Evaluation of Reference Genes in the Polyploid Complex Dianthus broteri (Caryophyllaceae) Using qPCR
Abstract
:1. Introduction
2. Results
2.1. Verification of Primer Specificity and Efficiency of Candidate Reference Genes
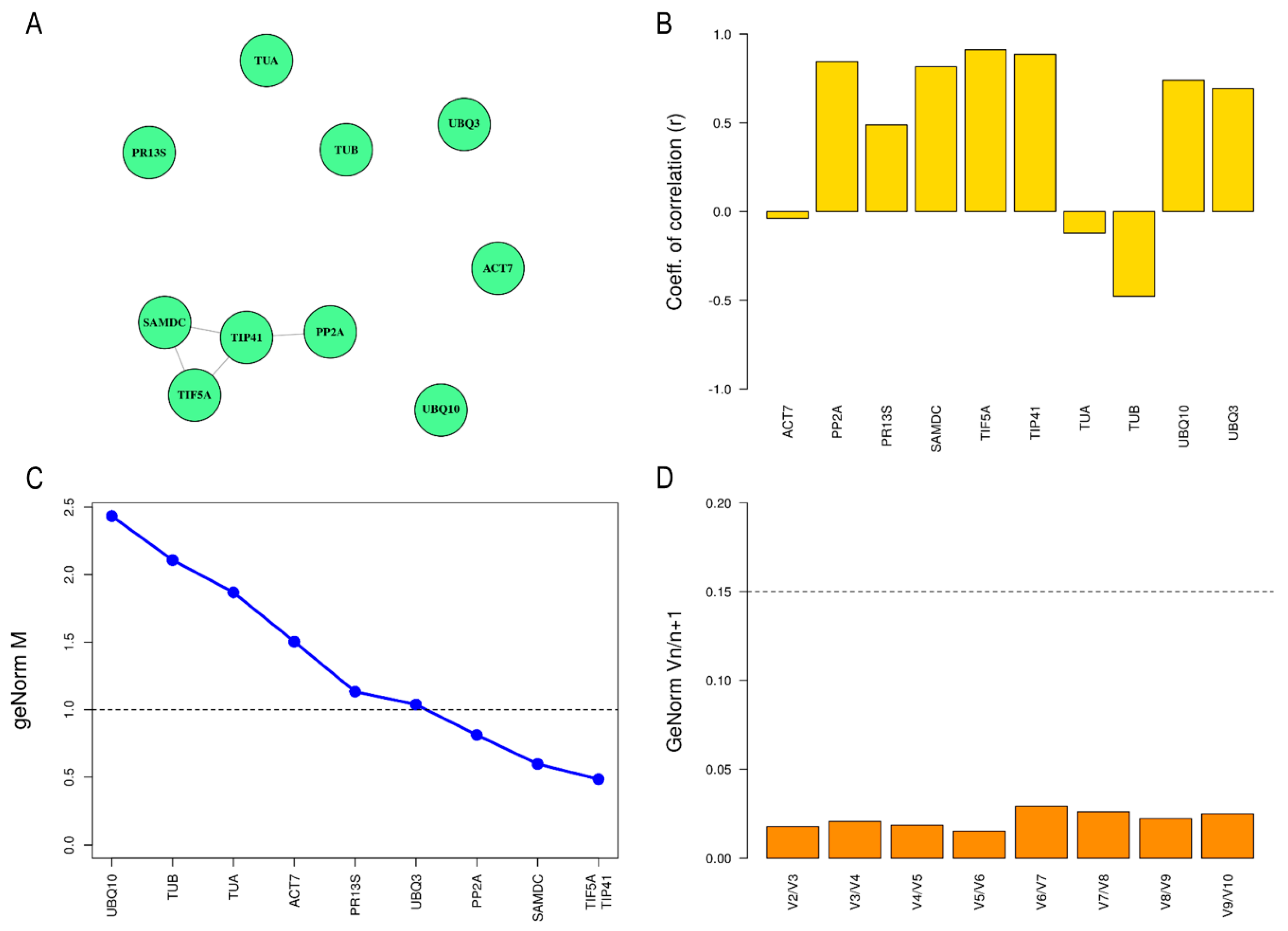
2.2. Determination of Expression Stability of Candidate Reference Genes in Leaf Tissue
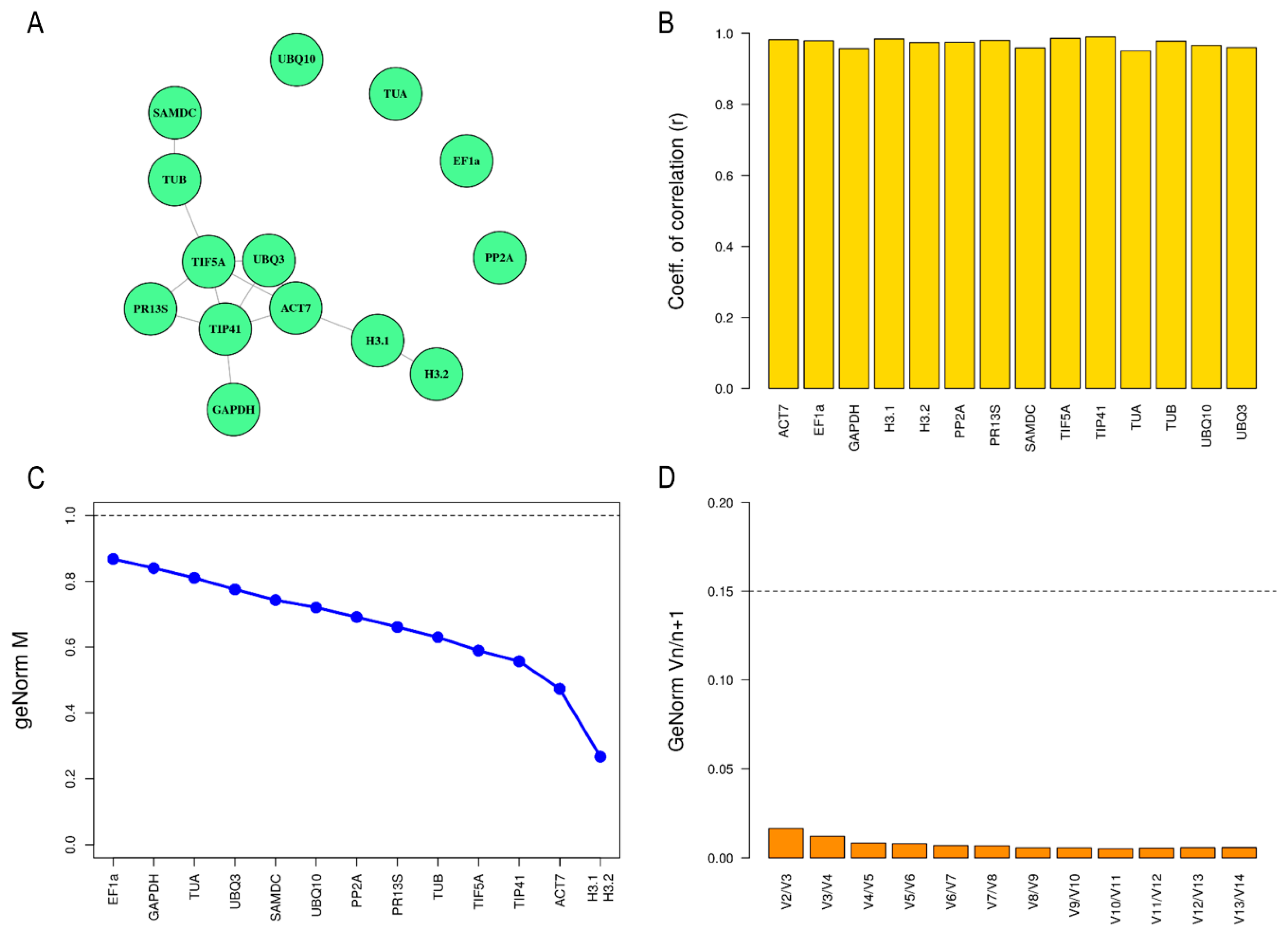
2.3. Determination of Expression Stability of Candidate Reference Genes in Petal Tissue
2.4. Determination of Optimal Number of Reference Genes
3. Discussion
3.1. Reference Gene Stability among Tissues and Ploidy Levels
3.2. Gene Expression Attenuation Enable the Comparison among the Four Ploidy Levels
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. RNA Extraction and cDNA Synthesis
4.3. Identification of Candidate Reference Genes and Efficiency Evaluation
4.4. Determination of Expression Stability of Reference Genes in Reproductive and Vegetative Tissues by Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lovén, J.; Orlando, D.A.; Sigova, A.A.; Lin, C.Y.; Rahl, P.B.; Burge, C.B.; Levens, D.L.; Lee, T.I.; Young, R.A. Revisiting global gene expression analysis. Cell 2012, 151, 476–482. [Google Scholar] [CrossRef] [Green Version]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Jeon, B.W.; Geng, S.; Yu, Y.; Balmant, K.; Chen, S.; Assmann, S.M. Plant Signal Transduction: Methods and Protocols. In Methods in Molecular Biology (Clifton, N.J.); Springer: New York, NY, USA, 2016; Volume 1363, pp. 9–12, 89–121. ISBN 9781493931156. [Google Scholar]
- Gopalam, R.; Rupwate, S.D.; Tumaney, A.W. Selection and validation of appropriate reference genes for quantitative real-time PCR analysis in Salvia hispanica. PLoS ONE 2017, 12, e0186978. [Google Scholar] [CrossRef] [Green Version]
- Monroy-Mena, S.; Chacón-Parra, A.L.; Farfán-Angarita, J.P.; Martínez-Peralta, S.T.; Ardila-Barrantes, H.D. Selection of reference genes for transcriptional analysis in the pathosystem carnation (Dianthus caryophyllus L.)-fusarium oxysporum f. sp. dianthi. Rev. Colomb. Quim. 2019, 48, 5–14. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Quantification Strategies in Real-time Polymerase Chain Reaction. In Polymerase Chain Reaction: Theory and Technology; International University Line (IUL): La Jolla, CA, USA, 2019; pp. 87–112. [Google Scholar]
- Koloušková, P.; Stone, J.D.; Štorchová, H. Evaluation of reference genes for reverse transcription quantitative real-time PCR (RT-qPCR) studies in Silene vulgaris considering the method of cDNA preparation. PLoS ONE 2017, 12, e0183470. [Google Scholar] [CrossRef] [Green Version]
- Kuang, J.; Yan, X.; Genders, A.J.; Granata, C.; Bishop, D.J. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 2018, 13, e0196438. [Google Scholar] [CrossRef]
- Yu, W.; Tao, Y.; Luo, L.; Hrovat, J.; Xue, A.; Luo, H. Evaluation of housekeeping gene expression stability in carnation (Dianthus caryophyllus). N. Z. J. Crop Hortic. Sci 2021, 49, 347–360. [Google Scholar] [CrossRef]
- Wang, H.; Chen, S.; Jiang, J.; Zhang, F.; Chen, F. Reference gene selection for cross-species and cross-ploidy level comparisons in Chrysanthemum spp. Sci. Rep. 2015, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Pérez, R.; Jouve, N.; De Bustos, A. Comparative analysis of gene expression among species of different ploidy. Mol. Biol. Rep. 2014, 41, 6525–6535. [Google Scholar] [CrossRef]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Van De Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017 187 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef] [Green Version]
- Wood, T.E.; Takebayashi, N.; Barker, M.S.; Mayrose, I.; Greenspoon, P.B.; Rieseberg, L.H. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 2009, 106, 13875–13879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltis, P.S.; Soltis, D.E. The Role of Hybridization in Plant Speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef] [Green Version]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; DePamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vyas, P.; Bisht, M.S.; Miyazawa, S.I.; Yano, S.; Noguchi, K.; Terashima, I.; Funayama-Noguchi, S. Effects of polyploidy on photosynthetic properties and anatomy in leaves of Phlox drummondii. Funct. Plant Biol. 2007, 34, 673–682. [Google Scholar] [CrossRef]
- Balao, F.; Casimiro-Soriguer, R.; Talavera, M.; Herrera, J.; Talavera, S. Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations. Ann. Bot. 2009, 104, 965–973. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [Green Version]
- Gottlieb, L.D. Plant polyploidy: Gene expression and genetic redundancy. Heredity 2003, 91, 91–92. [Google Scholar] [CrossRef]
- Osborn, T.C.; Chris Pires, J.; Birchler, J.A.; Auger, D.L.; Chen, Z.J.; Lee, H.S.; Comai, L.; Madlung, A.; Doerge, R.W.; Colot, V.; et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003, 19, 141–147. [Google Scholar] [CrossRef]
- Lopes, J.M.L.; de Matos, E.M.; de Queiroz Nascimento, L.S.; Viccini, L.F. Validation of reference genes for quantitative gene expression in the Lippia alba polyploid complex (Verbenaceae). Mol. Biol. Rep. 2021, 48, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Valente, L.M.; Savolainen, V.; Vargas, P. Unparalleled rates of species diversification in Europe. Proc. R. Soc. B Biol. Sci. 2010, 277, 1489–1496. [Google Scholar] [CrossRef] [Green Version]
- Carolin, R.C. Cytological and hybridization studies in the genus Dianthus. New Phytol. 1957, 56, 81–97. [Google Scholar] [CrossRef]
- Balao, F.; Valente, L.M.; Vargas, P.; Herrera, J.; Talavera, S. Radiative evolution of polyploid races of the Iberian carnation Dianthus broteri (Caryophyllaceae). New Phytol. 2010, 187, 542–551. [Google Scholar] [CrossRef] [PubMed]
- López-Jurado, J.; Mateos-Naranjo, E.; Balao, F. Niche divergence and limits to expansion in the high polyploid Dianthus broteri complex. New Phytol. 2019, 222, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Balao, F.; Herrera, J.; Talavera, S. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: A multivariate morphological approach. New Phytol. 2011, 192, 256–265. [Google Scholar] [CrossRef]
- López-Jurado, J.; Balao, F.; Mateos-Naranjo, E. Polyploidy-mediated divergent light-harvesting and photoprotection strategies under temperature stress in a Mediterranean carnation complex. Environ. Exp. Bot. 2020, 171, 103956. [Google Scholar] [CrossRef]
- Alonso, C.; Pérez, R.; Bazaga, P.; Medrano, M.; Herrera, C.M. MSAP markers and global cytosine methylation in plants: A literature survey and comparative analysis for a wild-growing species. Mol. Ecol. Resour. 2016, 16, 80–90. [Google Scholar] [CrossRef] [Green Version]
- Zhou, A.; Ma, H.; Feng, S.; Gong, S.; Wang, J. DsSWEET17, a tonoplast-localized sugar transporter from Dianthus spiculifolius, affects sugar metabolism and confers multiple stress tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1564. [Google Scholar] [CrossRef] [Green Version]
- Podevin, N.; Krauss, A.; Henry, I.; Swennen, R.; Remy, S. Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol. Breed. 2012, 30, 1237–1252. [Google Scholar] [CrossRef] [Green Version]
- Curis, E.; Nepost, C.; Grillault Laroche, D.; Courtin, C.; Laplanche, J.L.; Etain, B.; Marie-Claire, C. Selecting reference genes in RT-qPCR based on equivalence tests: A network based approach. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.B.; Shen, X.; Li, X.J.; Tian, Y.B.; Ouyang, H.J.; Huang, Y.M. Reference gene selection for expression studies in the reproductive axis tissues of Magang geese at different reproductive stages under light treatment. Sci. Rep. 2021, 11, 7573. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanGuilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008, 44, 619–626. [Google Scholar] [CrossRef] [Green Version]
- Giménez, M.J.; Pistón, F.; Atienza, S.G. Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 2011, 233, 163–173. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Jiang, J.; Chen, S.; Guan, Z.; Liao, Y.; Chen, F. Reference genes for normalizing transcription in diploid and tetraploid arabidopsis. Sci. Rep. 2014, 4, 6781. [Google Scholar] [CrossRef] [Green Version]
- Migocka, M.; Papierniak, A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2010, 28, 343–357. [Google Scholar] [CrossRef]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and Validation of Appropriate Reference Genes for Quantitative Real-Time PCR Analysis of Gene Expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of Reference Genes for Quantitative Real-Time PCR in Bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef] [Green Version]
- Fei, X.; Shi, Q.; Yang, T.; Fei, Z.; Wei, A. Expression Stabilities of Ten Candidate Reference Genes for RT-qPCR in Zanthoxylum bungeanum Maxim. Molecules 2018, 23, 802. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Yang, F.; Feng, J.; Wang, Y.; Lachenbruch, B.; Wang, J.; Wan, X. Genome-Wide Constitutively Expressed Gene Analysis and New Reference Gene Selection Based on Transcriptome Data: A Case Study from Poplar/Canker Disease Interaction. Front. Plant Sci. 2017, 8, 1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Gui, M.; Zhao, D.; Chen, M.; Ju, S.; Li, S.; Lu, Z.; Mo, X.; Wang, J. Study on reproductive barriers in 4x-2x crosses in Dianthus caryophyllus L. Euphytica 2013, 189, 471–483. [Google Scholar] [CrossRef]
- Coate, J.E.; Doyle, J.J. Quantifying Whole Transcriptome Size, a Prerequisite for Understanding Transcriptome Evolution Across Species: An Example from a Plant Allopolyploid. Genome Biol. Evol. 2010, 2, 534–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, W.; Liao, B.Y.; Chang, A.Y.F.; Zhang, J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010, 26, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Bekaert, M.; Edger, P.P.; Chris Pires, J.; Conant, G.C. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 2011, 23, 1719–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birchler, J.A.; Veitia, R.A. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 2012, 109, 14746–14753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, C.; Balao, F.; Bazaga, P.; Pérez, R. Epigenetic contribution to successful polyploidizations: Variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytol. 2016, 212, 571–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domínguez-Delgado, J.J.; López-Jurado, J.; Mateos-Naranjo, E.; Balao, F. Phenotypic diploidization in plant functional traits uncovered by synthetic neopolyploids in Dianthus broteri. J. Exp. Bot. 2021, 72, 5522–5533. [Google Scholar] [CrossRef]
- Feng, H.; Chen, Q.; Feng, J.; Zhang, J.; Yang, X.; Zuo, J. Functional Characterization of the Arabidopsis Eukaryotic Translation Initiation Factor 5A-2 That Plays a Crucial Role in Plant Growth and Development by Regulating Cell Division, Cell Growth, and Cell Death. Plant Physiol. 2007, 144, 1531–1545. [Google Scholar] [CrossRef]
- Punzo, P.; Ruggiero, A.; Possenti, M.; Nurcato, R.; Costa, A.; Morelli, G.; Grillo, S.; Batelli, G. The PP2A-interactor TIP41 modulates ABA responses in Arabidopsis thaliana. Plant J. 2018, 94, 991–1009. [Google Scholar] [CrossRef] [Green Version]
- Yagi, M.; Kosugi, S.; Hirakawa, H.; Ohmiya, A.; Tanase, K.; Harada, T.; Kishimoto, K.; Nakayama, M.; Ichimura, K.; Onozaki, T.; et al. Sequence Analysis of the Genome of Carnation (Dianthus caryophyllus L.). DNA Res. 2014, 21, 231–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. Relative quantification: Real time qPCR amplification efficiency. In Real-Time PCR; Dorak, T., Ed.; Taylor & Francis: London, UK, 2006; pp. 63–80. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Matz, M.V. MCMC.qpcr: Bayesian Analysis of qRT-PCR Data. R Package Version 1.2.4. 2020. Available online: https://CRAN.R-project.org/package=MCMC.qpcr (accessed on 22 December 2021).
- Matz, M.V.; Wright, R.M.; Scott, J.G. No control genes required: Bayesian analysis of qRT-PCR data. PLoS ONE 2013, 8, e71448. [Google Scholar] [CrossRef]
- Curis, E. SARP.compo: Network-Based Interpretation of Changes in Compositional Data. R Package Version 0.1.5. 2021. Available online: https://CRAN.R-project.org/package=SARP.compo (accessed on 22 December 2021).
- Zhong, S. ctrlGene: Assess the Stability of Candidate Housekeeping Genes. R Package Version 1.0.1. 2019. Available online: https://CRAN.R-project.org/package=ctrlGene (accessed on 22 December 2021).
- Etschmann, B.; Wilcken, B.; Stoevesand, K.; Von Der Schulenburg, A.; Sterner-Kock, A. Selection of reference genes for quantitative real-time PCR analysis in canine mammary tumors using the GeNorm algorithm. Vet. Pathol. 2006, 43, 934–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzáles, I.M.; Weiss, J.; Egea-cortines, M. Selección de Genes de Normalizacion Para RT-PCR Cuantitativa en Petunia hybrida. 2008, pp. 1–26. Available online: https://hdl.handle.net/10317/723 (accessed on 21 December 2021).
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Gong, H.; Sun, L.; Chen, B.; Han, Y.; Pang, J.; Wu, W.; Qi, R.; Zhang, T.M. Evaluation of candidate reference genes for RT-qPCR studies in three metabolism related tissues of mice after caloric restriction. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, D.H.; Siegfried, B.D. Selection of reference genes for normalization of RT-qPCR data in gene expression studies in Anthonomus eugenii Cano (Coleoptera: Curculionidae). Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]

| Gene | Gene Name | Accession No. (Carnation DB) | Primer Sequence | Efficiency | Product Size (bp) | Tm (°C) | References |
|---|---|---|---|---|---|---|---|
| EF1α | Elongation factor 1α | Dca5900.1 | F: ACCCCGACAAGATCCCATTT R: TGGTCAAGGGCCTCAAGTAG | 2.129 | 115 | 56.94 56.99 | [9] |
| TIP41 | Phosphatase activator | Dca43498.1 | F: GACACTCGTATGCATTGCGT R: CTCGAACTGATGACGCTTGG | 2.187 | 152 | 57.07 57.08 | [9] |
| UBQ10 | Ubiquitin 10 | Dca41829.1 | F: CCATTTGGTGTTGCGTCTCA R: TCGCTGCTCTCCACTTCC | 1.928 | 90 | 57.08 56.44 | [9] |
| UBQ 3 | Ubiquitin 3 | Dca119.1 | F: GCGTATGAGCAACGAGTCAG R: AGGATCTGCTTTACCCACCA | 1.953 | 150 | 57.17 56.27 | [5] |
| SAMDC | Adenosylmethionine decarboxylase gene | Dca28802.1 | F: AAACCAACTACGACGACCCT R: CCGATGCCTTCTCCTTGTCA | 2.021 | 72 | 56.95 57.75 | [9] |
| PP2A | Protein phosphatase 2A | Dca33231.1 | F: TCGAGCAGTTGATGGAGTGT R:ACTCTTCAACCAAAACCGCC | 1.992 | 87 | 59.03 58.97 | [9] |
| TIF5A | Translation initiation factor | Dca33327 | F: GGCGGGGAAAGACTTGATTC R: CTACTTGCCACCACTAACGT | 1.935 | 93 | 58.90 58.94 | [9] |
| H3.1 | Histone 3 | Dca5219.1 | F: GGAGGAGTGAAGAAGCCACA R: GTGCCAACACAGCATGACTC | 1.868 | 178 | 57.3 57.81 | [5] |
| H3.2 | Histone 3 | Dca5219.1 | F: CACAGGTACCGTCCTGGAAC R: GTGCCAACACAGCATGACTC | 1.882 | 160 | 58.06 57.81 | [5] |
| PR13S | Ribosomal protein | Dca22015.1 | F: AATCCCCGAGGACTTGTACC R:ACAAGAGTACTGGCGGTGGT | 2.051 | 201 | 56.77 59.09 | [5] |
| GAPDH | Glyceraldehyde 3 phosphate dehydrogenase | Dca8698.1 | F: GGCCAAGGTTATCAATGACAG R: CCTTCCACCTCTCCAGTCCT | 2.008 | 120 | 54.18 57.12 | [7] |
| ACT7 | Actin | Dca37612.1 | F: CGGTGGCTCTATCCTCGCTT R: TTCCTGTGGACGATTGACGG | 1.854 | 94 | 58.7 57.02 | [31] |
| TUA | Tubulin α | Dca60406.1 | F: ACATGGCTTGCTGTCTGATG R: TGGGGGCTGGTAGTTGATAC | 2.042 | 142 | 55.51 55.69 | [7] |
| TUB | Tubulin β | Dca39629.1 | F: TGTTGCATCCTGGTACTGCT R: GGCTTTCTTGCACTGGTACAC | 1.760 | 73 | 56.22 57.00 | [32] |
| BestKeeper | geNorm | |||
|---|---|---|---|---|
| Ranking | Gene | r Value | Gene | M Value |
| 1 | TIF5A | 0.893 | TIF5A | 0.471 |
| 2 | TIP41 | 0.868 | TIP41 | 0.471 |
| 3 | PP2A | 0.851 | SAMDC | 0.701 |
| 4 | SAMDC | 0.776 | PP2A | 0.845 |
| 5 | UBQ10 | 0.728 | UBQ3 | 1.042 |
| 6 | UBQ3 | 0.676 | PR13S | 1.137 |
| 7 | PR13S | 0.441 | ACT7 | 1.512 |
| 8 | ACT7 | −0.015 | TUB | 1.933 |
| 9 | TUA | −0.268 | UBQ10 | 2.308 |
| 10 | TUB | −0.431 | TUA | 2.648 |
| BestKeeper | geNorm | |||
|---|---|---|---|---|
| Ranking | Gene | r Value | Gene | M Value |
| 1 | TIP41 | 0.990 | H3.1 | 0.267 |
| 2 | TIF5A | 0.986 | H3.2 | 0.267 |
| 3 | H3.1 | 0.984 | ACT7 | 0.473 |
| 4 | ACT7 | 0.982 | TIP41 | 0.557 |
| 5 | PR13S | 0.980 | TIF5A | 0.589 |
| 6 | EF1a | 0.979 | TUB | 0.630 |
| 7 | TUB | 0.978 | PR13S | 0.661 |
| 8 | PP2A | 0.975 | PP2A | 0.691 |
| 9 | H3.2 | 0.974 | UBQ10 | 0.720 |
| 10 | UBQ10 | 0.966 | SAMDC | 0.743 |
| 11 | UBQ3 | 0.960 | UBQ3 | 0.776 |
| 12 | SAMDC | 0.959 | TUA | 0.810 |
| 13 | GAPDH | 0.957 | GAPDH | 0.840 |
| 14 | TUA | 0.950 | EF1a | 0.868 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Parra, A.; Picazo-Aragonés, J.; Balao, F. Evaluation of Reference Genes in the Polyploid Complex Dianthus broteri (Caryophyllaceae) Using qPCR. Plants 2022, 11, 518. https://doi.org/10.3390/plants11040518
Rodríguez-Parra A, Picazo-Aragonés J, Balao F. Evaluation of Reference Genes in the Polyploid Complex Dianthus broteri (Caryophyllaceae) Using qPCR. Plants. 2022; 11(4):518. https://doi.org/10.3390/plants11040518
Chicago/Turabian StyleRodríguez-Parra, Alba, Jesús Picazo-Aragonés, and Francisco Balao. 2022. "Evaluation of Reference Genes in the Polyploid Complex Dianthus broteri (Caryophyllaceae) Using qPCR" Plants 11, no. 4: 518. https://doi.org/10.3390/plants11040518







