SNORKEL Genes Relating to Flood Tolerance Were Pseudogenized in Normal Cultivated Rice
Abstract
:1. Introduction
2. Results
2.1. Sequence Comparison in the SK1/SK2 Region
2.2. Gene Prediction in the Region around the SK Genes and Phylogenetic Analysis
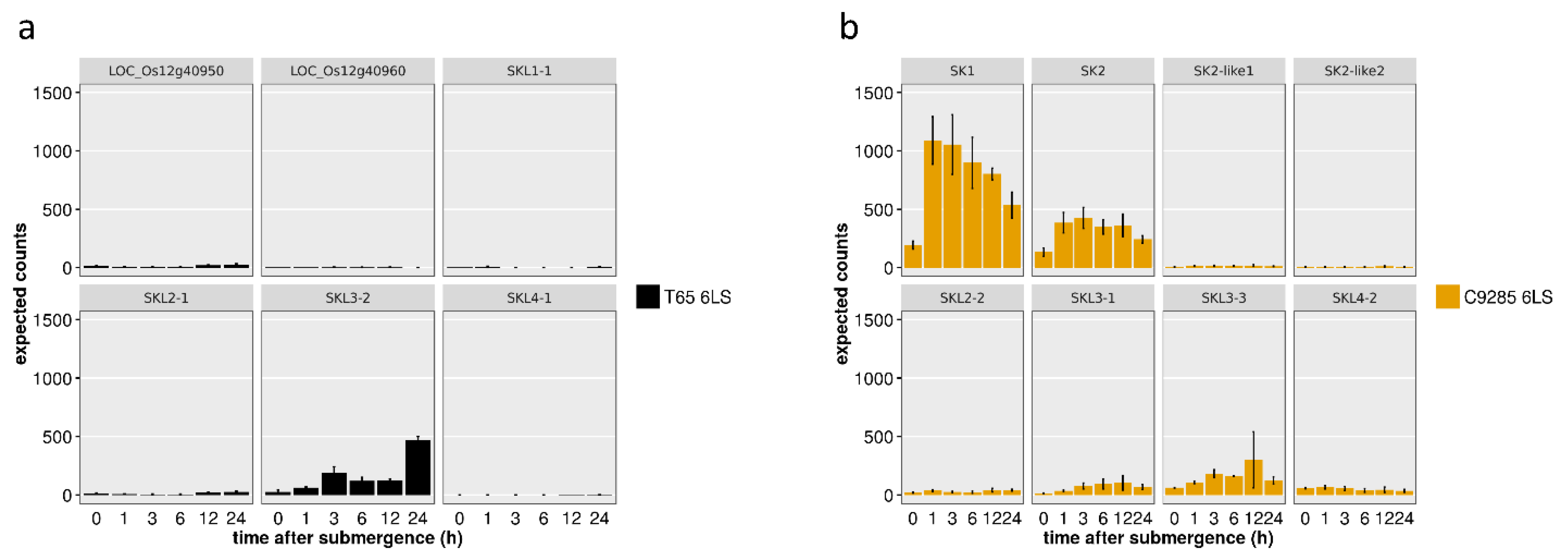
2.3. SK and SKL Gene Expression
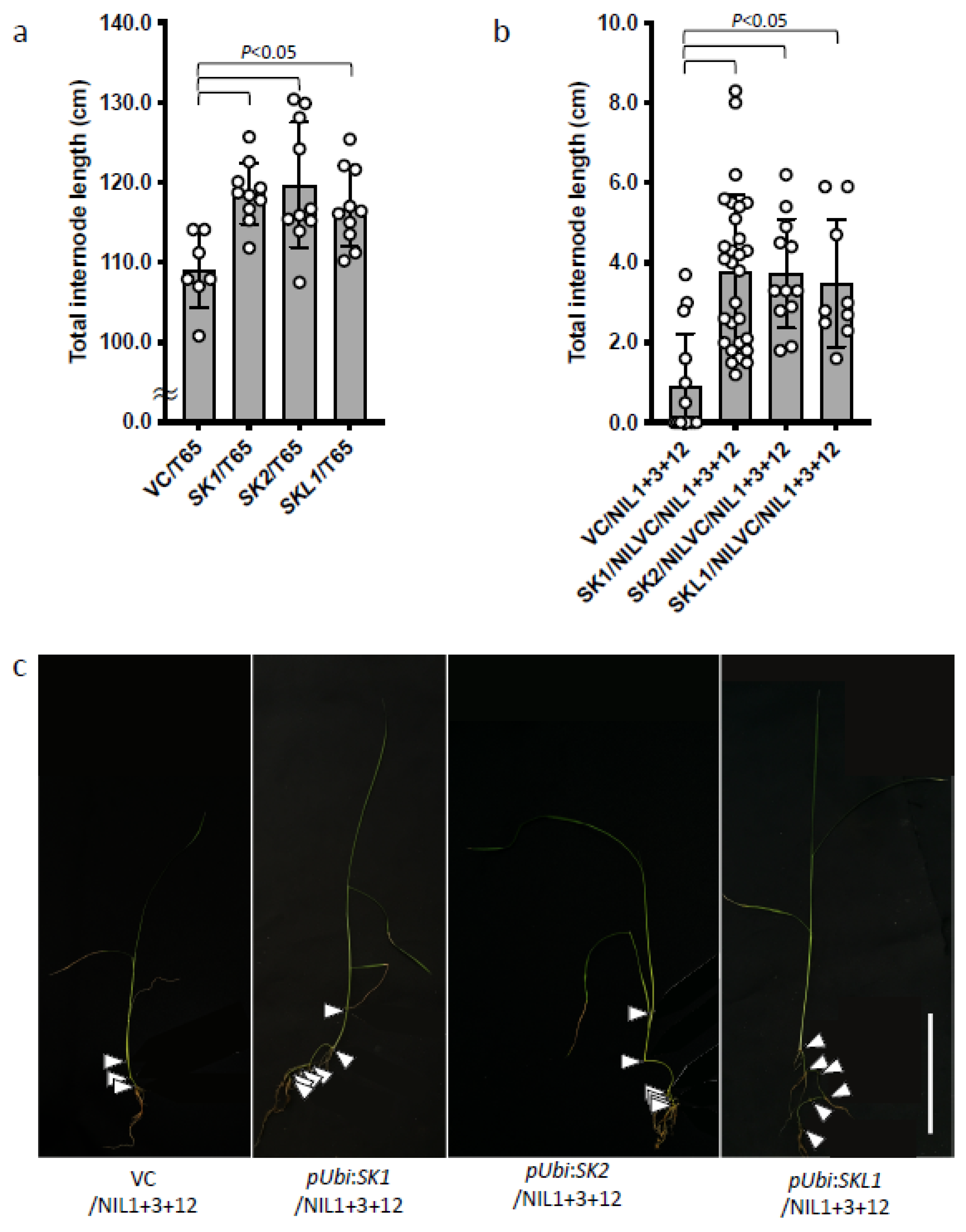
2.4. Effects of SKs and SKL1 on Internode Elongation
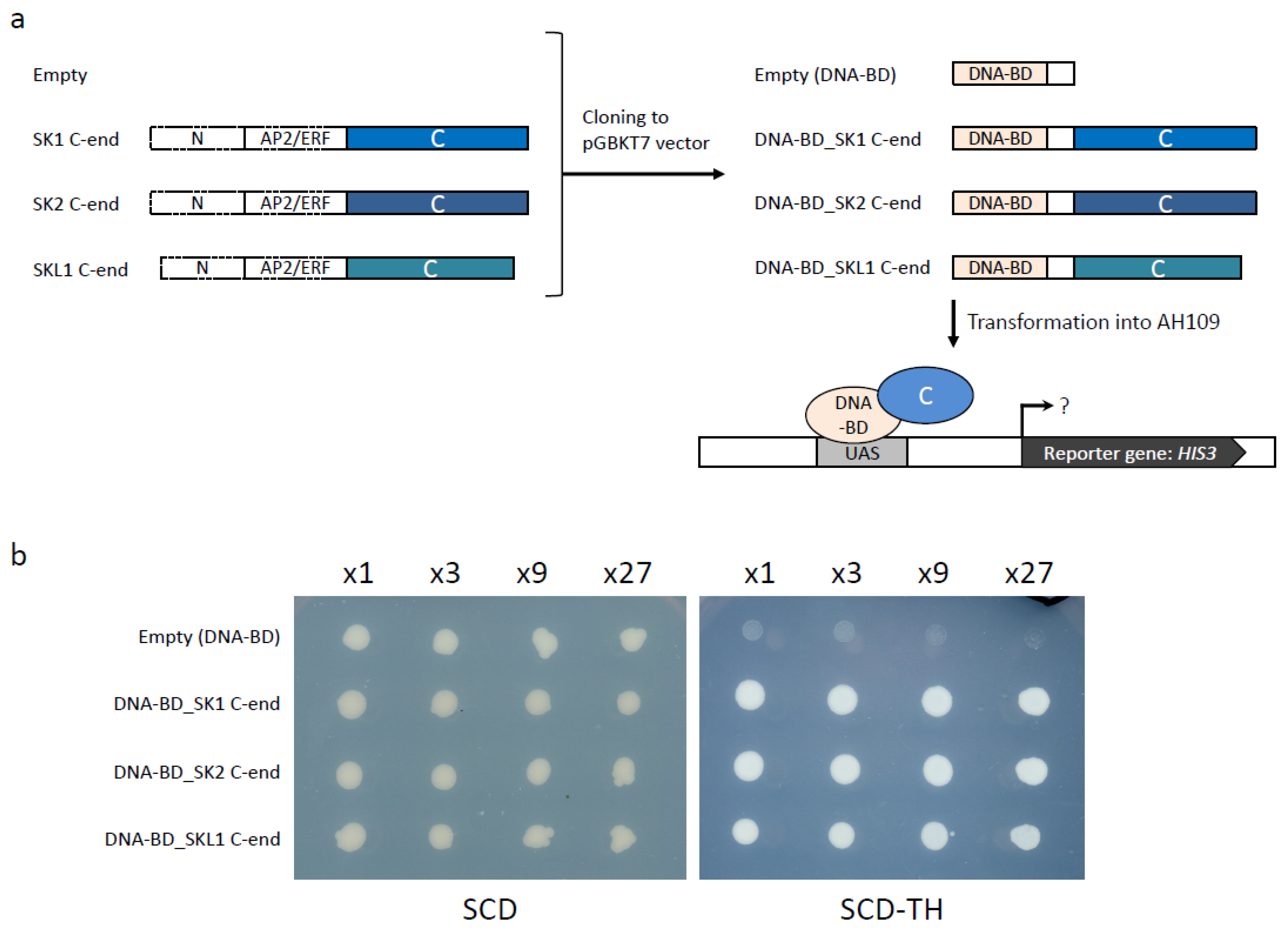
2.5. Validation of SKL1 Transcriptional Activity
3. Discussion
4. Materials and Methods
4.1. Construction of the BAC Library and Sequencing of BAC Clones
4.2. Gene Prediction and Sequence Comparison
4.3. Gene Expression Analysis
4.4. Production of Transgenic Plants
4.5. Plant Growth Conditions
4.6. Transcriptional Activity Assay
4.7. Statistics and Reproducibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, G.M.; Yandell, M.D.; Wortman, J.R.; Gabor Miklos, G.L.; Nelson, C.R.; Hariharan, I.K.; Fortini, M.E.; Li, P.W.; Apweiler, R.; Fleischmann, W.; et al. Comparative genomics of the eukaryotes. Science 2000, 287, 2204–2215. [Google Scholar] [CrossRef] [Green Version]
- Kent, W.J.; Baertsch, R.; Hinrichs, A.; Miller, W.; Haussler, D. Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 11484–11489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assis, R.; Bachtrog, D. Neofunctionalization of young duplicate genes in Drosophila. Proc. Natl. Acad. Sci. USA 2013, 110, 17409–17414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Zhang, J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 2005, 169, 1157–1164. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.Y.; González, J.M.; Ramachandran, S. Comparative Genomic and Transcriptomic Analysis of Tandemly and Segmentally Duplicated Genes in Rice. PLoS ONE 2013, 8, e63551. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Guo, Z.H.; Hao, P.P.; Wang, G.M.; Jin, Z.M.; Zhang, S.L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurokawa, Y.; Nagai, K.; Huan, P.D.; Shimazaki, K.; Qu, H.; Mori, Y.; Toda, Y.; Kuroha, T.; Hayashi, N.; Aiga, S.; et al. Rice leaf hydrophobicity and gas films are conferred by a wax synthesis gene (LGF1) and contribute to flood tolerance. New Phytol. 2018, 218, 1558–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, T.; Nakazono, M. Mechanisms of lysigenous aerenchyma formation under abiotic stress. Trends Plant Sci. 2021, 27, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.G.; Mori, Y.; Suzui, N.; Kurita, K.; Yamaguchi, M.; Miyoshi, Y.; Nagao, Y.; Ashikari, M.; Nagai, K.; Kawachi, N. Noninvasive imaging of hollow structures and gas movement revealed the gas partial-pressure-gradient-driven long-distance gas movement in the aerenchyma along the leaf blade to submerged organs in rice. New Phytol. 2021, 232, 1974–1984. [Google Scholar] [CrossRef]
- Mori, Y.; Kurokawa, Y.; Koike, M.; Malik, A.I.; Colmer, T.D.; Ashikari, M.; Pedersen, O.; Nagai, K. Diel O2 dynamics in partially and completely submerged deepwater rice: Leaf gas films enhance internodal O2 status, influence gene expression and accelerate stem elongation for ‘snorkelling’ during submergence. Plant Cell Physiol. 2019, 60, 973–985. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Hattori, Y.; Miura, K.; Asano, K.; Yamamoto, E.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. A major QTL confers rapid internode elongation in response to water rise in deepwater rice. Breed. Sci. 2007, 57, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Nagai, K.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. Mapping of three QTLs that regulate internode elongation in deepwater rice. Breed. Sci. 2008, 58, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 186, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minami, A.; Yano, K.; Gamuyao, R.; Nagai, K.; Kuroha, T.; Ayano, M.; Nakamori, M.; Koike, M.; Kondo, Y.; Niimi, Y.; et al. Time-course transcriptomics analysis reveals key responses of submerged deepwater rice to flooding. Plant Physiol. 2018, 176, 3081–3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, A. The fate of duplicated genes: Loss or new function? BioEssays 1998, 20, 785–788. [Google Scholar] [CrossRef]
- Liu, Z.; Tavares, R.; Forsythe, E.S.; André, F.; Lugan, R.; Jonasson, G.; Boutet-Mercey, S.; Tohge, T.; Beilstein, M.A.; Werck-Reichhart, D.; et al. Evolutionary interplay between sister cytochrome P450 genes shapes plasticity in plant metabolism. Nat. Commun. 2016, 7, 13026. [Google Scholar] [CrossRef] [Green Version]
- Asano, K.; Yamasaki, M.; Takuno, S.; Miura, K.; Katagiri, S.; Ito, T.; Doi, K.; Wu, J.; Ebana, K.; Matsumoto, T.; et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11034–11039. [Google Scholar] [CrossRef] [Green Version]
- Nagai, K.; Mori, Y.; Ishikawa, S.; Furuta, T.; Gamuyao, R.; Niimi, Y.; Hobo, T.; Fukuda, M.; Kojima, M.; Takebayashi, Y.; et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020, 584, 109–114. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Z.L.; Park, J.; Tyler, L.; Yusuke, J.; Qiu, K.; Nam, E.A.; Lumba, S.; Desveaux, D.; McCourt, P.; et al. The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 2016, 171, 2760–2770. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. A mutant gibberellin-synthesis gene in rice: New insight into the rice variant that helped to avert famine over thirty years ago. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Ashikari, M.; Sasaki, A.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Datta, S.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Loss-of-function of a Rice Gibberellin Biosynthetic Gene, GA20 oxidase (GA20ox-2), Led to the Rice ‘Green Revolution’. Breed. Sci. 2002, 52, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-B.; Zhao, X.; Ding, X.; Paterson, A.H.; Wing, R.A. Preparation of megabase-size DNA from plant nuclei. Plant J. 1995, 7, 175–184. [Google Scholar] [CrossRef] [Green Version]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, Y.; Ikeda-Ohtsubo, W.; Nagata, Y.; Tsuda, M. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinform. 2008, 9, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [Green Version]


| Gene Name | Gene Name | Gene Name | Gene Name | Gene Name | Gene Name | Gene Name |
|---|---|---|---|---|---|---|
| SK1 | C9285 | - | 771 | - | - | AB510478 |
| SK2 | C9285 | - | 777 | - | - | AB510479 |
| SKL1 | Nipponbare | LOC_Os12g40950 | 417 | 666 | - | GENSCAN |
| Nipponbare | LOC_Os12g40960 | 198 | - | |||
| SK2-like1 | C9285 | - | - | 726 | - | GENSCAN |
| SK2-like2 | C9285 | - | - | 831 | - | GENSCAN |
| SKL2-1 | Nibbonbare | LOC_Os12g41030 | 423 | - | OsERF#110 | - |
| SKL2-2 | C9285 | - | - | 423 | - | FGENESH |
| SKL3-1 | C9285 | - | - | 969 | - | FGENESH |
| SKL3-2 | Nipponbare | LOC_Os12g41060 | 972 | - | OsERF#112 | AK242027 |
| SKL3-3 | C9285 | - | - | 972 | - | FGENESH |
| SKL4-1 | Nipponbare | LOC_Os12g41040 * | 7020 | 594 | OsERF#111 | GENSCAN |
| SKL4-2 | C9285 | - | - | 762 | - | GENSCAN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagai, K.; Kurokawa, Y.; Mori, Y.; Minami, A.; Reuscher, S.; Wu, J.; Matsumoto, T.; Ashikari, M. SNORKEL Genes Relating to Flood Tolerance Were Pseudogenized in Normal Cultivated Rice. Plants 2022, 11, 376. https://doi.org/10.3390/plants11030376
Nagai K, Kurokawa Y, Mori Y, Minami A, Reuscher S, Wu J, Matsumoto T, Ashikari M. SNORKEL Genes Relating to Flood Tolerance Were Pseudogenized in Normal Cultivated Rice. Plants. 2022; 11(3):376. https://doi.org/10.3390/plants11030376
Chicago/Turabian StyleNagai, Keisuke, Yusuke Kurokawa, Yoshinao Mori, Anzu Minami, Stefan Reuscher, Jianzhong Wu, Takashi Matsumoto, and Motoyuki Ashikari. 2022. "SNORKEL Genes Relating to Flood Tolerance Were Pseudogenized in Normal Cultivated Rice" Plants 11, no. 3: 376. https://doi.org/10.3390/plants11030376
APA StyleNagai, K., Kurokawa, Y., Mori, Y., Minami, A., Reuscher, S., Wu, J., Matsumoto, T., & Ashikari, M. (2022). SNORKEL Genes Relating to Flood Tolerance Were Pseudogenized in Normal Cultivated Rice. Plants, 11(3), 376. https://doi.org/10.3390/plants11030376






