Meta-Analysis of Common and Differential Transcriptomic Responses to Biotic and Abiotic Stresses in Arabidopsis thaliana
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of DEGs in Arabidopsis thaliana in Response to Biotic/Abiotic Stresses
2.2. Gene Ontology
2.3. KEGG Analysis of DEGs
2.4. Differential Stress-Responsive Groups
2.5. Identification of DEGs Encoding Transcription Factors in Response to Biotic/Abiotic Stresses
2.6. Protein-Protein Interaction
2.7. Identification of miRNAs Targeting Downregulated Genes
3. Materials and Methods
3.1. Data Collection and Preprocessing
3.2. Meta-Analysis of an Expression Dataset
3.3. Gene Enrichment Analysis and Functional Analysis
3.4. Protein-Protein Interactions and Network Construction
3.5. Prediction of Potential miRNAs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhar, A.; Chakraborty, A.; Roy, A. Plant responses to biotic stress: Old memories matter. Plants 2022, 11, 84. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z.; Hua, J. A meta-analysis reveals opposite effects of biotic and abiotic stresses on transcript levels of Arabidopsis intracellular immune receptor genes. Front. Plant Sci. 2021, 12, 241. [Google Scholar] [CrossRef] [PubMed]
- Costa-Mattioli, M.; Walter, P. The integrated stress response: From mechanism to disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.G.; van der Linden, G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.I. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed]
- Nejat, N.; Mantri, N. Plant immune system: Crosstalk between responses to biotic and abiotic stresses the missing link in understanding plant defence. Curr. Issues Mol. Biol. 2017, 23, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, E.; Salinas, M.; Manzano-Agugliaro, F. Worldwide research on plant defense against biotic stresses as improvement for sustainable agriculture. Sustainability 2018, 10, 391. [Google Scholar] [CrossRef]
- Zhang, N.; Zhou, S.; Yang, D.; Fan, Z. Revealing shared and distinct genes responding to JA and SA signaling in Arabidopsis by meta-analysis. Front. Plant Sci. 2020, 11, 908. [Google Scholar] [CrossRef]
- Jiang, Z.; He, F.; Zhang, Z. Large-scale transcriptome analysis reveals Arabidopsis metabolic pathways are frequently influenced by different pathogens. Plant Mol. Biol. 2017, 94, 453–467. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2016, 444, 323–329. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
- Kissoudis, C.; Sunarti, S.; van de Wiel, C.; Visser, R.G.; van der Linden, C.G.; Bai, Y. Responses to combined abiotic and biotic stress in tomato are governed by stress intensity and resistance mechanism. J. Exp. Bot. 2016, 67, 5119–5132. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, D.; Bhardwaj, S.; Landi, M.; Sharma, A.; Ramakrishnan, M.; Sharma, A. The impact of drought in plant metabolism: How to exploit tolerance mechanisms to increase crop production. Appl. Sci. 2020, 10, 5692. [Google Scholar] [CrossRef]
- Tahmasebi, A.; Ashrafi-Dehkordi, E.; Shahriari, A.G.; Mazloomi, S.M.; Ebrahimie, E. Integrative meta-analysis of transcriptomic responses to abiotic stress in cotton. Prog. Biophys. Mol. Biol. 2019, 146, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi-Dehkordi, E.; Alemzadeh, A.; Tanaka, N.; Razi, H. Meta-analysis of transcriptomic responses to biotic and abiotic stress in tomato. PeerJ 2018, 6, e4631. [Google Scholar] [CrossRef]
- Bolton, M.D. Primary metabolism and plant defense—Fuel for the fire. Mol. Plant-Microbe Interact. 2009, 22, 487–497. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Bilgin, D.D.; Zavala, J.A.; Zhu, J.I.N.; Clough, S.J.; Ort, D.R.; Delucia, E.H. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 2010, 33, 1597–1613. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.J.; Wang, Z.L.; Zhang, J.W.; Liu, S.; He, Z.L.; He, M.R. Interaction effects of nitric oxideand salicylic acid in alleviating salt stress of Gossypium hirsutum L. J. Soil Sci. Plant Nutr. 2015, 15, 561–573. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, D.; Zhao, Y.; Wang, W.; Yang, H.; Tai, F.; Li, C.; Hu, X. The difference of physiological and proteomic changes in maize leaves adaptation to drought, heat, and combined both stresses. Front. Plant Sci. 2016, 7, 1471. [Google Scholar] [CrossRef]
- Jun, X.U.; Wang, X.Y.; Guo, W.Z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Schuler, M.A. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996, 112, 1411. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.B.; Song, B.; Li, H.P.; Xu, H.Q.; Qu, B.; Dang, F.J.; Liao, Y.C. Resistance to Fusarium head blight and seedling blight in wheat is associated with activation of a cytochrome P450 gene. Phytopathology 2010, 100, 183–191. [Google Scholar] [CrossRef]
- Mazarei, M.; Lennon, K.A.; Puthoff, D.P.; Rodermel, S.R.; Baum, T.J. Expression of an Arabidopsis phosphoglycerate mutase homologue is localized to apical meristems, regulated by hormones, and induced by sedentary plant-parasitic nematodes. Plant Mol. Biol. 2003, 53, 513–530. [Google Scholar] [CrossRef] [PubMed]
- Rozenzvieg, D.; Elmaci, C.; Samach, A.; Lurie, S.; Porat, R. Isolation of four heat shock protein cDNAs from grapefruit peel tissue and characterization of their expression in response to heat and chilling temperature stresses. Physiol. Plant. 2004, 121, 421–428. [Google Scholar] [CrossRef]
- Ma, W.; Zhao, T.; Li, J.; Liu, B.; Fang, L.; Hu, Y.; Zhang, T. Identification and characterization of the GhHsp20 gene family in Gossypium hirsutum. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.Y.; Li, M.W.; Yung, Y.L.; Wen, C.Q.; Lam, H.M. The unconventional P-loop NTPase OsYchF1 and its regulator OsGAP1 play opposite roles in salinity stress tolerance. Plant Cell Environ. 2013, 36, 2008–2020. [Google Scholar] [CrossRef]
- Shaar-Moshe, L.; Hübner, S.; Peleg, Z. Identification of conserved drought-adaptive genes using a cross-species meta-analysis approach. BMC Plant Biol. 2015, 15, 111. [Google Scholar] [CrossRef] [PubMed]
- La Camera, S.; Gouzerh, G.; Dhondt, S.; Hoffmann, L.; Fritig, B.; Legrand, M.; Heitz, T. Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2004, 198, 267–284. [Google Scholar] [CrossRef]
- Geng, D.; Shen, X.; Xie, Y.; Yang, Y.; Bian, R.; Gao, Y.; Li, P.; Sun, L.; Feng, H.; Ma, F.; et al. Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Hortic. Res. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Dubreuil-Maurizi, C.; Poinssot, B. Role of glutathione in plant signaling under biotic stress. Plant Signal. Behav. 2012, 7, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Ellgaard, L.; Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003, 4, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, J. Communications between the endoplasmic reticulum and other organelles during abiotic stress response in plants. Front. Plant Sci. 2019, 10, 749. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Park, C.J.; Park, J.M. Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front. Plant Sci. 2019, 10, 399. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Finkemeier, I. Mitochondrial energy and redox signaling in plants. Antioxid. Redox Signal. 2013, 18, 2122–2144. [Google Scholar] [CrossRef]
- Farhat, N.; Hichri, S.; Hildebrandt, T.M.; Debez, A.; Braun, H.P. Composition and stability of the oxidative phosphorylation system in the halophile plant Cakile maritima. Front. Plant Sci. 2019, 10, 1010. [Google Scholar] [CrossRef]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Mishra, D.; Shekhar, S.; Singh, D.; Chakraborty, S.; Chakraborty, N. Heat Shock Proteins and Abiotic Stress Tolerance in Plants. In Regulation of Heat Shock Protein Responses; Springer: Cham, Switzerland, 2018; pp. 41–69. [Google Scholar]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Singh, R.K.; Jaishankar, J.; Muthamilarasan, M.; Shweta, S.; Dangi, A.; Prasad, M. Genome-wide analysis of heat shock proteins in C4 model, foxtail millet identifies potential candidates for crop improvement under abiotic stress. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Sudisha, J.; Sharathchandra, R.G.; Amruthesh, K.N.; Kumar, A.; Shetty, H.S. Pathogenesis Related Proteins in Plant Defense Response. In Plant Defence: Biological Control; Springer: Dordrecht, Germany, 2012; pp. 379–403. [Google Scholar]
- Hepler, P.K. Calcium: A central regulator of plant growth and development. Plant Cell 2005, 17, 2142–2155. [Google Scholar] [CrossRef]
- Gust, A.A.; Pruitt, R.; Nurnberger, T. Sensing danger: Key to activating plant immunity. Trends Plant Sci. 2017, 22, 779–791. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant–pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Hamann, T. The plant cell wall integrity maintenance mechanismconcepts for organization and mode of action. Plant Cell Physiol. 2015, 56, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Durufle, H.; San Clemente, H.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. Cell wall proteome analysis of Arabidopsis thaliana mature stems. Proteomics 2017, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Burkart, R.C.; Stahl, Y. Dynamic complexity: Plant receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2017, 40, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Blümke, A.; Sode, B.; Ellinger, D.; Voigt, C.A. Reduced susceptibility to Fusarium head blight in Brachypodium distachyon through priming with the Fusarium mycotoxin deoxynivalenol. Mol. Plant Pathol. 2015, 16, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2017, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Duan, Y.; Yin, J.; Ye, S.; Zhu, J.; Zhang, F.; Lu, W.; Fan, D.; Luo, K. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 2014, 65, 6629–6644. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Deslandes, L.; Olivier, J.; Theulières, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef]
- Knoth, C.; Ringler, J.; Dangl, J.L.; Eulgem, T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant-Microbe Interact. 2007, 20, 120–128. [Google Scholar] [CrossRef]
- Murray, S.L.; Ingle, R.A.; Petersen, L.N.; Denby, K.J. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant Microbe Interact. 2007, 20, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Mzid, R.; Marchive, C.; Blancard, D.; Deluc, L.; Barrieu, F.; Corio-Costet, M.F.; Drira, N.; Hamdi, S.; Lauvergeat, V. Overexpression of VvWRKY2 in tobacco enhances broad resistance to necrotrophic fungal pathogens. Physiol. Plant. 2007, 131, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Yang, F.; Ma, Y.; Wu, Q.; Yi, K.; Zhang, D. Transcription factor WRKY30 mediates resistance to Cucumber mosaic virus in Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 517, 118–124. [Google Scholar] [CrossRef]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold- responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Qin, Q.L.; Liu, J.G.; Zhang, Z.; Peng, R.H.; Xiong, A.S.; Yao, Q.H.; Chen, J.M. Isolation, optimization, and functional analysis of the cDNA encoding transcription factor OsDREB1B in Oryza sativa L. Mol. Breed. 2007, 19, 329–340. [Google Scholar] [CrossRef]
- Wan, D.; Li, R.; Zou, B.; Zhang, X.; Cong, J.; Wang, R.; Xia, Y.; Li, G. Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 2012, 31, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Bethke, G.; Pecher, P.; Eschen-Lippold, L.; Tsuda, K.; Katagiri, F.; Glazebrook, J.; Scheel, D.; Lee, J. Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant-Microbe Interact. 2012, 25, 471–480. [Google Scholar] [CrossRef]
- Puig, S.; Andrés-Colás, N.; García-Molina, A.; Penarrubia, L. Copper and iron homeostasis in Arabidopsis: Responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ. 2007, 30, 271–290. [Google Scholar] [CrossRef]
- Burkhead, J.L.; Gogolin Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef]
- Yuan, M.; Chu, Z.; Li, X.; Xu, C.; Wang, S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 2010, 22, 3164–3176. [Google Scholar] [CrossRef]
- Borkow, G.; Gabbay, J. Putting copper into action: Copper-impregnated products with potent biocidal activities. FASEB J. 2004, 18, 1728–1730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, S.; Takano, T. Overexpression of a mitochondrial ATP synthase small subunit gene (AtMtATP6) confers tolerance to several abiotic stresses in Saccharomyces cerevisiae and Arabidopsis thaliana. Biotechnol. Lett. 2008, 30, 1289–1294. [Google Scholar] [CrossRef] [PubMed]
- Plomion, C.; Lalanne, C.; Claverol, S.; Meddour, H.; Kohler, A.; Bogeat-Triboulot, M.B.; Barre, A.; le Provost, G.; Dumazet, H.; Jacob, D.; et al. Mapping the proteome of poplar and application to the discovery of drought-stress responsive proteins. Proteomics 2006, 6, 6509–6527. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.; Chen, G.; Yoo, M.J.; Zhu, N.; Dufresne, D.; Erickson, J.E.; Shao, H.; Chen, S. Comparative proteomic analysis of Brassica napus in response to drought stress. J. Proteome Res. 2015, 14, 3068–3081. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q.; Dai, S. Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef]
- Sweetman, C.; Waterman, C.D.; Rainbird, B.M.; Smith, P.M.; Jenkins, C.D.; Day, D.A.; Soole, K.L. AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress. Plant Physiol. 2019, 181, 774–788. [Google Scholar] [CrossRef]
- Tahmasebi, A.; Khahani, B.; Tavakol, E.; Afsharifar, A.; Shahid, M.S. Microarray analysis of Arabidopsis thaliana exposed to single and mixed infections with Cucumber mosaic virus and turnip viruses. Physiol. Mol. Biol. Plants 2021, 27, 11–27. [Google Scholar] [CrossRef]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Sharma, D.; Tiwari, M.; Pandey, A.; Bhatia, C.; Sharma, A.; Trivedi, P.K. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 2016, 171, 944–959. [Google Scholar] [CrossRef]
- Gao, F.; Wang, N.; Li, H.; Liu, J.; Fu, C.; Xiao, Z.; Wei, C.; Lu, X.; Feng, J.; Zhou, Y. Identification of drought-responsive microRNAs and their targets in Ammopiptanthus mongolicus by using high-throughput sequencing. Sci. Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhu, P.; Kang, H.; Liu, L.; Cao, Q.; Sun, J.; Dong, T.; Zhu, M.; Li, Z.; Xu, T. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genom. 2020, 21, 1–16. [Google Scholar] [CrossRef]
- Wang, J.; Xu, M.; Li, Z.; Ye, Y.; Rong, H.; Xu, L.A. Tamarix microRNA profiling reveals new insight into salt tolerance. Forests 2018, 9, 180. [Google Scholar] [CrossRef]
- Barozai, M.Y.K.; Kakar, S.; Sarangzai, A.M. Profiling the carrot (Daucus carota L.) microRNAs and their targets. Pak. J. Bot. 2013, 45, 353–358. [Google Scholar]
- Wen, C.L.; Cheng, Q.; Zhao, L.Q.; Mao, A.J.; Yang, J.J.; Yu, S.C.; Weng, Y.; Xu, Y. Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072. [Google Scholar] [CrossRef]
- Liang, C.; Liu, H.; Hao, J.; Li, J.; Luo, L. Expression profiling and regulatory network of cucumber microRNAs and their putative target genes in response to cucumber green mottle mosaic virus infection. Arch. Virol. 2019, 164, 1121–1134. [Google Scholar] [CrossRef]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Bernsdorff, F.; Döring, A.C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and-independent pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef]
- Asai, S.; Rallapalli, G.; Piquerez, S.J.; Caillaud, M.C.; Furzer, O.J.; Ishaque, N.; Wirthmueller, L.; Fabro, G.; Shirasu, K.; Jones, J.D. Expression profiling during Arabidopsis/downy mildew interaction reveals a highly-expressed effector that attenuates responses to salicylic acid. PLoS Pathog. 2014, 10, e1004443. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Stephen, S.; Kazan, K.; Jin, G.; Fan, L.; Taylor, J.; Dennis, E.S.; Helliwell, C.A.; Wang, M.B. Characterization of the defense transcriptome responsive to Fusarium oxysporum-infection in Arabidopsis using RNA-seq. Gene 2013, 512, 259–266. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Cumbie, J.S.; Dharmawardhana, P.; Jaiswal, P.; Chang, J.H.; Palusa, S.G.; Refy, A.S.N.; Megraw, M.; Mockler, T.C. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant 2015, 8, 207–227. [Google Scholar] [CrossRef]
- Zorzatto, C.; Machado, J.P.B. Lopes, K.V.; Nascimento, K.J.; Pereira, W.A.; Brustolini, O.J.; Reis, P.A.B.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef]
- Lai, Z.; Schluttenhofer, C.M.; Bhide, K.; Shreve, J.; Thimmapuram, J.; Lee, S.Y.; Jun, D.-Y.; Mengiste, T. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 2014, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Howard, B.E.; Hu, Q.; Babaoglu, A.C.; Chandra, M.; Borghi, M.; Tan, X.; He, L.; Winter-Sederoff, H.; Gassmann, W.; Heber, S.; et al. High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS ONE 2013, 8, e74183. [Google Scholar] [CrossRef]
- Clauw, P.; Coppens, F.; Korte, A.; Herman, D.; Slabbinck, B.; Dhondt, S.; van Daele, T.; de Milde, L.; Vermeersch, M.; Inzé, D.; et al. Leaf growth response to mild drought: Natural variation in Arabidopsis sheds light on trait architecture. Plant Cell 2016, 28, 2417–2434. [Google Scholar] [CrossRef]
- Schlaen, R.G.; Mancini, E.; Sanchez, S.E.; Perez-Santángelo, S.; Rugnone, M.L.; Simpson, C.G.; Brown, J.W.S.; Zhang, X.; Chernomoretz, A.; Yanovsky, M.J.; et al. The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. USA 2015, 112, 9382–9387. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, J.; Gao, Z.; Lu, Y.; Yu, J.; Zheng, Q.; Yan, S.; Zhang, W.; He, H.; Zhu, Z.; et al. SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 2015, 8, 1038–1052. [Google Scholar] [CrossRef]
- Suzuki, N.; Bassil, E.; Hamilton, J.S.; Inupakutika, M.A.; Zandalinas, S.I.; Tripathy, D.; Luo, Y.; Dion, E.; Fukui, G.; Mittler, R.; et al. ABA is required for plant acclimation to a combination of salt and heat stress. PLoS ONE 2016, 11, e0147625. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hu, P.; Greenwood, C.M.; Beyene, J. Using the ratio of means as the effect size measure in combining results of microarray experiments. BMC Syst. Biol. 2009, 3, 106. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; von Mering, C.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
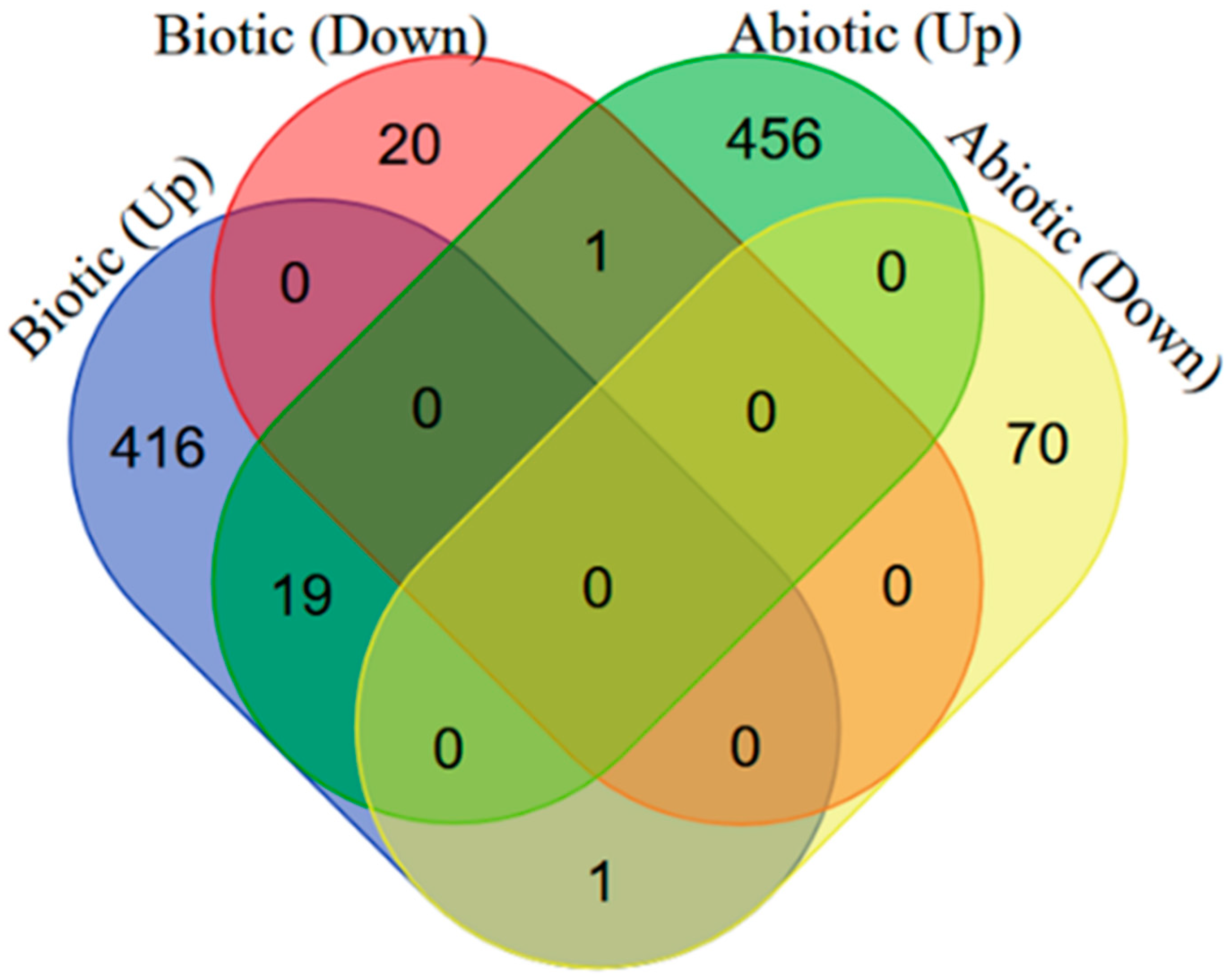
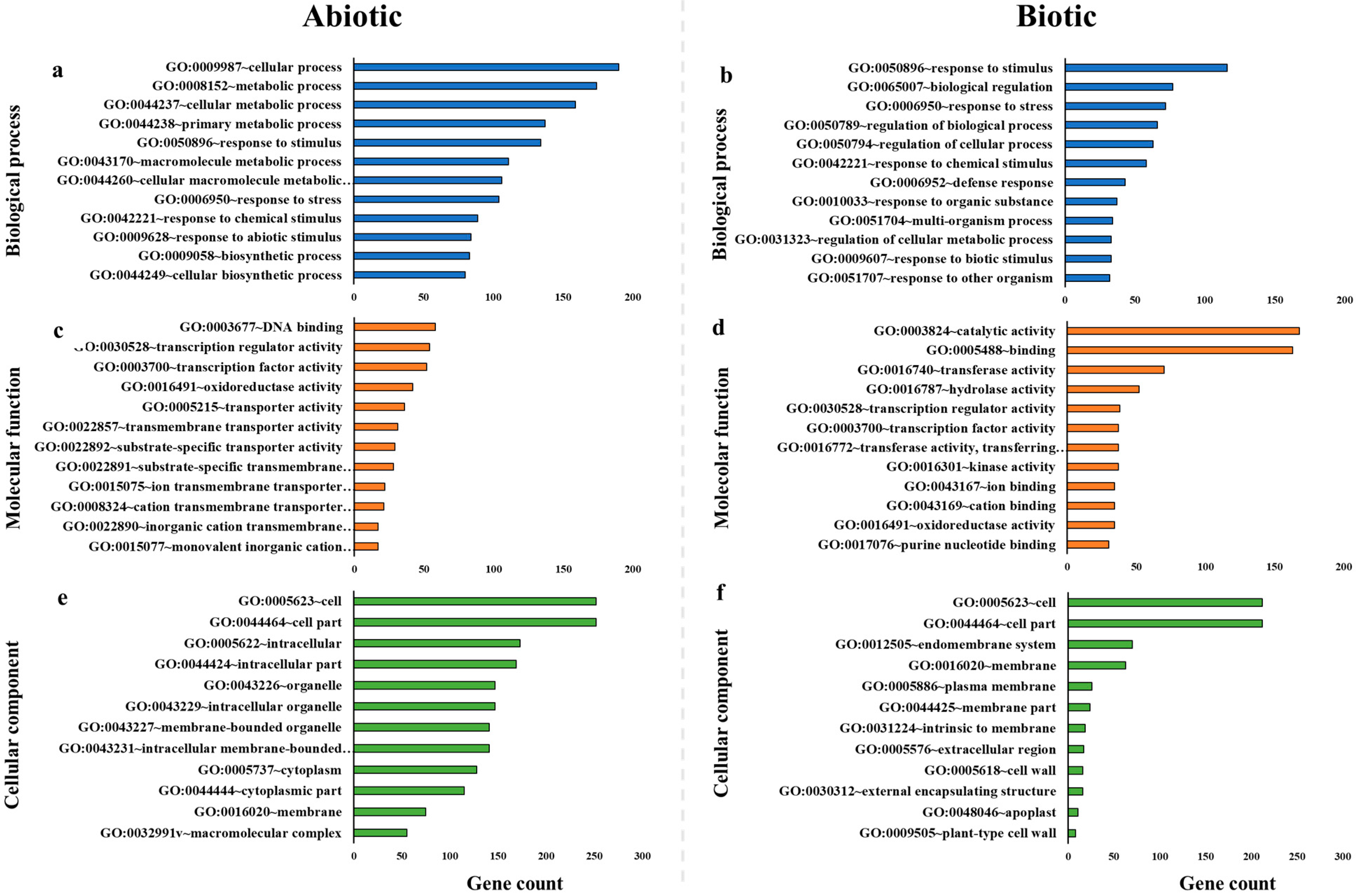


| Stress | Pathway | Gene Count | Adjusted p Value |
|---|---|---|---|
| Biotic | Plant-pathogen interaction | 11 | 0.000170 |
| Phenylpropanoid biosynthesis | 11 | 0.000026 | |
| Glutathione metabolism | 9 | 0.000021 | |
| Amino sugar and nucleotide sugar metabolism | 6 | 0.029237 | |
| Abiotic | Protein processing in the endoplasmic reticulum | 16 | 0.000002 |
| Plant hormone signal transduction | 12 | 0.003789 | |
| Oxidative phosphorylation | 7 | 0.049932 | |
| Carotenoid biosynthesis | 4 | 0.011602 |
| TF Family | No. of TF in Abiotic | No. of TF in Biotic |
|---|---|---|
| AP2-EREBP | 15 | 2 |
| bHLH | 2 | 1 |
| C2C2-CO-like | 1 | - |
| C2C2-Dof | 2 | - |
| C2H2 | 8 | 4 |
| C3H | 3 | 1 |
| CCAAT-HAP2 | 1 | 1 |
| G2-like | 1 | - |
| GRAS | 1 | - |
| Homeobox | 2 | - |
| HSF | 5 | 1 |
| MYB | 2 | 3 |
| NAC | 6 | 6 |
| RAV | 1 | - |
| REM | - | 1 |
| Trihelix | - | 1 |
| WRKY | 1 | 15 |
| Accession Number | Stress | Sample Number | Control Number | Related Article |
|---|---|---|---|---|
| E-MTAB-4151 | Pseudomonas syringae pv. maculicola | 24 | 12 | [92] |
| E-GEOD-53641 | Hyaloperonospora arabidopsidis | 204 | 72 | [93] |
| E-GEOD-34241 | Fusarium oxysporum | 8 | 4 | [94] |
| E-MTAB-4416 | Pseudomonas syringae | 6 | 3 | [95] |
| E-GEOD-56922 | Cabbage leaf curl virus | 8 | 4 | [96] |
| E-MTAB-4281 | Botrytis cinerea | 4 | 2 | [97] |
| E-MTAB-4450 | Pseudomonas syringae | 18 | 6 | [98] |
| E-MTAB-3908 | Drought | 21 | 11 | EBI |
| E-MTAB-5009 | Mild drought | 17 | 9 | [99] |
| E-GEOD-63406 | Temperature | 9 | 3 | [100] |
| PRJNA240248 | Salt | 4 | 2 | [101] |
| PRJNA295091 | Salt and heat stress | 12 | 3 | [102] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biniaz, Y.; Tahmasebi, A.; Afsharifar, A.; Tahmasebi, A.; Poczai, P. Meta-Analysis of Common and Differential Transcriptomic Responses to Biotic and Abiotic Stresses in Arabidopsis thaliana. Plants 2022, 11, 502. https://doi.org/10.3390/plants11040502
Biniaz Y, Tahmasebi A, Afsharifar A, Tahmasebi A, Poczai P. Meta-Analysis of Common and Differential Transcriptomic Responses to Biotic and Abiotic Stresses in Arabidopsis thaliana. Plants. 2022; 11(4):502. https://doi.org/10.3390/plants11040502
Chicago/Turabian StyleBiniaz, Yaser, Aminallah Tahmasebi, Alireza Afsharifar, Ahmad Tahmasebi, and Péter Poczai. 2022. "Meta-Analysis of Common and Differential Transcriptomic Responses to Biotic and Abiotic Stresses in Arabidopsis thaliana" Plants 11, no. 4: 502. https://doi.org/10.3390/plants11040502
APA StyleBiniaz, Y., Tahmasebi, A., Afsharifar, A., Tahmasebi, A., & Poczai, P. (2022). Meta-Analysis of Common and Differential Transcriptomic Responses to Biotic and Abiotic Stresses in Arabidopsis thaliana. Plants, 11(4), 502. https://doi.org/10.3390/plants11040502







