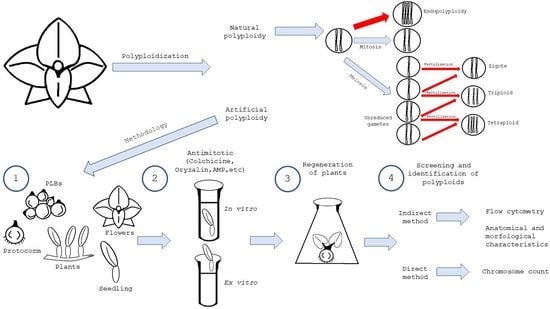
Polyploidization in Orchids: From Cellular Changes to Breeding Applications
Abstract
:1. Introduction
2. The Orchidaceae Family and Its Economic Importance in the World Floriculture
3. Orchid Karyotype
4. Natural Occurrence of Polyploid Cells in Orchids
4.1. Endopolyploidy
| Species/Cultivar | Plant Material | Nuclear DNA Content | References |
|---|---|---|---|
| Cattleya tigrina | Leaves, leaf bases, leaf tips, roots, Protocorm-like Bodies (PLBs) | 2C, 4C, 8C | Liz [76] |
| Cymbidium sp. | Embryo Parenchymal Cells | Nagl [73] | |
| Nine comercial hybrids of Cymbidium | Callus and PLBs | 2C, 4C, 8C, 16C | Teixeira et al. [58] |
| Cym. Twilight Moon ‘Day Light | Callus and PLBs | 2C, 4C, 8C, 16C | Teixeira da Silva; Singh; Tanaka [74] |
| Two comercial hybrids of Cymbidium and Cym. kanran | PLBs | 2C, 4C, 8C, 16C | Fukai; Hasegawa; Goi [72] |
| Dendrobium sp. | Root tips and new leaves | Jones; Kuehnle [64] | |
| Den. Chao Praya Smile | Seeds, Protocorms, Protocorms with leaves, stem tips, axillary buds and pseudobulbs, leaves, roots and flowers | 2C, 4C, 8C, 16C, 32C | Seah [65] |
| Doritaenopsis hybrid | Somatic embryos | 2C, 4C, 8C, 16C, 32C, 64C | Park; Paek [61] |
| Doritaenopsis | Somatic leaves, roots and embryos | 2C, 8C, 16, 64C | Park; Yeung; Paek [62] |
| Oncidium varicosum | Flowers | 2C, 4C, 8C, 16C | Lee et al. [69] |
| Onc. varicosum | Flowers | 2C, 4C, 8C, 16C | Lee et al. [59] |
| Phal. aphrodite subsp. formosana | sepals, petals, lip, columns, pollinia, pedicels, ovaries of fully open flowers, roots, protocorms, seedling leaves | 2C, 4C, 8C, 16C, 32C | Chen et al. [66] |
| Phal. aphrodite subsp. formosana | Ovarian tissue before/after pollination, seeds and protocorms | 2C, 4C, 8C, 16C | Jean et al. [57] |
| Phal. aphrodite subsp. formosana | Flowers | 2C, 4C, 8C, 16C | Lee et al. [69] |
| Phal. spp. | Protocorms | Chen et al. [56] | |
| Phal. spp. | Protocorms, PLBs and young leaves | 2C, 4C, 8C, 16C | Chen; Tang; Kao [68] |
| Phal. spp. | PLBs and young leaves | 2C, 4C, 8C | Chen; Tang [67] |
| Phal. spp. | Flowers, roots and leaves | 2C, 4C, 8C, 16C | Lin et al. [70] |
| Spatoglottis plicata | Leaves, roots, floral tissue, protocorms, young seedling leaves, roots | 2C, 4C, 8C, 16C | Yang; Loh [75] |
| Vanda Miss Joaquin | Leaves, buds, aerial and terrestrial roots, petals, sepals, pedicels, spine, sexual embryos | 2C, 4C, 8C, 16C, 32C, 64C | Lim; Loh [60] |
| V. sanderiana | Somatic embryos | 2C, 4C, 8C | Alvarez [63] |
| Vanilla planifolia | Roots | 2C, 4C, 8C, 16C, 32C | Kausch; Horner [71] |
4.2. Occurrence of Unreduced Gametes
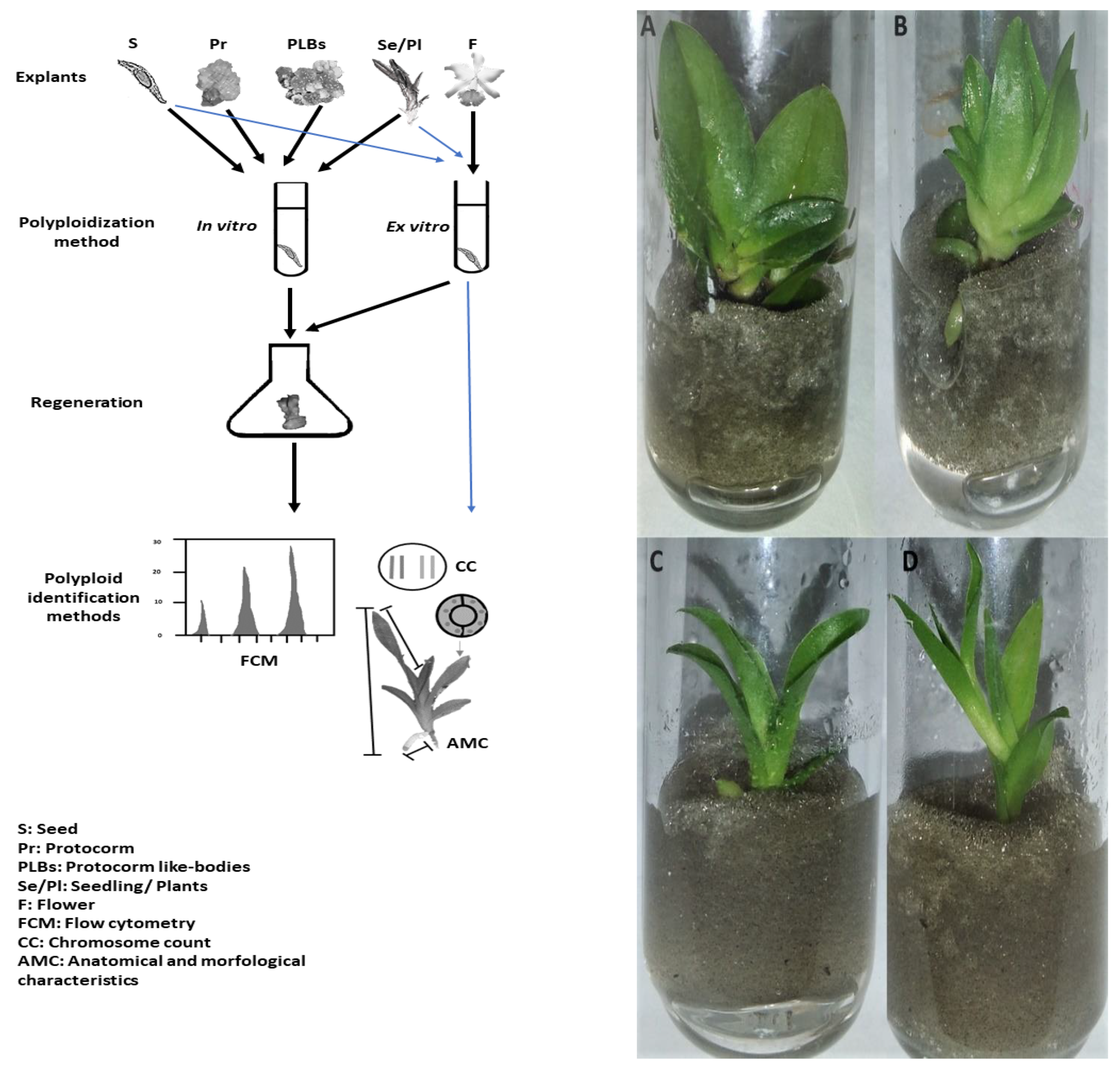
5. Artificial Induction of Polyploidy in Orchids
5.1. Cattleya Genus
5.2. Cymbidium Genus
5.3. Dendrobium Genus
5.4. Phalaenopsis Genus
5.5. Induction of Polyploidy in Oncidium, Vanda, and Others
6. Conclusions
7. Further Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; de Pamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masterson, J. Stomatal Size in Fossil Plants: Evidence for Polyploidy in Majority of Angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Salman-Minkov, A.; Sabath, N.; Mayrose, I. Whole-genome duplication as a key factor in crop domestication. Nat. Plants 2016, 2, 16115. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef] [Green Version]
- Brochmann, C.; Brysting, A.K.; Alsos, I.G.; Borgen, L.; Grundt, H.H.; Scheen, A.; Elven, R. Polyploidy in arctic plants. Biol. J. Linn. Soc. 2004, 82, 521–536. [Google Scholar] [CrossRef] [Green Version]
- Grant, V. Plant Speciation; Columbia University Press: New York, NY, USA, 1981. [Google Scholar]
- Maluszynska, J.; Kolano, B.; Sas-Nowosielska, H. Endopolyploidy in Plants. In Plant Genome Diversity; Springer: Vienna, Austria, 2013; Volume 2, pp. 99–119. [Google Scholar]
- Barow, M.; Meister, A. Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant Cell Environ. 2003, 26, 571–584. [Google Scholar] [CrossRef]
- Beemster, G.T.; De Vusser, K.; De Tavernier, E.; De Bock, K.; Inze, D. Variation in Growth Rate between Arabidopsis Ecotypes Is Correlated with Cell Division and A-Type Cyclin-Dependent Kinase Activity. Plant Physiol. 2002, 129, 854–864. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, D.W.; Harkins, K.R.; Knapp, S. Systemic Endopolyploidy in Arabidopsis thaliana. Plant Physiol. 1991, 96, 985–989. [Google Scholar] [CrossRef] [Green Version]
- Harlan, J.R.; Dewet, J.M.J. On, Ö. Winge and a Prayer: The origins of polyploidy. Bot. Rev. 1975, 41, 361–390. [Google Scholar] [CrossRef]
- Franke, R. Über Das Auftreten von Unreduzierten Gameten Bei Angiospermen; Arch. ZUchtungsforsch: Berlin, Germany, 1975; Volume 5. [Google Scholar]
- The Plant List. 2020. Available online: http://www.theplantlist.org/1.1/browse/A/Orchidaceae/#statistics (accessed on 28 August 2021).
- De, L.C.; Rao, A.N.; Rajeevan, P.K.; Pathak, P. Orchid improvement-an overview. J. Orchid. Soc. India 2014, 28, 35–45. [Google Scholar]
- De, L.C.; Pathak, P.; Rao, A.N.; Rajeevan, P.K. 2 Global Orchid Industry. In Commercial Orchids; De Gruyter Open Poland: Warsaw, Poland, 2015; pp. 13–19. [Google Scholar] [CrossRef]
- Cardoso, J.C.; Martinelli, A.P.; da Silva, J.A.T. A novel approach for the selection of Cattleya hybrids for precocious and season-independent flowering. Euphytica 2016, 210, 143–150. [Google Scholar] [CrossRef]
- Cardoso, J.C. Ionocidium Cerrado 101: Intergeneric orchid hybrid with high quality of blooming. Ornam. Hortic. 2017, 23, 351. [Google Scholar] [CrossRef] [Green Version]
- Griesbach, R.J. Colchicine-induced polyploidy in phalaenopsis orchids. Plant Cell Tissue Organ Cult. 1981, 1, 103–107. [Google Scholar] [CrossRef]
- Kaiser, R. The Scent of Orchids; Elsevier Science Publisher: Amsterdam, The Netherlands, 1993. [Google Scholar]
- Herman, D.E. The Species behind Standard Cattleyas. Orchids 1997, 66, 234–243. [Google Scholar]
- Kim, M.-S.; Cho, H.-R.; Rhee, H.-K.; Lim, J.-H.; Shin, H.-K. Development of New Cymbidium Variety ‘White Girl’ Medium Type, Red Lip Flower. Hortic. Environ. Biotechnol. 2010, 51, 235–238. [Google Scholar]
- Bhattacharjee, S.K.; De, L.C. Advanced Commercial Floriculture; Aavishkar Publishers: Rajasthan, India, 2003; Volume 1. [Google Scholar]
- Tang, C.-Y.; Chen, W.-H. Breeding and Development of New Varieties in Phalaenopsis. Orchid. Biotechnol. 2007, 57, 1–22. [Google Scholar] [CrossRef]
- Hsieh, K.-T.; Liu, S.-H.; Wang, I.-W.; Chen, L.-J. Phalaenopsis orchid miniaturization by overexpression of OsGA2ox6, a rice GA2-oxidase gene. Bot. Stud. 2020, 61, 10–11. [Google Scholar] [CrossRef]
- Lee, C. An economic analysis of orchid production under protected facilities in taiwan: Case of phalaenopsis. Acta Hortic. 2002, 30, 249–255. [Google Scholar] [CrossRef]
- Hinsley, A.; De Boer, H.J.; Fay, M.F.; Gale, S.W.; Gardiner, L.M.; Gunasekara, R.S.; Kumar, P.; Masters, S.; Metusala, D.; Roberts, D.; et al. A review of the trade in orchids and its implications for conservation. Bot. J. Linn. Soc. 2017, 186, 435–455. [Google Scholar] [CrossRef]
- Chen, C. The Fundamental Issue in the Phalaenopsis Industry in the Netherlands. J. Agric. For. 2020, 67, 89–100. [Google Scholar]
- Miguel, T.P.; Leonhardt, K.W. In vitro polyploid induction of orchids using oryzalin. Sci. Hortic. 2011, 130, 314–319. [Google Scholar] [CrossRef]
- Ranney, T.G. Polyploidy: From Evolution to New Plant Development. Comb. Proc. Int. Plant Propagators Soc. 2006, 56, 137–142. Available online: https://www.researchgate.net/profile/Thomas_Ranney/publication/228633988_Polyploidy_From_Evolution_to_New_Plant_DevelopmentC/links/0046352b095c13de6a000000.pdf (accessed on 3 September 2021).
- Felix, L.P.; Guerra, M. Variation in chromosome number and the basic number of subfamily Epidendroideae (Orchidaceae). Bot. J. Linn. Soc. 2010, 163, 234–278. [Google Scholar] [CrossRef]
- Bory, S.; Catrice, O.; Brown, S.; Leitch, I.J.; Gigant, R.; Chiroleu, F.; Grisoni, M.; Duval, M.-F.; Besse, P. Natural polyploidy in Vanilla planifolia (Orchidaceae). Genome 2008, 51, 816–826. [Google Scholar] [CrossRef]
- Jones, K.; Lim, K.Y.; Cribb, P.J. The Chromosomes of Orchids VII Dendrobium. Kew Bull. 1982, 37, 221. [Google Scholar] [CrossRef]
- Kao, Y.-Y.; Chang, S.-B.; Lin, T.-Y.; Hsieh, C.-H.; Chen, Y.-H. Differential Accumulation of Heterochromatin as a Cause for Karyotype Variation in Phalaenopsis Orchids. Ann. Bot. 2001, 87, 387–395. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-I.; Tseng, Y.; Chung, M.-C. Chromosome constitution and nuclear DNA content of Phalaenopsis hybrids. Sci. Hortic. 2019, 262, 109089. [Google Scholar] [CrossRef]
- Wilfret, G.J.; Kamemoto, H. Genome and Karyotype Relationships in the Genus Dendrobium (Orchidaceae). Cytologia 1971, 36, 604–613. [Google Scholar] [CrossRef] [Green Version]
- Wimber, J.; Donald, E. Cytogenetic Studies in the Genus Cymbidium (Tese Dee Doutorado); Claremont Graduate University: Claremont, CA, USA, 1956; Available online: https://scholarship.claremont.edu/cgi/viewcontent.cgi?article=1002&context=cgu_etd (accessed on 12 October 2021).
- Yu-Ge, L.I.; Wei-Hong, G.U.O.; Bo-Ji, W.U. A Karyological Study of Seven Species and One Variety of Cymbidium from China. J. Syst. Evol. 2002, 40, 406–413. [Google Scholar]
- Younis, A.; Ryu, K.B.; Hwang, Y.-J.; Jee, S.O.; Kim, M.-S.; Kim, C.K.; Lim, K.-B. Analysis of Chromosomes and Nuclear DNA Content in Nine Genotypes of Cymbidium. Flower Res. J. 2013, 21, 158–161. [Google Scholar] [CrossRef]
- Mohammadi, M.; Kaviani, B.; Sedaghathoor, S. In vivo polyploidy induction of Phalaenopsis amabilis in a bubble bioreactor system using colchicine. Ornam. Hortic. 2021, 27, 204–212. [Google Scholar] [CrossRef]
- Blumenschein, A. Uma Nova Espécie Do Gênero Cattleya Lindl. Publicação Científica Instituto de Genética Escola Superior de Agricultura Luiz de Queiroz. Genética Esc. 1961, 2, 23–33. [Google Scholar]
- Xie, L.; Zhou, S.; Wang, M.; Zeng, R.; Guo, H.; Zhang, Z. Creation and micropropagation of polyploids in Cymbidium hybridum. Acta Hortic. 2017, 16, 107–114. [Google Scholar] [CrossRef]
- Zhu, G.F.; Lü, F.B.; Wang, B.Q.; Chen, M.L. Chromosome Analysis of Hybrid Cymbidium. Acta Hortic. Sin. 2006, 33, 417–421. [Google Scholar]
- Lee, Y.-I.; Chung, M.-C.; Kuo, H.-C.; Wang, C.-N.; Lin, C.-Y.; Jiang, H.; Yeh, C.-H. The evolution of genome size and distinct distribution patterns of rDNA in Phalaenopsis (Orchidaceae). Bot. J. Linn. Soc. 2017, 185, 65–80. [Google Scholar] [CrossRef]
- Aoyama, M. Polyploidy in Phalaenopsis Hybrids; Bulletin of the Hiroshima Prefectural Agriculture Research Center: Higashihiroshima, Japan, 1993; Volume 57, pp. 55–62. [Google Scholar]
- Griesbach, R.J. Development of Phalaenopsis Orchids for the Mass-Market. In Trends in New Crops and New Uses; Janick, J., Whipkey, A., Eds.; ASHS Press: Alexandria, Egypt, 2002; pp. 458–465. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.454.6479&rep=rep1&type=pdf (accessed on 22 October 2021).
- Bolaños-Villegas, P.; Chen, F.-C. Cytological Identification of Chromosomal Rearrangements in Doritaenopsis and Phalaenopsis. J. Intl. Coop 2007, 2, 1–11. Available online: https://www.researchgate.net/profile/Pablo_Bolanos-Villegas2/publication/262373813_Cytological_identification_of_chromosomal_rearrangements_in_Doritaenopsis_and_Phalaenopsis/links/0c960538124efa044a000000/Cytological-identification-of-chromosomal-rearrang (accessed on 15 September 2021).
- Chuang, H.T.; Hsu, S.T.; Shen, T.M. Breeding Barriers in Yellow Phalaenopsis Orchids. J. Taiwan Soc. Hort. Sci. 2008, 54, 59–66. [Google Scholar]
- Jones, K. The Chromosomes of Orchids: II: Vandeae Lindl. Kew Bull. 1967, 21, 151. [Google Scholar] [CrossRef]
- Shindo, K.; Kamemoto, H. Karyotype analysis of some sarcanthine orchids. Am. J. Bot. 1963, 50, 73–79. [Google Scholar] [CrossRef]
- Storey, W.B. Chromosome Numbers of Some Vanda Species and Hybrids. Bull. Amer. Orchid Soc. 1952, 21, 801–806. [Google Scholar]
- Kamemoto, H.; Shindo, K. Meiosis in Interspecific and Intergeneric Hybrids of Vanda. Bot. Gaz. 1964, 125, 132–138. [Google Scholar] [CrossRef]
- Yip, M.-Y. Cytological Studies of Some Aranda (Arachnis × Vanda) Hybrids (Orchidaceae). Cytologia 1977, 42, 565–579. [Google Scholar] [CrossRef] [Green Version]
- Félix, L.P.; Guerra, M. Cytogenetics and cytotaxonomy of some Brazilian species of Cymbidioid orchids. Genet. Mol. Biol. 2000, 23, 957–978. [Google Scholar] [CrossRef]
- Chen, W.-H.; Kao, Y.-L.; Tang, C.-Y.; Jean, G.-T.; Chen, H.-H. Endopolyploidy in Phalaenopsis Orchids and Its Application in Polyploid Breeding. Orchid. Biotechnol. 2011, 10, 25–48. [Google Scholar] [CrossRef]
- Jean, G.-T.; Kao, Y.-L.; Tang, C.-Y.; Chen, W.-H. Distribution of Nuclei of Different Ploidy Levels during Ovule, Seed and Protocorm Development in Phalaenopsis aphrodite subsp. formosana (Orchidaceae). Am. J. Plant Sci. 2011, 2, 325–333. [Google Scholar] [CrossRef] [Green Version]
- da Silva, J.A.T.; Giang, D.T.T.; Dobránszki, J.; Zeng, S.; Tanaka, M. Ploidy analysis of Cymbidium, Phalaenopsis, Dendrobium and Paphiopedillum (Orchidaceae), and Spathiphyllum and Syngonium (Araceae). Biologia 2014, 69, 750–755. [Google Scholar] [CrossRef]
- Lee, H.-C.; Chen, Y.-J.; Markhart, A.H.; Lin, T.-Y. Temperature effects on systemic endoreduplication in orchid during floral development. Plant Sci. 2007, 172, 588–595. [Google Scholar] [CrossRef]
- Lim, W.L.; Loh, C.S. Endopolyploidy in Vanda Miss Joaquim (Orchidaceae). New Phytol. 2003, 159, 279–287. [Google Scholar] [CrossRef]
- Park, S.-Y.; Paek, K.-Y. Endoreduplication Pattern of Somatic Embryos and Variants Occurrence Affected by Pre-existed Endoreduplicated Cells in Doritaenopsis. J. Plant Biotechnol. 2006, 33, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-Y.; Yeung, E.C.; Paek, K.-Y. Endoreduplication in Phalaenopsis is affected by light quality from light-emitting diodes during somatic embryogenesis. Plant Biotechnol. Rep. 2010, 4, 303–309. [Google Scholar] [CrossRef]
- Alvarez, M.R. Quantitative changes in nuclear dna accompanying postgermination embryonic development in Vanda (Orchidaceae). Am. J. Bot. 1968, 55, 1036–1041. [Google Scholar] [CrossRef]
- Jones, W.E.; Kuehnle, A.R. Ploidy Identification Using Flow Cytometry in Tissues of Dendrobium Species and Cultivars. Lindleyana 1998, 13, 11–18. [Google Scholar]
- Seah, K.T. Endopolyploidy in Dendrobium Chao Praya Smile and Anthurium andraeanum Cv’ red Hot’ (Tese de Mestrado). 2009. Available online: https://core.ac.uk/download/pdf/48630159.pdf (accessed on 18 September 2021).
- Chen, W.-H.; Tang, C.-Y.; Lin, T.-Y.; Weng, Y.-C.; Kao, Y.-L. Changes in the endopolyploidy pattern of different tissues in diploid and tetraploid Phalaenopsis aphrodite subsp. formosana (Orchidaceae). Plant Sci. 2011, 181, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-H.; Tang, C.-Y. A Protocol for the Induction of Polyploids in Phalaenopsis Orchids by In Vitro Method Without Using Anti-microtubule Agents. In Orchid Propagation: From Laboratories to Greenhouses—Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 317–330. [Google Scholar] [CrossRef]
- Chen, W.H.; Tang, C.Y.; Kao, Y.L. Ploidy doubling by in vitro culture of excised protocorms or protocorm-like bodies in Phalaenopsis species. Plant Cell Tissue Organ Cult. 2009, 98, 229–238. [Google Scholar] [CrossRef]
- Lee, H.-C.; Chiou, D.-W.; Chen, W.-H.; Markhart, A.H.; Chen, Y.-H.; Lin, T.-Y. Dynamics of cell growth and endoreduplication during orchid flower development. Plant Sci. 2004, 166, 659–667. [Google Scholar] [CrossRef]
- Lin, S.; Lee, H.-C.; Chen, W.-H.; Chen, C.-C.; Kao, Y.-Y.; Fu, Y.-M.; Chen, Y.-H.; Lin, T.-Y. Nuclear DNA Contents of Phalaenopsis sp. and Doritis pulcherrima. J. Am. Soc. Hortic. Sci. 2001, 126, 195–199. [Google Scholar] [CrossRef]
- Kausch, A.P.; Horner, H.T. Increased Nuclear DNA Content in Raphide Crystal Idioblasts during Development in Vanilla planifolia L. (Orchidaceae). Eur. J. Cell Biol. 1984, 33, 7–12. Available online: https://www.researchgate.net/profile/Harry-Horner/publication/16498377_Increased_nuclear_DNA_content_in_raphide_crystal_idioblasts_during_development_in_Vanilla_planifolia_L_Orchidaceae/links/59dbbef5458515e9ab451ddf/Increased-nuclear-DNA-content-in-raphi (accessed on 22 September 2021).
- Fukai, S.; Hasegawa, A.; Goi, M. Polysomaty in Cymbidium. HortScience 2002, 37, 1088–1091. [Google Scholar] [CrossRef] [Green Version]
- Nagl, W. Evidence of DNA Amplification in the Orchid Cymbidium in Vitro. Cytobios 1972, 5, 145–154. [Google Scholar]
- Da Silva, J.A.T.; Singh, N.; Tanaka, M. Priming biotic factors for optimal protocorm-like body and callus induction in hybrid Cymbidium (Orchidaceae), and assessment of cytogenetic stability in regenerated plantlets. Plant Cell Tissue Organ Cult. 2006, 84, 135–144. [Google Scholar] [CrossRef]
- Yang, M.; Loh, C.S. Systemic endopolyploidy in Spathoglottis plicata (Orchidaceae) development. BMC Cell Biol. 2004, 5, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liz, R.D.D. Citometria de Fluxo, Análises Ultraestruturais e Bioquímica de Estruturas Semelhantes a Protocormos (ESPs) de Cattleya tigrina A. Richard (Tese de Mestrado). 2017. Available online: https://repositorio.ufsc.br/bitstream/handle/123456789/187571/PRGV0268-T.pdf?sequence=-1&isAllowed=y (accessed on 10 July 2021).
- Johnson, S.D.; Edwards, T.J. The structure and function of orchid pollinaria. Oesterreichische Bot. Z. 2000, 222, 243–269. [Google Scholar] [CrossRef]
- Teoh, S.B. Polyploid spore formation in diploid orchid species. Genetica 1984, 63, 53–59. [Google Scholar] [CrossRef]
- Zeng, R.-Z.; Zhu, J.; Xu, S.-Y.; Du, G.-H.; Guo, H.-R.; Chen, J.; Zhang, Z.-S.; Xie, L. Unreduced Male Gamete Formation in Cymbidium and Its Use for Developing Sexual Polyploid Cultivars. Front. Plant Sci. 2020, 11, 558. [Google Scholar] [CrossRef]
- Miduno, T. Chromosomenstudien an Orchidazeen. III. Cytologia 1940, 11, 156–177. [Google Scholar] [CrossRef] [Green Version]
- Hagerup, O. The Spontaneous Formation of Haploid, Polyploid, and Aneuploid Embryos in Some Orchids. K. Dansk. Videnskab. Selskab. Biol. Meddelel. 1947, 20, 1–22. [Google Scholar]
- Kamemoto, H.; Kasemsap, S.; Sagarik, R. Chromosome Numbers of Sarcanthine Orchid Species of Thailand. Nat. Hist. Bull. Siam. Soc. 1964, 10, 85. [Google Scholar]
- Germanà, M.A. Use of Irradiated Pollen to Induce Parthenogenesis and Haploid Production in Fruit Crops. In Plant Mutation Breeding and Biotechnology; Shu, Q.Y., Forester, B.P., Nakagawa, H., Eds.; CABI and FAO: Rome, Italy, 2012; pp. 411–421. [Google Scholar]
- Dhooghe, E.; Van Laere, K.; Eeckhaut, T.; Leus, L.; Van Huylenbroeck, J. Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tissue Organ Cult. 2010, 104, 359–373. [Google Scholar] [CrossRef]
- Eigsti, O.J.; Dustin, P. Colchicine in Agriculture, Medicine, Biology, Chemistry; Iowa University Press: Ames, Iowa, 1994. [Google Scholar]
- Randolph, L.F. Some Effects of High Temperature on Polyploidy and Other Variations in Maize. Proc. Natl. Acad. Sci. USA 1932, 18, 222. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1076195/pdf/pnas01731-0012.pdf (accessed on 25 September 2021). [CrossRef] [PubMed] [Green Version]
- Blakeslee, A.F.; Avery, A.G. Methods of inducing doubling of chromosomes in plants. J. Hered. 1937, 28, 393–411. [Google Scholar] [CrossRef]
- Eigsti, O.J. A Cytological Study of Colchicine Effects in the Induction of Polyploidy in Plants. Proc. Natl. Acad. Sci. USA 1938, 24, 56. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1077028/pdf/pnas01790-0006.pdf (accessed on 20 July 2021). [CrossRef] [PubMed] [Green Version]
- Murashige, T.; Nakano, R. Tissue Culture as a Potential Tool in Obtaining Polyploid Plants. J. Hered. 1966, 57, 115–118. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2015, 243, 281–296. [Google Scholar] [CrossRef]
- Eng, W.-H.; Ho, W.-S. Polyploidization using colchicine in horticultural plants: A review. Sci. Hortic. 2018, 246, 604–617. [Google Scholar] [CrossRef]
- Kuehnle, A.R. Orchids. In Flower Breeding and Genetics; Anderson, N.O., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 539–560. Available online: https://doi.org/https://doi.org/10.1007/978-1-4020-4428-1_20 (accessed on 18 August 2021). [CrossRef]
- Vichiato, M.R.M.; Vichiato, M.; Pasqual, M.; Castro, D.M.; Dutra, L.F. Indução e Identificação de Tetraplóides Em Dendrobium nobile Lindl. (Orchidaceae). Rev. Ciência Agronômica 2007, 38, 385–390. Available online: http://www.ccarevista.ufc.br/seer/index.php/ccarevista/article/view/98 (accessed on 14 May 2021).
- Wimber, D.E.; Wimber, D.R. Floral Characteristics of Diploid and Neotetraploid Cymbidiuns. Am. Orchid. Soc. Bull. 1967, 38, 572–576. [Google Scholar]
- Xu, L.; Najeeb, U.; Naeem, M.S.; Daud, M.K.; Cao, J.S.; Gong, H.J.; Shen, W.Q.; Zhou, W.J. Induction of tetraploidy in Juncus effusus by colchicine. Biol. Plant. 2010, 54, 659–663. [Google Scholar] [CrossRef]
- Silva, P.A.K.X.D.M.E.; Callegari-Jacques, S.; Bodanese-Zanettini, M.H. Induction and identification of polyploids in Cattleya intermedia Lindl. (orchidaceae) by in vitro techniques. Ciência Rural 2000, 30, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Menezes-Sá, T.S.A.; Arrigoni-Blank, M.D.F.; da Costa, A.S.; Santos-Serejo, J.D.A.; Blank, A.F.; Soares, C.A.; Moura, G.M.S. Chromosome doubling in Cattleya tigrina A. Rich. Sci. Plena 2019, 15, 202. [Google Scholar] [CrossRef] [Green Version]
- Menninger, E.D. Diary of a Colchicine-Induced Tetraploid Cymbidium. Am. Orchid Soc. Bull. 1963, 32, 885–887. [Google Scholar]
- Wimber, D.E.; van Cott, A. Artificially Induced Polyploidy in Cymbidiums. In Proceedings of the 5th World Orchid Conference, Long Beach, CA, USA, 19–17 March 1966. [Google Scholar]
- Kim, M.S.; Won, J.Y.; Song, C.H.; Eun, J.S.; Lee, D.W. Polyploidy Induction of Cymbidium kanran by Treatment of Colchicine in Vitro. RDA J. Hortic. Sci. 1997, 39, 73–76. [Google Scholar]
- Hwang, S.-H.; Kim, M.-S.; Park, S.-Y. Improvement of Chromosome Doubling Efficiency in Cymbidium Hybrids by Colchicine and Oryzalin Treatment. Korean J. Hortic. Sci. Technol. 2015, 33, 900–910. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.-J.; Gao, S.-P.; Zou, Z.-L.; Chen, G. Primary Studies on Polyploid Induction of Cymbidium Hybrid In Vitro. North. Hortic. 2009, 6, 915. [Google Scholar]
- Wang, M.; Zeng, R.; Xie, L.; Li, Y.; Zeng, F.; Zhang, Z. In Vitro Induction and Its Identification of Tetraploid Cymbidium hybridum. Acta Bot. Boreali-Occident. Sin. 2010, 30, 56–62. [Google Scholar]
- Xuejiao, L.; Zhilin, L.; Lipin, H. Induction and Identification of Polyploids in Wild Cymbidium lowianum. Chin. Agric. Sci. Bull. 2010, 13, 60. [Google Scholar]
- Ji, B.X.; Chen, D.W.; Zhang, C.C.; Min, D.; Huang, W.J.; Wang, Y. High Efficient Polyploid Induction of Cymbidium hybridum. Bull. Bot. Res. 2011, 31, 558–562. [Google Scholar]
- Mugui, W.; Ruizhen, Z.; Li, X.; Xuhua, G.; Zhisheng, Z. In Vitro Polyploid Induction and Its Identification in Cymbidium sinense. Chin. Agric. Sci. Bull. 2011, 27, 132–136. [Google Scholar]
- Song, L.; Yang, J.; Liu, D.; Li, Z.; Wang, Y. Polyploid Induction in Cymbidium sinense ‘Lv Mosu’ × Cymbidium hybridum Shijieheping F1 Generation. Guangxi Zhiwu Guihaia 2018, 38, 188–194. [Google Scholar]
- Grosso, V.; Farina, A.; Giorgi, D.; Nardi, L.; Diretto, G.; Lucretti, S. A high-throughput flow cytometry system for early screening of in vitro made polyploids in Dendrobium hybrids. Plant Cell Tissue Organ Cult. 2017, 132, 57–70. [Google Scholar] [CrossRef]
- Yenchon, S.; Te-Chato, S. Polyploidy induction of Dendrobium formosum by colchicine treatment in vitro. Acta Hortic. 2014, 12, 81–88. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, J. In vitro tetraploid induction from multigenotype protocorms and tetraploid regeneration in Dendrobium officinale. Plant Cell Tissue Organ Cult. 2020, 141, 289–298. [Google Scholar] [CrossRef]
- Tantasawat, P.; Khairum, A.; Chaowiset, W.; Wannajindaporn, A. Pronamide-induced polyploidy in rhynchostylis and dendrobium. Acta Hortic. 2012, 75, 615–620. [Google Scholar] [CrossRef]
- Kondo, H.; Phlaetita, W.; Mii, M.; Kikuchi, S.; Deguchi, A.; Miyoshi, K. Efficient Chromosome Doubling of an Interspecific Hybrid Dendrobium Stardust ‘Fire Bird’ by Treatment of Amiprofos-methyl to Protocorm-Like Body. In Vitro Cell. Dev. Biol. Plant 2020, 56, 738–749. [Google Scholar] [CrossRef]
- Chaicharoen, S.; Saejew, K. Autopolyploidy in Dendrobium phalaenopsis. J. Sci. Soc. Thail. 1981, 7, 25–32. Available online: http://scienceasia.org/1981.07.n1/v07_025_032.pdf (accessed on 21 October 2021).
- Atichart, P.; Bunnag, S. Polyploid Induction in Dendrobium secundum (Bl.) Lindl. by in Vitro Techniques. Thail. J. Agric. Sci. 2007, 40, 91–95. Available online: https://www.thaiscience.info/journals/Article/TJAS/10469513.pdf (accessed on 21 October 2021).
- Sarathum, S.; Hegele, M.; Tantiviwat, S.; Nanakorn, M. Effect of Concentration and Duration of Colchicine Treatment on Polyploidy Induction in Dendrobium scabrilingue L. Eur. J. Hortic. Sci. 2010, 75, 123–127. Available online: http://www.pubhort.org/ejhs/2010/file_1591098.pdf (accessed on 21 October 2021).
- Bunnag, S.; Hongthongkham, J. Polyploidy Induction of the Dendrobium Hybrid’Miss Singapore’through Tissue Culture. Acta Horticulturae. 2012, 953, 181–185. [Google Scholar] [CrossRef]
- Atichart, P. Polyploid Induction by Colchicine Treatments and Plant Regeneration of Dendrobium chrysotoxum. Thai J. Agric. Sci. 2013, 46, 59–63. Available online: https://www.researchgate.net/profile/Porntip_Atichart/publication/26555927_Agrobacterium-Mediated_Transformation_of_Dendrobium_secundum_Bl_Lindl_with_Antisense_ACC_Oxidase/links/0c96052436c218f654000000.pdf (accessed on 23 October 2021).
- Bunnag, S.; Hongthongkham, J. Polyploid induction of Dendrobium draconis rchb.f. Acta Hortic. 2015, 9, 445–451. [Google Scholar] [CrossRef]
- Zakizadeh, S.; Kaviani, B.; Hashemabadi, D. In vivo-induced polyploidy in Dendrobium ‘Sonia’ in a bubble bioreactor system using colchicine and oryzalin. Braz. J. Bot. 2020, 43, 921–932. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, J. Colchicine-induced tetraploidy in Dendrobium cariniferum and its effect on plantlet morphology, anatomy and genome size. Plant Cell Tissue Organ Cult. 2020, 144, 409–420. [Google Scholar] [CrossRef]
- Griesbach, R.J. Polypioidy in Phalaenopsis orchid improvement. J. Hered. 1985, 76, 74–75. [Google Scholar] [CrossRef]
- Della Rahayu, E.M.; Sukma, D.; Syukur, M. Rawati Induksi Poliploidi Phalaenopsis amabilis (L.) Blume dan Phalaenopsis amboinensis JJ Smith dengan Kolkisin dalam Kultur In Vitro. J. Agron. Indones. 2016, 43, 219. [Google Scholar] [CrossRef] [Green Version]
- Azmi, T.K.; Sukma, D.; Aziz, S.A.; Syukur, M. Polyploidy Induction of Moth Orchid (Phalaenopsis amabilis (L.) Blume) by Colchicine Treatment on Pollinated Flowers. J. Agric. Sci. 2016, 11, 2. Available online: http://repo.lib.sab.ac.lk:8080/xmlui/bitstream/handle/123456789/1013/8118-28584-2-PB.pdf?sequence=1&isAllowed=y (accessed on 29 June 2021). [CrossRef] [Green Version]
- Azmi, T.K.K.; Sukma, D.; Aziz, S.A.; Syukur, D.M. Morfologi dan Pertumbuhan Planlet Hasil Induksi Poliploidi melalui Perlakuan Kolkisin pada Kuncup Bunga Anggrek Bulan (Phalaenopsis amabilis (L.) Blume). J. Agron. Indones. 2016, 44, 68. [Google Scholar] [CrossRef] [Green Version]
- Rahayu, E.M.D.; Sukma, D.; Syukur, M.; Aziz, S.A.; Irawati, D. In Vivo Polyploid Induction Using Colchicine of Moth Orchid Seedling (Phalaenopsis amabilis (L.) Blume). Buletin Kebun Raya 2015, 18, 41–48. Available online: https://publikasikr.lipi.go.id/index.php/buletin/article/view/31/31 (accessed on 13 June 2021).
- Cui, G. Tetraploid of Phalaenopsis Induction via Colchicine Treatment from Protocorm like Bodies in Liquid Culture. Agric. Life Sci. 2010, 36, 49. [Google Scholar]
- Wongprichachan, P.; Huang, K.-L.; Chou, Y.-M.; Hsu, S.-T.; Liu, T.-Y.; Okubo, H. Induction of Unreduced Gamete in Phalaenopsis by N2O Treatments. J. Fac. Agric. Kyushu Univ. 2013, 58, 27–31. [Google Scholar] [CrossRef]
- Rungruchkanont, K.; Apisitwanich, S. Colchicine treatment: A method for genetic diversity induction of Doritis pulcherrima lindl. orchid of Thailand. Acta Hortic. 2015, 25, 175–180. [Google Scholar] [CrossRef]
- Zaker Tavallaie, F.; Kolahi, H. Induction of in Vitro Polyploidy in Ornamental Flowers of Orchid Species (Phalaenopsis amabilis). Iran. J. Rangel. Forests Plant Breed. Genet. Res. 2017, 25, 259–270. Available online: https://ijrfpbgr.areeo.ac.ir/article_113315_9ffa7b17d5edbc206df5e4be0d1d9ac1.pdf?lang=en (accessed on 13 October 2021).
- Unemoto, L.K.; De Faria, R.T.; Destro, D.; Barbosa, C.M.; Lone, A.B. Sobrevivência e diferenciação de protocormos de Oncidium flexuosum submetidos a tratamento com ácido peracético e colchicina. Acta Sci. Agron. 2009, 31, 503–508. [Google Scholar] [CrossRef]
- Cui, G.R.; Zhang, Z.X.; Zhang, C.Y.; Hu, N.B.; Sui, Y.H.; Li, J.Q. Polyploid Induction and Identification of Oncidium. Acta Prataculturae Sinica 2010, 19, 184–190. [Google Scholar]
- Nakasone, H.Y. Artificial Induction of Polyploidy in Orchids by the Use of Colchicine. 1960. Available online: https://scholarspace.manoa.hawaii.edu/bitstream/10125/11486/uhm_phd_6004660_r.pdf (accessed on 13 September 2021).
- Tuwo, M.; Indrianto, A. Improvement of Orchid Vanda Hybrid (Vanda limbata Blume X Vanda tricolor Lindl. var. suavis) By Colchicines Treatment In Vitro. Mod. Appl. Sci. 2016, 10, 83. [Google Scholar] [CrossRef] [Green Version]
- Kerdsuwan, N.; Te-chato, S. Effects of Colchicine on Survival Rate, Morphological, Physiological and Cytological Characters of Chang Daeng Orchid (Rhynchostylis gigantea var. rubrum sagarik) in vitro. J. Agric. Technol. 2012, 8, 1451–1460. Available online: http://ijat-aatsea.com/pdf/v8_n4_12_July/25_IJAT_2012_8_4__NattapornKerdsuwan-plantscience-accepted.pdf (accessed on 13 September 2021).
- Chung, M.Y.; Kim, C.Y.; Min, J.S.; Lee, D.-J.; Naing, A.H.; Chung, J.D.; Kim, C.K. In Vitro Induction of Tetraploids in an Interspecific Hybrid of Calanthe (Calanthe discolor × Calanthe sieboldii) through Colchicine and Oryzalin Treatments. Plant Biotechnol. Rep. 2014, 8, 251–257. Available online: https://link.springer.com/content/pdf/10.1007/s11816-014-0317-4.pdf (accessed on 14 June 2021). [CrossRef]
- Huy, N.P.; Tam, D.T.T.; Luan, V.Q.; Tung, H.T.; Hien, V.T.; Ngan, H.T.M.; Duy, P.N.; Nhut, D.T. In vitro polyploid induction of Paphiopedilum villosum using colchicine. Sci. Hortic 2019, 252, 283–290. [Google Scholar] [CrossRef]
- Suhaila, A.S.; Norwati, M.; Yap, B.J.W.; Mahani, M.C.; Kandasamy, K.I.; Faridah, Q.Z.; Fadelah, A.A.; Hasnida, H.N.; Nazirah, A.; Haliza, I.; et al. The Potential of Paphiopedilum callosum Polyploids towards the Development of an Improved Native Malaysian Slipper Orchid. In Proceedings of the Conference on Forestry and Forest Products Research 2014, Kahramanmaraş, Turkey, 8 May 2014; Available online: https://www.researchgate.net/profile/Nor-Fadilah-WOOK/publication/275884534_Production_of_high_quality_planting_materials_of_Eurycoma_longifolia_and_Labisia_pumila_in_FRIM_A_Mohd_Zaki_MA_Farah_Fazwa_N_Lokmal_S_Norhayati_SB_Syafiqah_Nabilah_W_Norfadilah/li (accessed on 16 June 2021).
- CDC. Colchicine: Biotoxine. Centers of Disease Control and Prevention. 2011. Available online: https://www.cdc.gov/niosh/ershdb/emergencyresponsecard_29750016.html (accessed on 12 October 2021).
| Species/ Cultivar | Type of Explant | Treatment | Polyploidy Assessment Method | Polyploidy Efficiency | Anatomical and Morphological Features of Polyploids | References |
|---|---|---|---|---|---|---|
| Cattleya intermedia Lind. | PLBs | Colchicine 0.05% for 4 days for Clon 114 Colchicine 0.1% for 4 days for Clon 121 | Chromosome count Stomatal density | Clon 114 Mixoploides (42.0%) Tetraploides (29.0%) Clon 121 Mixoploides (33.0%) Tetraploides (22.0%) | Higher stomatal density | Silva, Callegari-Jacques, and Bodanese-Zanettini [96] |
| Cattleya tigrina Lind. | Plantlets | Colchicine 12.5 mM for 48-h | Flow cytometry Stomatal density Stomatal functionality Stomatal index | 72.5% polyploids | Greater stomatal functionality, lower stomatal density, lower stomatal index | Menezes-Sá et al. [97] |
| Species/Cultivar | Type of Explant | Treatment | Polyploidy Assessment Method | Polyploidy Efficiency | Anatomical and Morphological Features of Polyploids | References |
|---|---|---|---|---|---|---|
| Cymbidium ‘Promised Land’ | PLBs | Oryzalin 14.4, 28.9 and 57.7_M | - | - | - | Miguel and Leonhardt [30] |
| Cym. hydridum | PLBs | Colchicine 0.1% for 3 days | Chromosome count Morphological characterization | 27.6% polyploids | Shorter length and number of roots, larger root diameter | Xie et al. [43] |
| Hybrids: Cym. Showgirls “Sirly” and Cym. Mystery Island “Silk Road” | PLBs | Colchicine 50 mg L−1 for 7 days Oryzalin 5 mg L−1 for 2 weeks Colchicine 50 mg L−1 for 7 days Oryzalin 10 mg L−1 for 3 days | Flow cytometry Morphological characterization Anatomical characterization | Cymbidium Showgirls “Sirly” (60.0% with colchicine and 47.0% with oryzalin) Cymbidium Mystery Island “Silk Road” (16.7% with colchicine and 6.7% with oryzalin) | Shorter and wider dark green leaves, less growth. Greater length and width of guard cells, lower stomatal density. | Hwang, Kim, and Park [101] |
| Cym. Ruby Shower ‘Murasakin Okimi’ | PLBs | Colchicine 300 mg L−1 for 15 days | Chromosome count | 30.0% polyploids | Yang et al. [102] | |
| Cym. hybridum | PLBs | Colchicine 0.05% for 5 days | Morphological characterization Stomatal density Anatomical characterization | 23.7% polyploids | Darker, compact and resistant leaves | Wang et al. [103] |
| Cym. lowianum | Seedlings | Colchicine 0.04% for 72 h | Morphological characterization Anatomical characterization | 60.0% polyploids | Short stem, obscure leaves, greater width, less growth. Larger size of stomata, smaller number of stomata | Xuejiao, Zhilin, and Lipin [104] |
| Cym. hybridum | Young shoots | Colchicine 0.05% por 24 h | Morphological characterization Anatomical characterization | 28.2% polyploids | Ji et al. [105] | |
| Cym. sinense ‘QiJianBaiMo’ | Rhizome | Colchicine 0.01% for 3 days | 11.1% polyploids | Hard leaves and thickened roots | Mugui et al. [106] | |
| Hybrid: Cym. sinenthese ‘Lv mosu’ × Cym. hybridum ‘Shijieheping’ | Protocorms | Colchicine 0.03% for 72 h | Flow cytometry Morphological characterization Anatomical characterization | 36.0% polyploids | Wider green leaves, thicker roots and less growth. Greater length and width of guard cells, lower stomatal density. | Song et al. [107] |
| Species | Types of Explant | Treatment | Polyploidy Assessment Method | Polyploidy Efficiency | Anatomical and Morphological Features of Polyploids | References |
|---|---|---|---|---|---|---|
| Dendrobium‘Gatton Sun Ray’ | PLBs | Oryzalin 14.4 μM (5 mg L−1) for 6 days | Anatomical characterization | Longer stomata length | Miguel and Leonhardt [30] | |
| Den. nobile | Seedlings | Colchicine 0.1% for 96 h | Chromosome count Morphological characterization | 29.2% polyploids | Smaller height, smaller diameter of pseudobulbs, smaller leaf length, larger leaf width. | Vichiato et al. [93] |
| Den. phalaenopsis × Den. loddigesii | PLBs | Colchicine 0.05% (5 mg L−1) for 3 days | Flow cytometry Anatomical characterization | 80.0% polyploids | Longer stomata length, lower stomatal density. | Grosso et al. [108] |
| Den. formosum | PLBs | Colchicine 0.2% (20 mg L−1) for 48 h | Morphological characterization Anatomical characterization Stomatal density | 75.0% polyploids | Longer stem, thick green leaves—obscure. Larger stomata size, lower stomatal density. | Yenchon and Te-chato [109] |
| Den. officinale | Protocorms | Oryzalin 14.4 μM (5 mg L−1) for 24 h | Flow cytometry Chromosome count | 37.4% polyploids | Smaller height, smaller leaf length, smaller root length, larger stem and root diameter, larger lip and gynostemium width. | Zhang and Gao [110] |
| Den.‘Burana Jade’ | Protocorms | Pronamide 100 μM for 2 days | Morphological characterization Anatomical characterization | 33.3% polyploids | Thickened leaves, shorter length, short and thickened stem. Greater number of stomata. | Tantasawat et al. [111] |
| Den. stardust “Fire Bird” | PLBs | Amiprofosfo-metil (AMP) 160 mg for 12 | Flow cytometry Chromosome count Morphological characterization | 80.0% amphydiploids | Larger stem size and number of leaves. | Kondo et al. [112] |
| Den. phalaenopsis | Protocorms | Colchicine 0.05% for 9 days | Chromosome count Morphological characterization | 50.0% polyploids | Thick and dark green leaves, greater number of flowers/inflorescences. Greater guard cell length, greater guard cell width. | Chaicharoen and Saejew [113] |
| Den. secundum | Protocorms | Colchicine 0.05% for 1 day | Flow cytometry Chlorophyll Content Anatomical characterization | Greater thickness of roots, stem and leaves, greater flower size. Greater length of guard cells. | Atichart and Bunnag [114] | |
| Den. srabrilingue | PLBs | Colchicine 0.075% (7.5 mg L−1) for 14 days | Flow cytometry | 43.1% polyploids | Larger diameter of stem and roots, dark green leaves. | Sarathum et al. [115] |
| Den. “Miss Singapore” | Protocorms | Colchicine 0.01% (1 mg L−1) for 2 days | Flow cytometry Chromosome count | Bunnag and Hongthongkham [116] | ||
| Den. chrysotoxum | PLBs | Colchicine 0.04% (4 mg L−1) for 24 h | Flow cytometry | 47.0% tetraploids | Atichart [117] | |
| Den. draconis | Protocorms | Colchicine 0.05% (5 mg L−1) for 3 days | Flow cytometry Morphological characterization Anatomical characterization | 43.0% polyploids | Smaller stem size; large, thick, dark green leaves. Smaller guard cell number/area unit, larger guard cell size. | Bunnag and Hongthongkham [118] |
| Den. ‘Sonia’ | PLBs | Colchicine 0.15% (15 mg L−1) for 3 days | Flow cytometry Chromosome count Morphological characterization Anatomical characterization | 26.6% polyploids | Greater mass and smaller width of the bulb, greater length and width of leaves. Longer stomata length. | Zakizadeh, Kaviani, and Hashemabadi [119] |
| Den. cariniferum | Protocorms | Colchicine 0.05% (5 mg L−1) for 24 h | Flow cytometry Morphological characterization Anatomical characterization | 33.0% polyploids | Greater width and thickening of leaves, greater diameter of stem and root. Greater length of stomata, lower stomatal density, greater number of chloroplasts, thickened spongy tissue, larger leaf veins, smaller cells of the adaxial epidermis and trichomes | Zhang and Gao [120] |
| Species | Type of Explant | Treatment | Polyploidy Assessment Method | Polyploidy Efficiency | Anatomical and Morphological Features of Polyploids | References |
|---|---|---|---|---|---|---|
| Phal. equestris, Phal. fasciata, Phal. Betty Hausermann | Protocorms | Colchicine 50 mg L−1 for 10 days | Chromosome count | 46.0% polyploids | Griesbach [20] | |
| Phal. bellina | Protocorms | Oryzalin 14.4 μM (5 mg L−1) for 3 days | Anatomical characterization | Longer stomata length | Miguel and Leonhardt [30] | |
| Phal. amabilis var. grandiflora | In vitro plantlets from PLBs | 0.15% colchicine for 72 h under bubble bioreactor | Flow cytometry, morphological and cytological measurements | Reduction in plantlet length and number, and stomatal density; Increases in leaf number and width, guard cells and chloroplast number | Mohammadi, Kaviani, and Sedaghathoor [41] | |
| Phal. Goden Sands “Canary” | Protocorms | Colchicine 0.5 mg L−1 for 10 days | 50.0% polyploids | More obscure flowers, larger and larger in diameter | Griesbach [121] | |
| Phal. amabilis; Phal. amboinensis | Protocorms | Colchicine 50 mg L−1 for 10 days | Chromosome count Anatomical characterization Stomatal density | 33.3% polyploids of Phal. amabilis 40.0% polyploids of Phal. amboinensis | Thick dark green leaves. Longer stomatal length, lower stomatal density | Rahayu et al. [122] |
| Phal. amabilis | Pollinated flowers | Colchicine 50 mg L−1 for 3 or 5 days Colchicine 500 mg L−1 for 5 days | Morphological characterization Anatomical characterization | 60.0% polyploids 100.0% polyploids | Shorter leaf length, shorter length and larger diameter of roots, longer length and diameter of the basal organ of the protocorm. Longer stomatal length | Azmi et al. [123] |
| Phal. amabilis | Bud flowers | Colchicine for 3 days at 50, 500 or 1000mg L−1 | Morphological characterization | 71.2% polyploids * 86.7% polyploids * 100.0% polyploids * | Smaller root size, larger number of roots, larger root thickness, larger stem diameter, larger length of the basal organ of the protocorm. | Azmi et al. [124] |
| Phal. amabilis | Seedlings | Colchicine 5000 mg/L | Morphological characterization Anatomical characterization Stomatal density | 50.0% polyploids | Greatest height Greater length and width of stomata, lower stomatal density | Rahayu et al. [125] |
| Phalaenopsis | PLBs | Colchicine 500, 1000 and 2000 mg L−1 for 1, 3 e 7 days | Cui G. [126] | |||
| Phal. amabilis | Pollen grains | Nitrous oxide (N2O) for 24 h Nitrous oxide (N2O) for 48 h | Chromosome count Flow cytometry | Triploids (36.0%) Tetraploids (7.0%) Tetraploids (5.0%) | - | Wongprichachan et al. [127] |
| Phal. pulcherrima (ex Doritis pulcherrima) | Protocorms | Colchicine 100 mg L−1 for 10 days | Chromosome count | 25.0% polyploids | Rungruchkanont and Apisitwanich [128] | |
| Phal. amabilis | Auxiliary gems | Colchicine 0.2% for 48 h | Chromosome count Flow cytometry Anatomical characterization Morphological characterization | 70.0% polyploids | Increase in size and decrease in density of guard cells | Zaker Tavallaie and Kolahi [129] |
| Species | Explant | Treatment | Polyploidy Assessment Method | Polyploidy Efficiency | Anatomical and Morphological Features of Polyploids | References |
|---|---|---|---|---|---|---|
| Epidendrum‘Helen’s Pride’ | Protocorms | Oryzalin 57.7 μM (20 mg L−1) for 6 days | Anatomical characterization | 2 polyploids | Longer stomata length | Miguel and Leonhard [30] |
| Odontioda ‘Emma Sander’ | Protocorms | Oryzalin 28.9 μM (10 mg L−1), 14,4 μM (5 mg L−1) or 57.7 μM (20 mg L−1) for 6 dias | Anatomical characterization | 3 polyploids 2 polyploids 3 polyploids | Longer stomata length | Miguel and Leonhardt [30] |
| Rynchostylis gigantea ‘K3.0124W’ Rynchostylis gigantea ‘K3.0131W’ | Seeds | Pronamide 200 μM for 4 days | Chromosome count Morphological characterization Stomatal density Anatomical characterization | Rynchostylis gigantea ‘K3.0124W’ (23.0%) Rynchostylis gigantea ‘K3.0131W’ (35.0%) | Thick, rounded leaves. Anatomic: lower stomatal density, larger stomata size. | Tantasawat et al. [104] |
| Vanda “Miss Joaquin” | Young shoots | Colchicine 0.5% (50 mg L−1) and 1.5% (150 mg L−1) for 6 days | Chromosome count Anatomical characterization | Longer stomatal length | Nakasone [132] | |
| Hybrid Vanda limbiata Blume X Vanda tricolor Lindl. var. suavis | Protocorms | Colchicine 0.5% (50 mg L−1) for 6 h | Flow cytometry Chromosome count Morphological characterization Anatomical characterization | Smaller number of roots, smaller root length, smaller number of leaves, smaller leaf width. Anatomical: Larger stomatal size, smaller stomatal index. | Tuwo and Indrianto [133] | |
| Rynchostylis gigantea var. rubrum sagarik | PLBs | Colchicine 20 mg L−1 for 72 h | Chromosome count Morphological characterization Stomatal density Number of chloroplasts | 60.0% polyploids | Smaller size of seedlings and roots, larger number of leaves, smaller leaf size. Higher stomatal density, lower number of chloroplasts | Kerdsuwan and Te-chato [134] |
| Hybrid Calanthe discolor X Calanthe sieboldii | Seeds | Colchicine 0.1% (10 mg L−1) for 7 days Oryzalin 0.003% (0,3 mg L−1) for 1 days | Flow cytometry Size of stomata Stomatal density | 81.0% polyploids 86.0% poliploids | Smaller size, thick dark green rounded leaves, larger leaf width, larger stem and root diameter. Lower stomatal density, larger stomatal size | Chung et al. [135] |
| Paphiopedilum villosum | Sprouts | Colchicine 50 μM (20 mg L−1) for 6 days | Flow cytometry Chromosome count Anatomical Characterization Stomatal density | 19.9% polyploids | Greater leaf length, greater leaf width. Longer guard cell length, lower stomatal density | Huy et al. [136] |
| Paphiopedilum callosum | Seeds | Colchicine 1000 mg L−1 for 54 h | Chromosome count Morphological characterization Anatomical characterization | Larger seedling size | Suhaila Siti et al. [137] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vilcherrez-Atoche, J.A.; Iiyama, C.M.; Cardoso, J.C. Polyploidization in Orchids: From Cellular Changes to Breeding Applications. Plants 2022, 11, 469. https://doi.org/10.3390/plants11040469
Vilcherrez-Atoche JA, Iiyama CM, Cardoso JC. Polyploidization in Orchids: From Cellular Changes to Breeding Applications. Plants. 2022; 11(4):469. https://doi.org/10.3390/plants11040469
Chicago/Turabian StyleVilcherrez-Atoche, Joe Abdul, Carla Midori Iiyama, and Jean Carlos Cardoso. 2022. "Polyploidization in Orchids: From Cellular Changes to Breeding Applications" Plants 11, no. 4: 469. https://doi.org/10.3390/plants11040469
APA StyleVilcherrez-Atoche, J. A., Iiyama, C. M., & Cardoso, J. C. (2022). Polyploidization in Orchids: From Cellular Changes to Breeding Applications. Plants, 11(4), 469. https://doi.org/10.3390/plants11040469