Development and Validation of a One-Step Reverse Transcription Real-Time PCR Assay for Simultaneous Detection and Identification of Tomato Mottle Mosaic Virus and Tomato Brown Rugose Fruit Virus
Abstract
:1. Introduction
2. Results
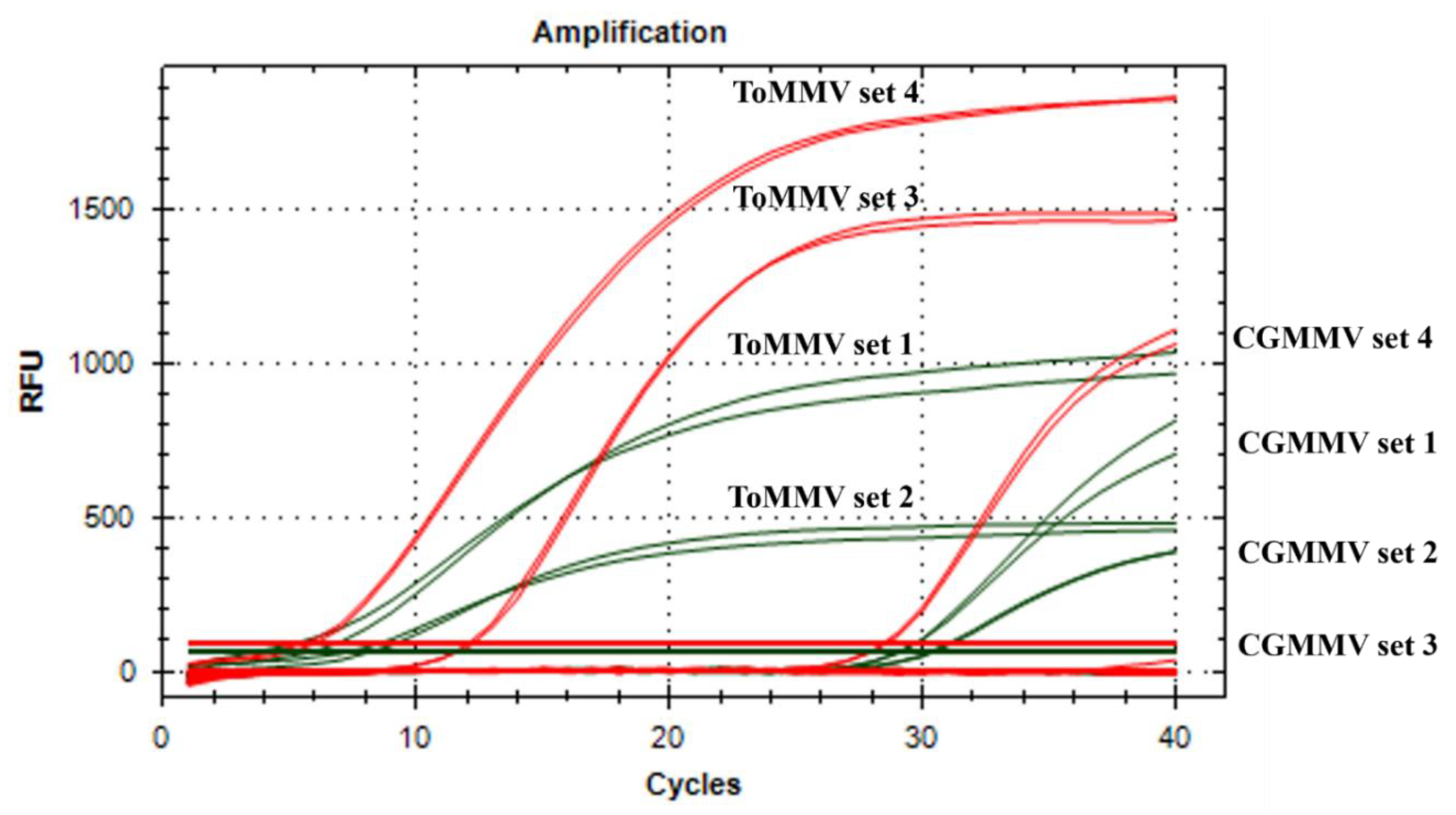
2.1. Primers/Probe Design and Evaluation
2.2. Duplex Assay Validation
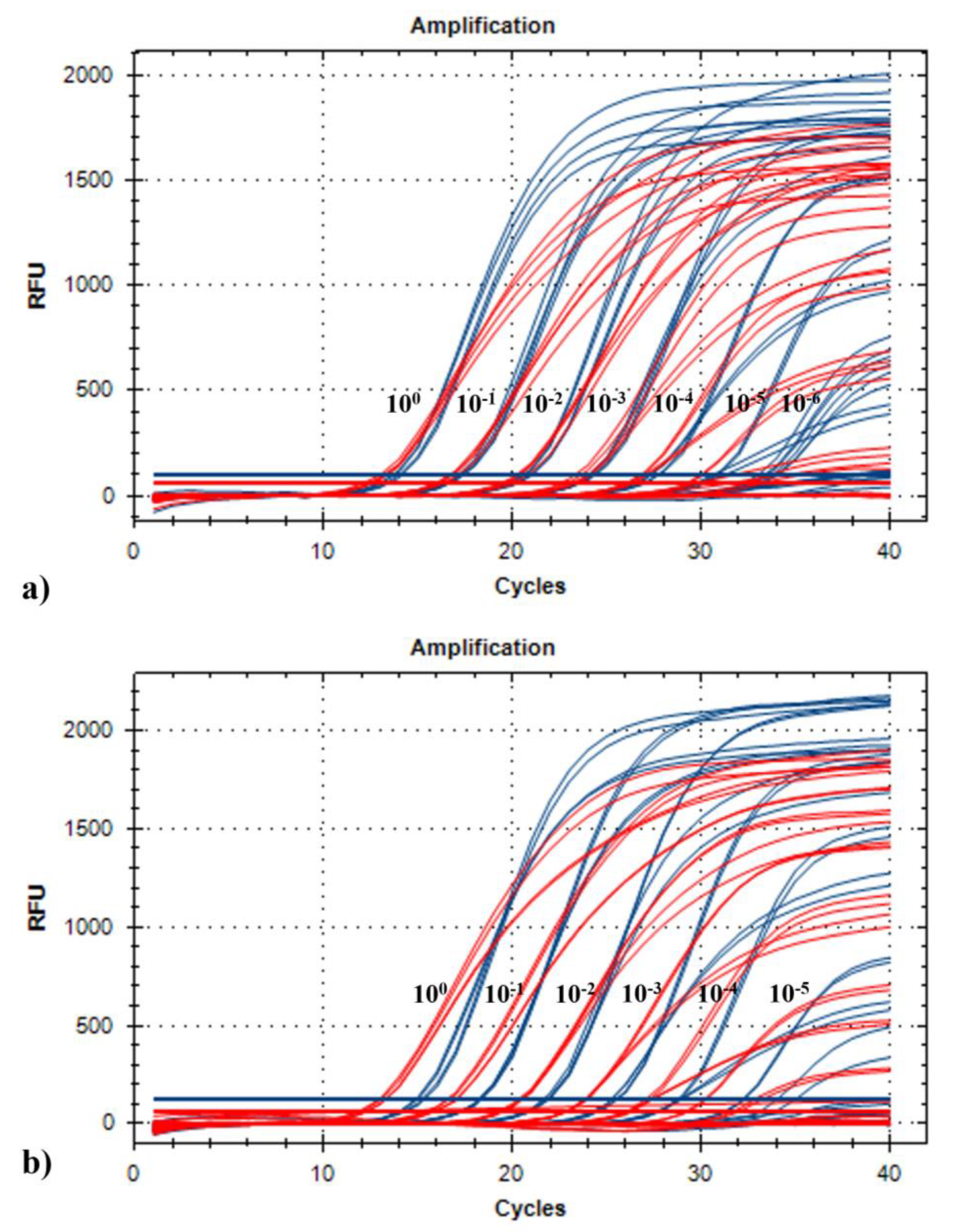
2.2.1. Analytical Sensitivity
2.2.2. Analytical Specificity
2.2.3. Selectivity
2.2.4. Repeatability and Reproducibility
2.3. Use of the Validated Duplex Assay in Diagnosis of Seed Samples
3. Discussion
4. Material and Methods
4.1. Plant Material and RNA Extraction
4.2. Primer Design and Reaction Set Up
4.3. Duplex Assay Validation
4.3.1. Analytical Sensitivity, Limit of Detections and Efficiencies
4.3.2. Analytical Specificity
4.3.3. Selectivity
4.3.4. Repeatability and Reproducibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. FAOSTAT; Food and Agriculture Organization of the United Nations: Rome, Italy, 2021. [Google Scholar]
- Viruses of Plants. In Descriptions and Lists from the VIDE Database; Brunt, A.; Crabtree, K.; Dallwitz, M.; Gibbs, A.; Watson, L.; Zurcher, E. (Eds.) Australian National University: Canberra, Australia, 1997. [Google Scholar]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging Viral Diseases of Tomato Crops. Mol. Plant-Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dombrovsky, A.; Smith, E. Seed Transmission of Tobamoviruses: Aspects of Global Disease Distribution. In Advances in Seed Biology; InTech: London, UK, 2017. [Google Scholar]
- Maule, A.; Wang, D. Seed Transmission of Plant Viruses: A Lesson in Biological Complexity. Trends Microbiol. 1996, 4, 153–158. [Google Scholar] [CrossRef]
- Broadbent, L.; Fletcher, J.T. The Epidemiology of Tomato Mosaic. Ann. Appl. Biol. 1963, 52, 233–241. [Google Scholar] [CrossRef]
- Roberts, A.G. Plant Viruses: Soil-borne. In eLS; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A.; et al. A New Israeli Tobamovirus Isolate Infects Tomato Plants Harboring Tm-22 Resistance Genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef] [Green Version]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A New Tobamovirus Infecting Tomato Crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef]
- EPPO EPPO Global Database. Available online: https://gd.eppo.int/taxon/TOBRFV/distribution (accessed on 16 December 2021).
- Panno, S.; Caruso, A.G.; Davino, S. First Report of Tomato Brown Rugose Fruit Virus on Tomato Crops in Italy. Plant Dis. 2019, 103, 1443. [Google Scholar] [CrossRef]
- Skelton, A.; Buxton-Kirk, A.; Ward, R.; Harju, V.; Frew, L.; Fowkes, A.; Long, M.; Negus, A.; Forde, S.; Adams, I.P.; et al. First Report of Tomato Brown Rugose Fruit Virus in Tomato in the United Kingdom. New Dis. Rep. 2019, 40, 12. [Google Scholar] [CrossRef] [Green Version]
- EPPO First Report of Tomato Brown Rugose Fruit Virus in the Netherlands. EPPO Reporting Service. 2019. Available online: https://gd.eppo.int/reporting/article-6639 (accessed on 6 December 2021).
- EPPO First Report of Tomato Brown Rugose Fruit Virus in Greece. Available online: https://gd.eppo.int/reporting/article-6640 (accessed on 6 December 2021).
- EPPO First Report of Tomato Brown Rugose Fruit Virus in Spain. Available online: https://gd.eppo.int/reporting/article-6668 (accessed on 6 December 2021).
- Cambrón-Crisantos, J.M.; Rodríguez-Mendoza, J.; Valencia-Luna, J.B.; Alcasio-Rangel, S.; García-Ávila, C.D.J.; López-Buenfil, J.A.; Ochoa-Martínez, D.L. Primer Reporte de Tomato Brown Rugose Fruit Virus (ToBRFV) En Michoacán, México. Rev. Mex. Fitopatol. 2018, 37, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Ling, K.-S.; Tian, T.; Gurung, S.; Salati, R.; Gilliard, A. First Report of Tomato Brown Rugose Fruit Virus Infecting Greenhouse Tomato in the United States. Plant Dis. 2019, 103, 1439. [Google Scholar] [CrossRef]
- Fidan, H.; Sarikaya, P.; Calis, O. First Report of Tomato Brown Rugose Fruit Virus on Tomato in Turkey. New Dis. Rep. 2019, 39, 18. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.-Y.; Ma, H.-Y.; Han, S.-L.; Geng, C.; Tian, Y.-P.; Li, X.-D. First Report of Tomato Brown Rugose Fruit Virus Infecting Tomato in China. Plant Dis. 2019, 103, 2973. [Google Scholar] [CrossRef]
- PM 7/146 (1) Tomato Brown Rugose Fruit Virus. EPPO Bull. 2021, 51, 178–197. [CrossRef]
- Li, R.; Gao, S.; Fei, Z.; Ling, K.-S. Complete Genome Sequence of a New Tobamovirus Naturally Infecting Tomatoes in Mexico. Genome Announc. 2013, 1, e00794-13. [Google Scholar] [CrossRef] [Green Version]
- Zhan, B.; Cao, N.; Wang, K.; Zhou, X. Detection and Characterization of an Isolate of Tomato Mottle Mosaic Virus Infecting Tomato in China. J. Integr. Agric. 2018, 17, 1207–1212. [Google Scholar] [CrossRef] [Green Version]
- Sui, X.; Zheng, Y.; Li, R.; Padmanabhan, C.; Tian, T.; Groth-Helms, D.; Keinath, A.P.; Fei, Z.; Wu, Z.; Ling, K.-S. Molecular and Biological Characterization of Tomato Mottle Mosaic Virus and Development of RT-PCR Detection. Plant Dis. 2017, 101, 704–711. [Google Scholar] [CrossRef] [Green Version]
- Ambrós, S.; Martínez, F.; Ivars, P.; Hernández, C.; de la Iglesia, F.; Elena, S.F. Molecular and Biological Characterization of an Isolate of Tomato Mottle Mosaic Virus (ToMMV) Infecting Tomato and Other Experimental Hosts in Eastern Spain. Eur. J. Plant Pathol. 2017, 149, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Webster, C.G.; Rosskopf, E.N.; Lucas, L.; Mellinger, H.C.; Adkins, S. First Report of Tomato Mottle Mosaic Virus Infecting Tomato in the United States. Plant Health Prog. 2014, 15, 151–152. [Google Scholar] [CrossRef]
- Turina, M.; Geraats, B.P.J.; Ciuffo, M. First Report of Tomato Mottle Mosaic Virus in Tomato Crops in Israel. New Dis. Rep. 2016, 33, 1. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.Y.; Wang, C.L.; Xiang, D.; Li, R.H.; Liu, Y.; Li, F. First Report of Tomato Mottle Mosaic Virus Infection of Pepper in China. Plant Dis. 2014, 98, 1447. [Google Scholar] [CrossRef]
- Nagai, A.; Duarte, L.M.L.; Chaves, A.L.R.; Alexandre, M.A.v.; Ramos-González, P.L.; Chabi-Jesus, C.; Harakava, R.; dos Santos, D.Y.A.C. First Complete Genome Sequence of an Isolate of Tomato Mottle Mosaic Virus Infecting Plants of Solanum Lycopersicum in South America. Genome Announc. 2018, 6, e00427-18. [Google Scholar] [CrossRef] [Green Version]
- EPPO. First Report of Tomato Mottle Mosaic Virus in the Czech Republic. EPPO Reporting Service. 2020. Available online: https://gd.eppo.int/reporting/article-6930 (accessed on 10 December 2021).
- Lovelock, D.A.; Kinoti, W.M.; Bottcher, C.; Wildman, O.; Dall, D.; Rodoni, B.C.; Constable, F.E. Tomato Mottle Mosaic Virus Intercepted by Australian Biosecurity in Capsicum Annuum Seed. Australas. Plant Dis. Notes 2020, 15, 8. [Google Scholar] [CrossRef] [Green Version]
- Mut Bertomeo, M. Detección de Tomato Mottle Mosaic Virus En Semilla Comercial de Tomate y Pimiento. 2021. Available online: https://riunet.upv.es/handle/10251/173932 (accessed on 15 December 2021).
- Li, Y.; Wang, Y.; Hu, J.; Xiao, L.; Tan, G.; Lan, P.; Liu, Y.; Li, F. The Complete Genome Sequence, Occurrence and Host Range of Tomato Mottle Mosaic Virus Chinese Isolate. Virol. J. 2017, 14, 15. [Google Scholar] [CrossRef] [Green Version]
- Nagai, A.; Duarte, L.M.L.; Chaves, A.L.R.; Peres, L.E.P.; dos Santos, D.Y.A.C. Tomato Mottle Mosaic Virus in Brazil and Its Relationship with Tm-22 Gene. Eur. J. Plant Pathol. 2019, 155, 353–359. [Google Scholar] [CrossRef]
- JKI Express PRA on Tomato Mottle Mosaic Virus (in German). 2020. Available online: https://pflanzengesundheit.julius-kuehn.de/dokumente/upload/ToMMV_expr-pra.pdf (accessed on 10 December 2021).
- Dutch NPPO Tomato Mottle Mosaic Virus Quick Scan. 2020. Available online: https://english.nvwa.nl/documents/plant/plant-health/pest-risk-analysis/documents/quick-scan-tomato-mottle-mosaic-virus (accessed on 10 December 2021).
- EPPO PM 7/76 (5) Use of EPPO Diagnostic Standards. EPPO Bull. 2018, 48, 373–377. [CrossRef] [Green Version]
- Dovas, C.I.; Efthimiou, K.; Katis, N.I. Generic Detection and Differentiation of Tobamoviruses by a Spot Nested RT-PCR-RFLP Using DI-Containing Primers along with Homologous DG-Containing Primers. J. Virol. Methods 2004, 117, 137–144. [Google Scholar] [CrossRef]
- EPPO PM 7/98 (4) Specific Requirements for Laboratories Preparing Accreditation for a Plant Pest Diagnostic Activity. EPPO Bull. 2019, 49, 530–563. [CrossRef] [Green Version]
- CABI Cucumber Green Mottle Mosaic Virus Datasheet. Available online: https://www.cabi.org/isc/datasheet/16951 (accessed on 20 December 2021).
- Panno, S.; Ruiz-Ruiz, S.; Caruso, A.G.; Alfaro-Fernandez, A.; Font San Ambrosio, M.I.; Davino, S. Real-Time Reverse Transcription Polymerase Chain Reaction Development for Rapid Detection of Tomato Brown Rugose Fruit Virus and Comparison with Other Techniques. PeerJ 2019, 7, e7928. [Google Scholar] [CrossRef] [Green Version]
- Bernabé-Orts, J.M.; Torre, C.; Méndez-López, E.; Hernando, Y.; Aranda, M.A. New Resources for the Specific and Sensitive Detection of the Emerging Tomato Brown Rugose Fruit Virus. Viruses 2021, 13, 1680. [Google Scholar] [CrossRef]
- Menzel, W.; Jelkmann, W.; Maiss, E. Detection of Four Apple Viruses by Multiplex RT-PCR Assays with Coamplification of Plant MRNA as Internal Control. J. Virol. Methods 2002, 99, 81–92. [Google Scholar] [CrossRef]
- Weller, S.A.; Elphinstone, J.G.; Smith, N.C.; Boonham, N.; Stead, D.E. Detection of Ralstonia Solanacearum Strains with a Quantitative, Multiplex, Real-Time, Fluorogenic PCR (TaqMan) Assay. Appl. Environ. Microbiol. 2000, 66, 2853–2858. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]


| Set | Primer/Probe ID | 5′-3′ Sequence | Labeling | Genome Position (nt) | Coding Region |
|---|---|---|---|---|---|
| 1 | ToMMV MWAT Fw | GGAACAGGTTTCTACAACCAGAG | 6124–6146 | CP | |
| ToMMV MWAT Rv | CGTACGCACCACGTGTATTT | 6235–6254 | |||
| ToMMV MWAT Pr1 | TTCTGCGCCAGCGTCCTAAGTAAT | HEX/BHQ1 | 6180–6203 | ||
| 2 | ToMMV MWAT Fw | GGAACAGGTTTCTACAACCAGAG | 6124–6146 | CP | |
| ToMMV MWAT Rv | CGTACGCACCACGTGTATTT | 6235–6254 | |||
| ToMMV MWAT Pr2 | GGCCTGGACTTCTGCGCCA | HEX/BHQ1 | 6171–6189 | ||
| 3 | ToMMV CataAT Fw | CAGCATCTGCTTGGTCGATAA | 5173–5193 | MP | |
| ToMMV CataAT Rv | GGAACGATCTTAAACTGGAACCT | 5255–5277 | |||
| ToMMV CataAT Pr | AATGCAAAGAGCGGATGAAGCGAC | Texas Red/BHQ2 | 5197–5220 | ||
| 4 | ToMMV CSPAT Fw | GGAACAGGTTTCTACAACCAGAG | 6124–6146 | CP | |
| ToMMV CSPAT Rv | CGTACGCACCACGTGTATTT | 6235–6254 | |||
| ToMMV CSPAT Pr | TTCTGCGCCAGCGTCCTAAGTAAT | Texas Red/BHQ2 | 6180–6203 |
| Duplex A | Duplex B | Triplex A | Triplex B | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ToBFRV M&W | ToMMV Set 1 | ToBRFV M&W | ToMMV Set 3 | ToBRFV ISHI-Veg | ToMMV Set 3 | ToBRFV ISHI-Veg | ToMMV Set 4 | |||
| CP-FAM | CP-HEX | CP-FAM | MP-TexasRed | CP-FAM | MP-HEX | MP-TexasRed | CP-FAM | MP-HEX | CP-TexasRed | |
| HL | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| HS | 37.8 * | NA | NA | NA | 36.0 * | 36.5 * | NA | 35.4 * | 36.1 * | NA |
| ToBRFV IL | 16.2 | NA | 17.5 | NA | 18.2 | 18.4 | NA | 18.7 | 18.8 | NA |
| ToBRFV IS | 27.9 | NA | 25.8 | NA | 28.4 | 28.2 | NA | 28.5 | 28.1 | NA |
| ToMMV IL | NA | 9.2 | NA | 12.3 | NA | NA | 13.6 | NA | NA | 8.8 |
| Mixed sample IL | 15.2 | 10.1 | 16.6 | 12.3 | 18.6 | 19.2 | 14.4 | 18.2 | 18.8 | 9.6 |
| NAC1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NAC2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Leaves | Seeds | |||||||
|---|---|---|---|---|---|---|---|---|
| ToBRFVs | ToBRFVd | ToMMVs | ToMMVd | ToBRFVs | ToBRFVd | ToMMVs | ToMMVd | |
| 100 | 14.1 | 13.7 | 12.2 | 12.5 | 15.2 | 14.9 | 12.0 | 12.3 |
| 10−1 | 17.4 | 17.2 | 16.0 | 16.1 | 18.3 | 18.2 | 15.5 | 15.9 |
| 10−2 | 21.1 | 20.8 | 19.5 | 19.8 | 21.8 | 21.8 | 19.4 | 19.5 |
| 10−3 | 24.4 | 24.2 | 22.7 | 23.2 | 25.4 | 25.2 | 22.9 | 23.0 |
| 10−4 | 27.8 | 27.4 | 26.1 | 26.3 | 28.8 | 28.8 | 26.3 | 26.2 |
| 10−5 | 30.8 | 31.0 | 29.1 | 30.2 | 32.2 | 31.9 | 29.3 | 29.3 |
| 10−6 | 34.5 | 34.1 | 32.8 | NA | 34.5 | NA | 31.9 | NA |
| 10−7 | 38.8 * | NA | NA | NA | NA | NA | NA | NA |
| 10−8 | 39.4 * | NA | NA | NA | NA | NA | NA | NA |
| R2 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 |
| E | 97.63 | 95.67 | 98.03 | 95.20 | 95.83 | 95.42 | 97.33 | 96.57 |
| NIB | CREA | |||||||
|---|---|---|---|---|---|---|---|---|
| Single Assay | Duplex | Single Assay | Duplex | |||||
| Virus | Collection | ID | ToBRFV M&W | ToBRFV (Cq) | ToMMV (Cq) | ToBRFV M&W | ToBRFV (Cq) | ToMMV (Cq) |
| BPeMV | DSMZ | BN-4708 | nt | nt | nt | NA | NA | NA |
| nt | nt | nt | NA | NA | NA | |||
| nt | nt | nt | NA | NA | NA | |||
| CGMMV | NIB | NIB V 271 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| CGMMV | NIB | NIB V 320 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| CGMMV | nt | nt | nt | NA | NA | NA | ||
| DSMZ | PV-0375 | nt | nt | nt | NA | NA | NA | |
| nt | nt | nt | NA | NA | NA | |||
| ObPV | DSMZ | PV-1176 | 38.4 * | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| ORSV | DSMZ | PV-1048 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| PaMMV | DSMZ | PV-0606 (2020) ** | 33.9 | 33.5 | 31.5 | nt | nt | nt |
| 33.8 | 33.5 | 31.4 | nt | nt | nt | |||
| 33.4 | 33.7 | 31.4 | nt | nt | nt | |||
| PaMMV | DSMZ | PV-0606 (2021) ** | 33.8 | 34.2 | 29.6 | nt | nt | nt |
| 35.0 | 34.3 | 29.5 | nt | nt | nt | |||
| 34.0 | 33.7 | 29.3 | nt | nt | nt | |||
| PMMoV | CREA | CREA-552 | nt | nt | nt | NA | NA | NA |
| nt | nt | nt | NA | NA | NA | |||
| nt | nt | nt | NA | NA | NA | |||
| PMMoV | DSMZ | PV-0165 | nt | nt | nt | NA | NA | NA |
| nt | nt | nt | NA | NA | NA | |||
| nt | nt | nt | NA | NA | NA | |||
| RMV | DSMZ | PV-0145 | 33.9 | 36.9 | NA | nt | nt | nt |
| 34.4 | 35.2 | NA | nt | nt | nt | |||
| 34.8 | 37.4 | NA | nt | nt | nt | |||
| SFBV | DSMZ | PV-1058 | NA | NA | NA | nt | nt | nt |
| NA | 38.2 * | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| SHMV | DSMZ | PV-0156 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| ToMV | NIB | NIB V 036 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| ToMV | NIB | NIB V 049 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| ToMV | NIB | NIB V 072 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| ToMV | NIB | NIB V 104 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| ToMV | DSMZ | PV-0141 | nt | nt | nt | NA | NA | NA |
| nt | nt | nt | NA | NA | NA | |||
| nt | nt | nt | NA | NA | NA | |||
| TMGMV | DSMZ | PV-0124 | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | |||
| NA | NA | NA | NA | NA | NA | |||
| TMV | NIB | NIB V 037 | NA | NA | NA | nt | nt | nt |
| NA | NA | NA | nt | nt | nt | |||
| NA | NA | NA | nt | nt | nt | |||
| TMV | DSMZ | PV-1252 | nt | nt | nt | NA | NA | NA |
| nt | nt | nt | NA | NA | NA | |||
| nt | nt | nt | NA | NA | NA | |||
| TMV | DSMZ | PV-0137 | NA | NA | NA | NA | NA | NA |
| 38.5* | NA | NA | NA | NA | NA | |||
| NA | NA | NA | NA | NA | NA | |||
| TMV | DSMZ | PV-0943 | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | |||
| NA | NA | NA | NA | NA | NA | |||
| YMoV | DSMZ | PV-0527 | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | |||
| NA | NA | NA | NA | NA | NA | |||
| Single | Duplex | ||||
|---|---|---|---|---|---|
| Virus | Collection | ID | ToBRFV M&W (Cq) | ToBRFV M&W (Cq) | ToMMV (Cq) |
| ToMMV | DSMZ, DE | PV-1267 | 36.9 * | NA | 8.4 |
| 38.9 * | NA | 8.0 | |||
| 37.6 * | NA | 8.1 | |||
| ToMMV | IBMCP; SP | S1 | nt | NA | 12.7 |
| nt | NA | 12.8 | |||
| nt | NA | 12.9 | |||
| ToMMV | IBMCP; SP | S2 | nt | NA | 15.9 |
| nt | NA | 15.7 | |||
| nt | NA | 15.7 | |||
| ToBRFV | CREA, IT | MR50 (100,000 × dilution) | 25.5 25.6 25.3 | 25.7 26.1 26.0 | NA NA NA |
| ToBRFV | Volcani center; IS | S21 | nt | 6.7 | NA |
| nt | 6.8 | NA | |||
| nt | 5.1 | NA | |||
| ToBRFV | Volcani center; S | S22 | nt | 6.8 | NA |
| nt | 4.7 | NA | |||
| nt | 4.6 | NA | |||
| ToBRFV (Cq) | ToMMV (Cq) | ToBRFV (Cq) | ToMMV (Cq) | ToBRFV (Cq) | ToMMV (Cq) | ToBRFV (Cq) | ToMMV (Cq) | ToBRFV (Cq) | ToMMV (Cq) | ToBRFV (Cq) | ToMMV (Cq) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run ID Samples | 1 | 2 | 3 | 4 | 5 | 6 | |||||||
| NIB | ToBRFV PC1 | 25.6 | NA | 26.7 | NA | 24.7 | NA | 25.6 | NA | 25.3 | NA | 25.5 | NA |
| ToBRFV PC2 | 31.8 | NA | 32.5 | NA | 31.2 | NA | 30.3 | NA | 31.0 | NA | 30.4 | NA | |
| ToMMV PC1 | NA | 26.2 | NA | 26.6 | NA | 25.7 | NA | 27.8 | NA | 27.2 | NA | 27.3 | |
| ToMMV PC2 | NA | 32.9 | NA | 33.0 | NA | 32.9 | NA | 35.4 | NA | 34.2 | NA | 35.6 | |
| NAC1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| NAC2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| CREA | ToBRFV PC1 | 23.4 | NA | 23.6 | NA | 23.3 | NA | 23.4 | NA | nt | nt | nt | nt |
| ToBRFV PC2 | 30.8 | NA | 30.8 | NA | 31.1 | NA | 31.0 | NA | nt | nt | nt | nt | |
| ToMMV PC1 | NA | 24.4 | NA | 24.9 | NA | 24.8 | NA | 25.2 | nt | nt | nt | nt | |
| ToMMV PC2 | NA | 31.4 | NA | 31.2 | NA | 31.2 | NA | 32.0 | nt | nt | nt | nt | |
| NAC1 | NA | NA | NA | NA | NA | NA | NA | NA | nt | nt | nt | nt | |
| NAC2 | NA | NA | NA | NA | NA | NA | NA | NA | nt | nt | nt | nt | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiberini, A.; Manglli, A.; Taglienti, A.; Vučurović, A.; Brodarič, J.; Ferretti, L.; Luigi, M.; Gentili, A.; Mehle, N. Development and Validation of a One-Step Reverse Transcription Real-Time PCR Assay for Simultaneous Detection and Identification of Tomato Mottle Mosaic Virus and Tomato Brown Rugose Fruit Virus. Plants 2022, 11, 489. https://doi.org/10.3390/plants11040489
Tiberini A, Manglli A, Taglienti A, Vučurović A, Brodarič J, Ferretti L, Luigi M, Gentili A, Mehle N. Development and Validation of a One-Step Reverse Transcription Real-Time PCR Assay for Simultaneous Detection and Identification of Tomato Mottle Mosaic Virus and Tomato Brown Rugose Fruit Virus. Plants. 2022; 11(4):489. https://doi.org/10.3390/plants11040489
Chicago/Turabian StyleTiberini, Antonio, Ariana Manglli, Anna Taglienti, Ana Vučurović, Jakob Brodarič, Luca Ferretti, Marta Luigi, Andrea Gentili, and Nataša Mehle. 2022. "Development and Validation of a One-Step Reverse Transcription Real-Time PCR Assay for Simultaneous Detection and Identification of Tomato Mottle Mosaic Virus and Tomato Brown Rugose Fruit Virus" Plants 11, no. 4: 489. https://doi.org/10.3390/plants11040489
APA StyleTiberini, A., Manglli, A., Taglienti, A., Vučurović, A., Brodarič, J., Ferretti, L., Luigi, M., Gentili, A., & Mehle, N. (2022). Development and Validation of a One-Step Reverse Transcription Real-Time PCR Assay for Simultaneous Detection and Identification of Tomato Mottle Mosaic Virus and Tomato Brown Rugose Fruit Virus. Plants, 11(4), 489. https://doi.org/10.3390/plants11040489









