||
| LC | MDP0000197743 | LTPG20 | Bifunctional inhibitor/lipid-transfer protein | 0.87 | |||||||||
| LC | MDP0000293049 | LTPG20 | Bifunctional inhibitor/lipid-transfer protein | 1.48 | |||||||||
| LC | MDP0000525641 | FLA1 | FASCICLIN-like arabinogalactan 1 | 0.33 | 0.58 | 0.34 | 0.24 | 0.50 | 0.24 | 1.19 | 0.34 | ||
| LC | MDP0000904458 | FLA2 | FASCICLIN-like arabinogalactan 2 | 1.29 | 0.67 | 1.37 | 1.38 | 0.65 | 0.92 | 1.01 | 2.05 | 4.01 | 0.83 |
| LC | MDP0000658332 | FLA11 | FASCICLIN-like arabinogalactan-protein 10 | 1.26 | 0.48 | 0.74 | 0.38 | 0.44 | 1.27 | 0.63 | 1.09 | 1.72 | 0.15 |
| LC | MDP0000301828 | PA2, PRX53 | peroxidase 2 | 0.52 | 0.83 | 0.56 | 0.45 | 1.08 | 0.48 | 1.90 | 3.37 | 0.59 | |
| LC | MDP0000208152 | PA2, PRX53 | peroxidase 2 | 0.62 | 0.18 | 0.23 | 0.71 | 0.35 | 0.79 | 1.94 | 0.26 | ||
| LC | MDP0000209189 | Peroxidase superfamily protein | 1.65 | 1.32 | 2.12 | 1.74 | 2.05 | 3.08 | 1.73 | 5.45 | 8.17 | 2.61 | |
| LC | MDP0000207215 | Peroxidase superfamily protein | 1.50 | 1.25 | 1.88 | 1.00 | 0.98 | 2.07 | 1.38 | 2.69 | 4.74 | 1.31 | |
| LC | MDP0000283650 | Peroxidase superfamily protein | 0.62 | 0.66 | 1.27 | 0.43 | 0.49 | 0.73 | 0.65 | 1.26 | 2.26 | 0.66 | |
| LC | MDP0000519318 | WAK3 | wall associated kinase 3 | 0.68 | 0.38 | ||||||||
| LC | MDP0000323987 | XTH5 | xyloglucan endotransglucosylase/hydrolase 5 | 0.26 | 0.53 | ||||||||
| LC | MDP0000269483 | XTH6 | xyloglucan endotransglucosylase/hydrolase 6 | 0.35 | 0.63 | ||||||||
| Contig Code | Gene Symbol | TAIR Code | Annotation Description | Log2 Ratio (RPKM CG/RPKM CB) | ||||
|---|---|---|---|---|---|---|---|---|
| 57 DAFB | 78 DAFB | 99 DAFB | 120 DAFB | 150 DAFB | ||||
| Suberin synthesis | ||||||||
| MDP0000433567 | KCS2 | AT1G04220.1 | 3-ketoacyl-CoA synthase 2 | 0.49 | 2.04 | 1.59 | 0.90 | −0.60 |
| MDP0000922301 | KCS4 | AT1G19440.1 | 3-ketoacyl-CoA synthase 4 | 3.28 | 2.84 | 2.62 | 1.18 | 1.48 |
| MDP0000150502 | GPAT5 | AT3G11430.1 | glycerol-3-phosphate acyltransferase 5 | 4.00 | 3.24 | 2.49 | 1.23 | −0.18 |
| MDP0000923760 | CYP86A1 | AT5G58860.1 | cytochrome P450, family 86, subfamily A, polypeptide 1 | 4.02 | 3.01 | 2.38 | 1.08 | −0.21 |
| MDP0000306273 | CYP86B1 | AT5G23190.1 | cytochrome P450, family 86, subfamily B, polypeptide 1 | 3.81 | 3.34 | 2.66 | 1.43 | −0.07 |
| MDP0000138841 | FAR5 | AT3G44550.1 | fatty acid reductase 5 | 2.77 | 3.41 | 2.57 | 1.20 | −0.39 |
| MDP0000312412 | FATB | AT1G08510.1 | fatty acyl-ACP thioesterases B | 4.24 | 3.42 | 2.54 | 1.33 | −0.17 |
| Phenylpropanoid | ||||||||
| MDP0000219895 | C4H | AT2G30490.1 | cinnamate-4-hydroxylase | 0.16 | 2.32 | 2.77 | 0.79 | 0.47 |
| MDP0000260512 | 4CL2 | AT3G21240.1 | 4-coumarate: CoA ligase 2 | 1.67 | 2.06 | 1.56 | 0.94 | −0.35 |
| MDP0000287919 | CHS, TT4 | AT5G13930.1 | Chalcone and stilbene synthase family protein | −2.87 | 2.63 | 2.53 | 0.67 | 2.41 |
| MDP0000274127 | CHI, TT5 | AT3G55120.1 | Chalcone-flavanone isomerase family protein | 2.26 | 2.06 | 1.89 | 0.60 | 0.15 |
| MDP0000264424 | HCT | AT5G48930.1 | shikimate O-hydroxycinnamoyltransferase | 0.66 | 2.80 | 2.33 | 0.55 | 0.11 |
| MDP0000479113 | OMT1 | AT5G54160.1 | O-methyltransferase 1 | 3.80 | 3.44 | 2.79 | 1.26 | −0.26 |
| MDP0000246535 | CCoAOMT1 | AT4G34050.1 | Caffeoyl CoA methyltransferase | 2.59 | 3.33 | 2.71 | 1.13 | −0.24 |
| MDP0000164173 | CYP84A1, FAH1 | AT4G36220.1 | ferulic acid 5-hydroxylase 1 | 3.34 | 3.15 | 2.63 | 1.68 | 0.40 |
| Transport | ||||||||
| MDP0000265619 | ABCG2 | AT2G37360.1 | ABC-2 type transporter family protein | 1.88 | 2.01 | 1.92 | 1.06 | −0.05 |
| MDP0000299379 | ABCG6 | AT5G13580.1 | ABC-2 type transporter family protein | 3.17 | 3.08 | 2.67 | 0.90 | 0.00 |
| MDP0000731415 | ABCG23 | AT5G19410.1 | ABC-2 type transporter family protein | 4.06 | 3.20 | 2.40 | 1.26 | −0.25 |
| MDP0000193438 | ABCG11 | AT1G17840.1 | white-brown complex homolog protein 11 | −1.84 | 2.43 | 2.01 | 0.74 | −0.45 |
| MDP0000940078 | LTP1 | AT2G38540.1 | lipid transfer protein 1 | −2.01 | 2.79 | 1.72 | 1.90 | 0.76 |
| MDP0000197743 | LTPG20 | AT3G22620.1 | Bifunctional inhibitor/lipid-transfer protein | 4.23 | 3.60 | 2.46 | 1.02 | −0.28 |
| MDP0000304463 | LTPG16, EDA4 | AT2G48140.1 | Bifunctional inhibitor/lipid-transfer protein | 4.14 | 3.40 | 2.67 | 1.29 | −0.18 |
| MDP0000137283 | LTPG5 | AT3G22600.1 | Bifunctional inhibitor/lipid-transfer protein | 1.96 | 2.17 | 1.98 | 1.05 | −0.11 |
| Pentacyclic triterpene synthesis | ||||||||
| MDP0000207731 | AT1G78950.1 | beta amyrin synthase | 3.69 | 3.18 | 3.01 | 1.50 | 1.81 | |
| MDP0000661381 | CYP96A2 | AT4G32170.1 | cytochrome P450, family 96, subfamily A, polypeptide 2 | 1.91 | 2.55 | 2.12 | 0.64 | 1.54 |
| MDP0000266125 | MdOSC5 | AT1G78955.1 | Oxidosqualene 5/lupeol Synthase | 3.74 | 3.17 | 3.04 | 1.59 | 1.64 |
| MDP0000212688 | LUP1 | AT1G78970.1 | lupeol synthase 1 | 4.19 | 3.08 | 3.27 | 1.40 | 2.20 |
| MDP0000151814 | LUP2 | AT1G78960.1 | lupeol synthase 2 | 2.84 | 3.15 | 2.69 | 1.42 | 1.44 |
| MDP0000634676 | SQE1 | AT1G58440.1 | FAD/NAD(P)-binding oxidoreductase family protein | −2.70 | 1.15 | 1.99 | −0.50 | 0.08 |
| Cell wall metabolism | ||||||||
| MDP0000139485 | XTH30 | AT1G32170.1 | xyloglucan endotransglucosylase/hydrolase 30 | 1.34 | 2.68 | 1.59 | 0.86 | −0.16 |
| MDP0000311765 | XTH32 | AT2G36870.1 | xyloglucan endotransglucosylase/hydrolase 32 | 1.99 | 3.01 | 2.51 | 1.23 | −0.24 |
| MDP0000398765 | XTH5 | AT5G13870.1 | xyloglucan endotransglucosylase/hydrolase 5 | 2.23 | 3.14 | 2.32 | 0.80 | 0.01 |
| MDP0000320017 | XTH23 | AT4G25810.1 | xyloglucan endotransglycosylase 6 | 1.69 | 3.48 | 1.98 | 1.49 | −0.48 |
| MDP0000205889 | AGP16 | AT2G46330.1 | arabinogalactan protein 16 | −0.14 | 2.13 | 1.02 | 0.94 | −0.89 |
| MDP0000287357 | AGP30 | AT2G33790.1 | arabinogalactan protein 30 | 1.81 | 2.10 | 2.39 | 1.54 | 0.17 |
| MDP0000165381 | AGP31 | AT1G28290.1 | arabinogalactan protein 31 | 0.63 | 1.83 | 2.35 | 1.11 | −0.02 |
| MDP0000133529 | PEL | AT3G53190.1 | Pectin lyase-like superfamily protein | 2.78 | 3.31 | 1.83 | 0.94 | −0.61 |
| MDP0000297071 | PMEI | AT5G09760.1 | Pectin methylesterase inhibitor | 0.85 | 2.69 | 1.71 | 0.74 | −0.26 |
| MDP0000560112 | EXPA1 | AT1G69530.1 | expansin A1 | 0.35 | 2.05 | 2.50 | 4.62 | 2.58 |
| MDP0000195798 | EXPA4 | AT2G39700.1 | expansin A4 | 1.96 | 2.31 | 1.02 | 0.85 | −1.22 |
| MDP0000290170 | EXPB2 | AT1G65680.1 | expansin B2 | 1.43 | 3.04 | 1.34 | 0.90 | 0.86 |
| MDP0000208152 | PA2, PRX53 | AT5G06720.1 | peroxidase 2 | 1.96 | 1.60 | 2.72 | 1.33 | 0.74 |
| MDP0000312095 | PRX72 | AT5G66390.1 | Peroxidase superfamily protein | 1.63 | 2.71 | 1.83 | 0.78 | 0.77 |
| MDP0000306867 | PRX52 | AT5G05340.1 | Peroxidase superfamily protein | 2.49 | 2.79 | 0.87 | −0.44 | −0.90 |
| MDP0000126274 | LAC7 | AT3G09220.1 | laccase 7 | 1.10 | 4.16 | 1.27 | 0.09 | −1.68 |
| MDP0000262848 | LAC14 | AT5G09360.1 | laccase 14 | 1.24 | 2.26 | 2.29 | 0.09 | 0.51 |
| MDP0000655646 | LAC15 | AT5G48100.1 | laccase 15 | 0.72 | 2.16 | 1.60 | 0.18 | 0.28 |
| Transcription factor | ||||||||
| MDP0000211677 | MYB4 | AT4G38620.1 | myb domain protein 4 | 0.81 | 1.37 | 2.10 | 1.39 | 1.18 |
| MDP0000210851 | MYB7 | AT2G16720.1 | myb domain protein 7 | 2.13 | 3.11 | 1.82 | 0.90 | 0.58 |
| MDP0000124049 | MYB36 | AT5G57620.1 | myb domain protein 36 | 3.52 | 3.35 | 2.67 | 1.26 | 1.20 |
| MDP0000787808 | MYB42 | AT4G12350.1 | myb domain protein 42 | 0.32 | 2.01 | 1.28 | −0.29 | −0.20 |
| MDP0000291518 | MYB52 | AT1G17950.1 | myb domain protein 52 | 3.57 | 3.52 | 2.67 | 1.42 | 0.79 |
| MDP0000133542 | MYB58 | AT1G16490.1 | myb domain protein 58 | 3.70 | 3.40 | 1.86 | 0.74 | −0.81 |
| MDP0000157506 | MYB67 | AT3G12720.1 | myb domain protein 67 | 4.11 | 3.24 | 2.90 | 1.63 | 1.18 |
| MDP0000786507 | MYB68 | AT5G65790.1 | myb domain protein 68 | 3.87 | 3.39 | 3.29 | 1.42 | 0.80 |
| MDP0000682032 | MYB85 | AT4G22680.1 | myb domain protein 85 | 3.28 | 2.91 | 2.46 | 1.12 | 0.11 |
| MDP0000320772 | MdMYB93 | AT1G34670.1 | myb domain protein 93 | 4.11 | 3.19 | 2.54 | 1.34 | −0.15 |
| MDP0000197283 | MYB102 | AT4G21440.1 | myb domain protein 102 | 2.12 | 2.26 | 0.82 | 0.11 | −1.20 |
| MDP0000262032 | ANAC072, RD26 | AT4G27410.2 | NAC domain containing protein 72 | 1.17 | 1.12 | 2.01 | 1.07 | 0.68 |
| MDP0000129335 | NAC100 | AT5G61430.1 | NAC domain containing protein 100 | 1.45 | 2.19 | 1.06 | 0.31 | −0.36 |
| MDP0000690168 | NAC038 | AT2G24430.1 | NAC domain containing protein 38 | 3.23 | 3.08 | 2.64 | 1.22 | 0.84 |
| MDP0000235213 | NAC058 | AT3G18400.1 | NAC domain containing protein 58 | 3.59 | 3.44 | 2.49 | 1.19 | 0.00 |
| MDP0000124509 | NAC075 | AT4G29230.1 | NAC domain containing protein 75 | 3.29 | 3.20 | 2.11 | 1.27 | 0.39 |
| MDP0000126517 | NAC083, VNI2 | AT5G13180.1 | NAC domain containing protein 83 | 2.73 | 3.11 | 2.52 | 1.18 | 0.71 |
| Acyl-transferases | ||||||||
| MDP0000698860 | AT4G13840.1 | HXXXD-type acyl-transferase family protein | 3.87 | 3.28 | 2.52 | 1.35 | 0.01 | |
| MDP0000593263 | AT1G32910.1 | HXXXD-type acyl-transferase family protein | 3.05 | 2.92 | 2.50 | 1.22 | −0.10 | |
| MDP0000478556 | AT4G13840.1 | HXXXD-type acyl-transferase family protein | 4.01 | 2.68 | 2.01 | 1.21 | −0.08 | |
| MDP0000312405 | AT5G41040.1 | HXXXD-type acyl-transferase family protein | 4.03 | 3.24 | 2.92 | 1.60 | 0.82 | |
| MDP0000253113 | AT3G26040.1 | HXXXD-type acyl-transferase family protein | 1.36 | 2.08 | 2.08 | 0.93 | 0.87 | |
| MDP0000258308 | RWP1, FCT, FHT | AT5G41040.1 | Feruloyl-hydroxycinnamoyl acytransfearase | 4.18 | 3.22 | 2.53 | 1.24 | −0.18 |
| Miscellaneous | ||||||||
| MDP0000660239 | AT1G74460.1 | GDSL-like Lipase/Acylhydrolase superfamily protein | 4.45 | 3.52 | 2.54 | 1.32 | −0.41 | |
| MDP0000174332 | AT1G75900.1 | GDSL-like Lipase/Acylhydrolase superfamily protein | 3.01 | 2.91 | 2.64 | 1.21 | 0.23 | |
| MDP0000172849 | AT3G03990.1 | alpha/beta-Hydrolases superfamily protein | 1.06 | 2.05 | 1.94 | 1.24 | 1.29 | |
| MDP0000594621 | AT1G47480.1 | alpha/beta-Hydrolases superfamily protein | 3.47 | 3.04 | 2.54 | 1.06 | 0.01 | |
| MDP0000930224 | AT2G18360.1 | alpha/beta-Hydrolases superfamily protein | 3.37 | 3.07 | 2.79 | 1.09 | 0.68 | |
| MDP0000188790 | AT5G06570.1 | alpha/beta-Hydrolases superfamily protein | 1.66 | 1.20 | 2.59 | 0.51 | 0.47 | |
| MDP0000927926 | AT4G18550.1 | alpha/beta-Hydrolases superfamily protein | 3.48 | 2.81 | 3.04 | 1.13 | 0.85 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
André, C.M.; Guerriero, G.; Lateur, M.; Charton, S.; Leclercq, C.C.; Renaut, J.; Hausman, J.-F.; Legay, S. Identification of Novel Candidate Genes Involved in Apple Cuticle Integrity and Russeting-Associated Triterpene Synthesis Using Metabolomic, Proteomic, and Transcriptomic Data. Plants 2022, 11, 289. https://doi.org/10.3390/plants11030289
André CM, Guerriero G, Lateur M, Charton S, Leclercq CC, Renaut J, Hausman J-F, Legay S. Identification of Novel Candidate Genes Involved in Apple Cuticle Integrity and Russeting-Associated Triterpene Synthesis Using Metabolomic, Proteomic, and Transcriptomic Data. Plants. 2022; 11(3):289. https://doi.org/10.3390/plants11030289
Chicago/Turabian StyleAndré, Christelle M., Gea Guerriero, Marc Lateur, Sophie Charton, Celine C. Leclercq, Jenny Renaut, Jean-Francois Hausman, and Sylvain Legay. 2022. "Identification of Novel Candidate Genes Involved in Apple Cuticle Integrity and Russeting-Associated Triterpene Synthesis Using Metabolomic, Proteomic, and Transcriptomic Data" Plants 11, no. 3: 289. https://doi.org/10.3390/plants11030289
APA StyleAndré, C. M., Guerriero, G., Lateur, M., Charton, S., Leclercq, C. C., Renaut, J., Hausman, J.-F., & Legay, S. (2022). Identification of Novel Candidate Genes Involved in Apple Cuticle Integrity and Russeting-Associated Triterpene Synthesis Using Metabolomic, Proteomic, and Transcriptomic Data. Plants, 11(3), 289. https://doi.org/10.3390/plants11030289










