Thiophenes—Naturally Occurring Plant Metabolites: Biological Activities and In Silico Evaluation of Their Potential as Cathepsin D Inhibitors
Abstract
:1. Introduction
2. Structural Characterization of Thiophenes
3. Biosynthesis of Thiophenes
4. Biological Activities of Thiophenes
4.1. Anti-Inflammatory Activity
| Compound Name | Biological Activity | Assay, Organism, or Cell Line | Biological Results | Ref. | |
|---|---|---|---|---|---|
| Compound | Positive Control | ||||
| Foetithiophene F (6) | Antimicrobial | Broth microdilution/B. cereus | 50 µg/mL (MIC) | Gentamicin 10 µg/mL (MIC) | [25] |
| 5-Propinyl-thiophene-2-carboxylic acid (7) | In vitro anti-inflammatory/NO | LPS-stimulated production in BV-2 microglial cells | 79.5 µM (IC50) | Quercetin 16.3 µM (IC50) | [26] |
| 3-Hydroxy-5-propinyl-2-acetyl-thiophene (8) | In vitro anti-inflammatory/NO | LPS-stimulated production in BV-2 microglial cells | 98.5 µM (IC50) | Quercetin 16.3 µM (IC50) | [26] |
| 2-(3,4-Dihydroxybut-1-ynyl)-5-(penta-1,3-diynyl)thiophene (9) | In vitro anti-inflammatory/NO | LPS-stimulated production in the RAW 264.7 cell line | 2.5 µg/mL (IC50) | Indomethacin 65.4 µg/mL (IC50) | [28] |
| Cytotoxicity | Resazurin reduction/CEM/ADR5000 | 21.09 µM (IC50) | Doxorubicin 195.12 µM (IC50) | [30] | |
| Cytotoxicity | Resazurin reduction/CCRF-CEM | 46.96 µM (IC50) | Doxorubicin 0.20 µM (IC50) | [30] | |
| Antimicrobial | INT/E. coli | 64.0 µg/mL (MIC) | Chloramphenicol 64.0 µg/mL (MIC) | [29] | |
| Antimicrobial | INT/E. aerogenes | 64.0 µg/mL (MIC) | Chloramphenicol 16.0 µg/mL (MIC) | [29] | |
| Antimicrobial | INT/K. pneumoniae | 64.0 µg/mL (MIC) | Chloramphenicol 16.0 µg/mL (MIC) | [29] | |
| Antimicrobial | INT/P. stuartil | 64.0 µg/mL (MIC) | Chloramphenicol 128.0 µg/mL (MIC) | [29] | |
| Antimicrobial | INT/E. cloacae | 256.0 µg/mL (MIC) | Chloramphenicol 256.0 µg/mL (MIC) | [29] | |
| Antimicrobial | INT/P. aeruginosa | 256.0 µg/mL (MIC) | Chloramphenicol 16.0 µg/mL (MIC) | [29] | |
| 2-(Penta-1,3-diyn-1-yl)-5–(4-acetoxy-3-hydroxybuta-1-yn-1-yl) thiophene (11) | CYP2A6 inhibition | Enzymatic reconstitution | 6.43 µM (IC50) | Methoxsalen 0.19 µM (IC50) | [31] |
| CYP2A13 inhibition | Enzymatic reconstitution | 6.18 µM (IC50) | Methoxsalen 0.43 µM (IC50) | [31] | |
| Echinothiophene A (15) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 0.42 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 1.44 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/Fusarium solani | 64.0 µg/mL (MIC) | Carbendazim 0.5 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. vasinfectum | 16.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/F. oxysporum f. sp. niveum | 8.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Phytophthora infestans | 128.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 16.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 4.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Echinothiophene B (16) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 2.65 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 9.23 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/Fusarium solani | 32.0 µg/mL (MIC) | Carbendazim 0.5 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. vasinfectum | 64.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/F. oxysporum f. sp. niveum | 16.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Phytophthora infestans | 256.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 8.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternataalternata | 8.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Echinothiophene C (17) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 16.55 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 18.17 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/F. oxysporum f. sp. vasinfectum | 128.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. niveum | 256.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 128.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternataalternata | 32.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| 2-(Pro-1-ynyl)-5-(5,6-dihydroxypenta-1,3-diynyl) thiophene (PYDDT) (18) | CYP2A6 inhibition | Enzymatic reconstitution | 3.90 µM (IC50) | Methoxsalen 0.19 µM (IC50) | [31] |
| CYP2A13 inhibition | Enzymatic reconstitution | 2.40 µM (IC50) | Methoxsalen 0.43 µM (IC50) | [31] | |
| 5-(1,2-Dihydroxyethyl)-2-(E)-hept-5-ene-1,3-diynylthiophene (19) | In vitro anti-inflammatory/NO | LPS-stimulated production in the RAW 264.7 cell line | 28.2 µM (IC50) | -Indomethacin 13.2 µM (IC50) -Aminoguanidine 24.2 µM (IC50) | [34] |
| 5-(1,2-Dihydroxy-ethyl)-2-(Z)-hept-5-ene-1,3-diynylthiophene (20) | In vitro anti-inflammatory/NO | LPS-stimulated production in the RAW 264.7 cell line | 12.8 µM (IC50) | -Indomethacin 13.2 µM (IC50) -Aminoguanidine 24.2 µM (IC50) | [34] |
| 2-(Prop-1-inyl)-5-(6-acetoxy-5-hydroxyhexa-1,3-diinyl) thiophene (22) | CYP2A6 inhibition | Enzymatic reconstitution | 4.44 µM (IC50) | Methoxsalen 0.19 µM (IC50) | [31] |
| CYP2A13 inhibition | Enzymatic reconstitution | 2.94 µM (IC50) | Methoxsalen 0.43 µM (IC50) | [31] | |
| 3′′R-Pluthiophenol (23) | In vitro anti-inflammatory/NO | LPS-stimulated production in RAW 264.7 macrophages cells | 84.5 (NRC % inhibition) | Dexamethasone 62.2 (NRC % inhibition) | [35] |
| 3′′R-Pluthiophenol-4′′-acetate (24) | In vitro anti-inflammatory/NO | LPS-stimulated production in RAW 264.7 macrophages cells | 83.4 (NRC % inhibition) | Dexamethasone 62.2 (NRC % inhibition) | [35] |
| 3′′-Ethoxy-3′′S-pluthiophenol (25) | In vitro anti-inflammatory/NO | LPS-stimulated production in RAW 264.7 macrophages cells | 86.9 (NRC % inhibition) | Dexamethasone 62.2 (NRC % inhibition) | [35] |
| 3′′-Ethoxy-3′′S-pluthiophenol-4′′-acetate (26) | In vitro anti-inflammatory/NO | LPS-stimulated production in RAW 264.7 macrophages cells | 90.1 (NRC % inhibition) | Dexamethasone 62.2 (NRC % inhibition) | [35] |
| Rupestriene B (27) | In vitro anti-inflammatory/NO | LPS-stimulated production in BV-2 microglial cells | 8.5 µM (IC50) | Quercetin 4.3 µM (IC50) | [36] |
| Rupestriene C (28) | In vitro anti-inflammatory/NO | LPS-stimulated production in BV-2 microglial cells | 5.3 µM (IC50) | Quercetin 4.3 µM (IC50) | [36] |
| 5-(3,4-Dihydroxybut-1-ynyl)-2,2′-bithiophene (33) | In vitro anti-inflammatory/NO | LPS-stimulated production in the RAW 264.7 cell line | 20.0 µg/mL (IC50) | Indomethacin 65.4 µg/mL (IC50) | [28] |
| 5-(But-3-en-1-ynyl)-2,2′-bithiophene (5-BBT) (37) | Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 7.81 µg/mL (MFC) in light | Amphotericin B 0.50 µg/mL (MFC) Fluconazole ˃ 64 µg/mL (MFC) Itraconazole ˃ 16 µg/mL (MFC) | [40] |
| Larvicidal | Larval mortality/Aedes albopictus | 0.34 µg/mL (LC50) | Rotenone 3.75 µg/mL (LC50) | [41] | |
| Larvicidal | Larval mortality/Aedes albopictus | 0.72 µg/mL (LC95) | Rotenone 9.45 µg/mL (LC95) | [41] | |
| Larvicidal | Larval mortality/Anopheles sinensis | 1.36 µg/mL (LC50) | Rotenone 1.25 µg/mL (LC50) | [41] | |
| Larvicidal | Larval mortality/Anopheles sinensis | 1.93 µg/mL (LC95) | Rotenone 2.24 µg/mL (LC95) | [41] | |
| Larvicidal | Larval mortality/Culex pipiens pallens | 0.12 µg/mL (LC50) | Rotenone 1.88 µg/mL (LC50) | [41] | |
| Fungicidal | Larval mortality/Culex pipiens pallens | 0.18 µg/mL (LC95) | Rotenone 3.74 µg/mL (LC95) | [41] | |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 62.50 µg/mL (MFC) in low oxygen and light | - | [59] | |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 7.81 µg/mL (MFC) in normal oxygen and light | - | [59] | |
| 5-(4-Isovaleroyloxybut-1-ynyl)-2,2′-bithiophene (5-IBT) (38) | Larvicidal | Larval mortality/Aedes albopictus | 0.45 µg/mL (LC50) | Rotenone 3.75 µg/mL (LC50) | [41] |
| Larvicidal | 0.66 µg/mL (LC95) | Rotenone 9.45 µg/mL (LC95) | [41] | ||
| Larvicidal | Larval mortality/Anopheles sinensis | 5.36 µg/mL (LC50) | Rotenone 1.25 µg/mL (LC50) | [41] | |
| Larvicidal | 11.26 µg/mL (LC95) | Rotenone 2.24 µg/mL (LC95) | [41] | ||
| Larvicidal | Larval mortality/Culex pipiens pallens | 0.33 µg/mL (LC50) | Rotenone 1.88 µg/mL (LC50) | [41] | |
| Larvicidal | 0.54 µg/mL (LC95) | Rotenone 3.74 µg/mL (LC95) | [41] | ||
| 5-(4-Hydroxy-1-butynyl)-2,2′-bithiophene (43) | In vitro anti-inflammatory/NO | LPS-stimulated production in the RAW 264.7 cell line | 6.7 µg/mL (IC50) | Indomethacin 65.4 µg/mL (IC50) | [28] |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 3.90 µg/mL (MFC) in light | -Amphotericin B 0.50 µg/mL (MFC) -Fluconazole ˃ 64 µg/mL (MFC) -Itraconazole ˃ 16 µg/mL (MFC) | [40] | |
| Antimicrobial | Broth microdilution/S. aureus ATCC 2592 | 8.0 µg/mL (MIC) | Levofloxacin 8.0 μg/mL (MIC) | [27] | |
| Antimicrobial | Broth microdilution/E. coli ATCC 25922 | 64.0 µg/mL (MIC) | Levofloxacin 16.0 μg/mL (MIC) | [27] | |
| Antimicrobial | Broth microdilution/C. albicans ATCC2002 | 64.0 µg/mL (MIC) | Levofloxacin 64.0 μg/mL (MIC) | [27] | |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 250.0 µg/mL (MFC) in low oxygen and light | - | [59] | |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 3.90 µg/mL (MFC) in normal oxygen and light | - | [59] | |
| Anti-inflammatory | Colorimetric/5-LOX | 41.82 µM (IC50) | Indomethacin 0.89 µM (IC50) | [43] | |
| 5-(4-Acetoxy-1-butynl)-2,2′-bithiophene (44) | Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 7.81 µg/mL (MFC) in light | -Amphotericin B 0.50 µg/mL (MFC) -Fluconazole ˃ 64 µg/mL (MFC) -Itraconazole ˃ 16 µg/mL (MFC) | [40] |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 62.50 µg/mL (MFC) in low oxygen and light | - | [59] | |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 7.81 µg/mL (MFC) in normal oxygen and light | - | [59] | |
| 6-Methoxy-arctinol-b (48) | In vitro anti-inflammatory/NO | LPS-stimulated production in the RAW 264.7 cell line | 30.6 µM (IC50) | -Indomethacin 13.2 µM (IC50) -Aminoguanidine 24.2 µM (IC50) | [34] |
| Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 5.83 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] | |
| 7.05 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/Fusarium solani | 128.0 µg/mL (MIC) | Carbendazim 0.5 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. vasinfectum | 256.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/F. oxysporum f. sp. niveum | 128.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternataalternata | 32.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Arctinol-b (49) | In vitro anti-inflammatory/NO | LPS-stimulated production in the RAW 264.7 cell line | 48.7 µM (IC50) | -Indomethacin 13.2 µM (IC50) -Aminoguanidine 24.2 µM (IC50) | [34] |
| Antimicrobial | Broth microdilution/S. aureus ATCC 2592 | 8.0 µg/mL (MIC) | Levofloxacin 8.0 μg/mL (MIC) | [27] | |
| Antimicrobial | Broth microdilution/E. coli ATCC 25922 | 64.0 µg/mL (MIC) | Levofloxacin 16.0 μg/mL (MIC) | [27] | |
| Antimicrobial | Broth microdilution/C. albicans ATCC2002 | 64.0 µg/mL (MIC) | Levofloxacin 64.0 μg/mL (MIC) | [27] | |
| Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 13.48 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] | |
| 14.72 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/F. oxysporum f. sp. vasinfectum | 256.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. niveum | 64.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Phytophthora infestans | 128.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 64.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Arctinone-b (50) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 1.14 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 2.00 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/F. oxysporum f. sp. vasinfectum | 256.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | |
| Broth microdilution/Colletotrichum gloeosporioides | 64.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 128.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Arctinol (51) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 15.90 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 17.82 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/F. oxysporum f. sp. vasinfectum | 256.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. niveum | 128.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Phytophthora infestans | 128.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 16.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Arctinal (52) | Antimicrobial | Broth microdilution/S. aureus ATCC 2592 | 32.0 µg/mL (MIC) | Levofloxacin 8.0 μg/mL (MIC) | [19] |
| Antimicrobial | Broth microdilution/E. coli ATCC 25922 | 64.0 µg/mL (MIC) | Levofloxacin 16.0 μg/mL (MIC) | [19] | |
| Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 2.62 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] | |
| 8.75 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/F. oxysporum f. sp. vasinfectum | 64.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. niveum | 128.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 64.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Arctinol A (53) | Antimicrobial | Broth microdilution/S. aureus ATCC 2592 | 8.0 µg/mL (MIC) | Levofloxacin 8.0 μg/mL (MIC) | [27] |
| Antimicrobial | Broth microdilution/E. coli ATCC 25922 | 64.0 µg/mL (MIC) | Levofloxacin 16.0 μg/mL (MIC) | [27] | |
| 5′-(3,4-Dihydroxybut-1-yn-1-yl)-[2,2′-bithiophene]-5-carbaldehyde (57) | Antimicrobial | Broth microdilution/S. aureus ATCC 2592 | 128.0 µg/mL (MIC) | Levofloxacin 8.0 μg/mL (MIC) | [27] |
| Antimicrobial | Broth microdilution/E. coli ATCC 25922 | 256.0 µg/mL (MIC) | Levofloxacin 16.0 μg/mL (MIC) | [27] | |
| Antimicrobial | Broth microdilution/C. albicans ATCC2002 | 256.0 µg/mL (MIC) | Levofloxacin 64.0 μg/mL (MIC) | [27] | |
| 4-Hydroxy-1-(5′-methyl-[2,2′-bithiophen]-5-yl)butan-1-one (58) | Antimicrobial | Broth microdilution/S. aureus ATCC 2592 | 8.0 µg/mL (MIC) | Levofloxacin 8.0 μg/mL (MIC) | [27] |
| Antimicrobial | Broth microdilution/E. coli ATCC 25922 | 32.0 µg/mL (MIC) | Levofloxacin 16.0 μg/mL (MIC) | [27] | |
| Antimicrobial | Broth microdilution/C. albicans ATCC2002 | 32.0 µg/mL (MIC) | Levofloxacin 64.0 μg/mL (MIC) | [27] | |
| 5′-(3,4-Dihydroxybut-1-yn-1-yl)-[2,2′-bithiophene]-5-carboxylic acid (59) | Antimicrobial | Broth microdilution/S. aureus ATCC 2592 | 256.0 µg/mL (MIC) | Levofloxacin 8.0 μg/mL (MIC) | [27] |
| Echinothiophene D (61) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 2.57 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 1.80 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/Fusarium solani | 32.0 µg/mL (MIC) | Carbendazim 0.5 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. vasinfectum | 128.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/F. oxysporum f. sp. niveum | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Phytophthora infestans | 256.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 8.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 16.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Echinothiophene E (62) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 8.28 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 9.12 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/Fusarium solani | 64.0 µg/mL (MIC) | Carbendazim 0.5 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. vasinfectum | 32.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/F. oxysporum f. sp. niveum | 128.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Phytophthora infestans | 256.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 16.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Echinothiophene F (63) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 20.13 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 18.41 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/F. oxysporum f. sp. niveum | 128.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | |
| Broth microdilution/Colletotrichum gloeosporioides | 256.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 64.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| 2-Prop-1-inyl-5′-(2-hydroxy-3-chloropropyl) dithiophene (64) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 0.91 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] |
| 0.86 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| Antifungal | Broth microdilution/Fusarium solani | 64.0 µg/mL (MIC) | Carbendazim 0.5 µg/mL (MIC) | [19] | |
| Broth microdilution/F. oxysporum f. sp. vasinfectum | 32.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/F. oxysporum f. sp. niveum | 4.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Phytophthora infestans | 32.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 4.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [19] | ||
| Broth microdilution/Alternaria alternata | 4.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [19] | ||
| Ecliprostin A (65) | Antibacterial | Broth microdilution/S. aureus | 25.0 µM (MIC) | Penicillin 0.156 µM (MIC) | [18] |
| Ecliprostin B (66) | Antibacterial | Broth microdilution/S. aureus | 6.25 µM (MIC) | Penicillin 0.156 µM (MIC) | [18] |
| Ecliprostin C (67) | Antibacterial | Broth microdilution/S. aureus | 25.0 µM (MIC) | Penicillin 0.156 µM (MIC) | [18] |
| Echinbithiophenedimer A (68) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 16.53 µg/mL (LC50) in light | Ethoprophos 36.15 (LC50) in light α-Terthienyl 0.62 (LC50) in light | [17] |
| 18.17 µg/mL (LC50) in dark | Ethoprophos 31.94 (LC50) in dark α-Terthienyl 2.23 (LC50) in dark | [17] | |||
| Antifungal | Broth microdilution/Alternaria alternata; | 16.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [17] | |
| Broth microdilution/Pyricularia oryzae | 16.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Fusarium oxysporum | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 64.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Phytophthora infestans | 128.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [17] | ||
| Echinbithiophenedimer B (69) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 13.88 µg/mL (LC50) in light | Ethoprophos 36.15 (LC50) in light α-Terthienyl 0.62 (LC50) in light | [17] |
| 16.28 µg/mL (LC50) in dark | Ethoprophos 31.94 (LC50) in dark α-Terthienyl 2.23 (LC50) in dark | [17] | |||
| Antifungal | Broth microdilution/Alternaria alternata | 16.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [17] | |
| Broth microdilution/Pyricularia oryzae | 16.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Fusarium oxysporum | 16.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 32.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Phytophthora infestans | 128.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [17] | ||
| Echinbithiophenedimer C (70) | Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 8.73 µg/mL (LC50) in light | Ethoprophos 36.15 (LC50) in light α-Terthienyl 0.62 (LC50) in light | [17] |
| 9.39 µg/mL (LC50) in dark | Ethoprophos 31.94 (LC50) in dark α-Terthienyl 2.23 (LC50) in dark | [17] | |||
| Antifungal | Broth microdilution/Alternaria alternata; | 8.0 µg/mL (MIC) | Carbendazim 16.0 µg/mL (MIC) | [17] | |
| Broth microdilution/Pyricularia oryzae | 8.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Fusarium oxysporum | 32.0 µg/mL (MIC) | Carbendazim 8.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Colletotrichum gloeosporioides | 32.0 µg/mL (MIC) | Carbendazim 2.0 µg/mL (MIC) | [17] | ||
| Broth microdilution/Phytophthora infestans | 128.0 µg/mL (MIC) | Carbendazim 256.0 µg/mL (MIC) | [17] | ||
| 4-(5′-(hydroxymethyl)-[2,2′-bithiophene]-5-yl)but-3-yn-1-ol) (Thio1) (74) | Anthelmintic | Larval development test/Haemonchus contortus | 0.3243 mg/mL (EC50) | Levamisole 1.88 mg/mL (EC50) | [46] |
| Anthelmintic | Fecal egg count reduction test/Haemonchus contortus | 0.1731 mg/mL (EC50) | Levamisole 1.88 mg/mL (EC50) | [46] | |
| 2,2′:5′,2′′-Terthiophene (α-Terthienyl) (75) | Cytotoxicity | MTT/SKOV3 | 77.23 µM (IC50) | Cisplatin 11.25 µM (IC50) | [39] |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 0.24 µg/mL (MFC) in light | Amphotericin B 0.50 µg/mL (MFC) Fluconazole ˃ 64 µg/mL (MFC) Itraconazole ˃ 16 µg/mL (MFC) | [40] | |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 7.81 µg/mL (MFC) in low oxygen and light | - | [59] | |
| Fungicidal | Broth microdilution/C. albicans ATCC 10231 | 0.24 µg/mL (MFC) in normal oxygen and light | - | [59] | |
| Larvicidal | Larval mortality/Aedes albopictus | 1.41 µg/mL (LC50) | Rotenone 3.75 µg/mL (LC50) | [41] | |
| Larvicidal | 2.19 µg/mL (LC95) | Rotenone 9.45 µg/mL (LC95) | [41] | ||
| Larvicidal | Larval mortality/Anopheles sinensis | 1.79 µg/mL (LC50) | Rotenone 1.25 µg/mL (LC50) | [41] | |
| Larvicidal | 2.54 µg/mL (LC95) | Rotenone 2.24 µg/mL (LC95) | [41] | ||
| Larvicidal | Larval mortality/Culex pipiens pallens | 1.38 µg/mL (LC50) | Rotenone 1.88 µg/mL (LC50) | [41] | |
| Larvicidal | 2.15 µg/mL (LC95) | Rotenone 3.74 µg/mL (LC95) | [41] | ||
| Nematicidal | Nematode Mortality/J2s of Meloidogyne incognita | 0.56 µg/mL (LC50) in light | Abamectin 8.73 (LC50) in light | [19] | |
| 1.77 µg/mL (LC50) in dark | Abamectin 9.38 (LC50) in dark | [19] | |||
| 5-Formyl-2,2′:5′,2′′-terthiophene (Ecliptal) (76) | Cytotoxicity | MTT/SKOV3 | 24.57 µM (IC50) | Cisplatin 11.25 µM (IC50) | [39] |
| Cytotoxicity | MTT/Hec1A | 12.00 µM (IC50) | Cisplatin 120.42 µM (IC50) | [47] | |
| Cytotoxicity | MTT/Ishikawa | 2.20 µM (IC50) | Cisplatin 10.11 µM (IC50) | [47] | |
| Anti-inflammatory | Colorimetric/5-LOX | 26.18 µM (IC50) | Indomethacin 0.89 µM (IC50) | [43] | |
| Antibacterial | Broth microdilution/S. aureus | 25.0 µM (MIC) | Penicillin G 0.156 µM (MIC) | [38] | |
| 5-Hydroxymethyl-2,2′:5′,2′′-terthiophene (α-Terthienylmethanol) (77) | Cytotoxicity | MTT/SKOV3 | 7.73 µM (IC50) | Cisplatin 11.25 µM (IC50) | [39] |
| Cytotoxicity | MTT/Hec1A | 0.38 µM (IC50) | Cisplatin 120.42 µM (IC50) | [47] | |
| Cytotoxicity | MTT/Ishikawa | 0.35 µM (IC50) | Cisplatin 10.11 µM (IC50) | [47] | |
| Cytotoxicity | MTT/A2780 | 1.18 µM (IC50) | Cisplatin 10.80 µM (IC50) | [20] | |
| Cytotoxicity | MTT/SKOV3 | 15.51 µM (IC50) | Cisplatin 43.05 µM (IC50) | [20] | |
| Cytotoxicity | MTT/OVCAR3 | 0.20 µM (IC50) | Cisplatin 35.46 µM (IC50) | [20] | |
| Cytotoxicity | MTT/ES2 | 18.82 µM (IC50) | Cisplatin 29.58 µM (IC50) | [20] | |
| Antibacterial | Broth microdilution/S. aureus | 25.0 µM (MIC) | Penicillin G 0.156 µM (MIC) | [38] | |
| 5-Hydroxymethyl-(2,2′:5′,2′′)-terthienyl angelate (79) | Cytotoxicity | MTT/Hec1A | 129.85 µM (IC50) | Cisplatin 120.42 µM (IC50) | [47] |
| Cytotoxicity | MTT/Ishikawa | 6.87 µM (IC50) | Cisplatin 10.11 µM (IC50) | [47] | |
| 5-Hydroxymethyl-(2,2′:5′,2′′)-terthienyl tiglate (80) | Cytotoxicity | MTT/Hec1A | 2.66 µM (IC50) | Cisplatin 120.42 µM (IC50) | [47] |
| Cytotoxicity | MTT/Ishikawa | 9.68 µM (IC50) | Cisplatin 10.11 µM (IC50) | [47] | |
| 5-Methoxy-(2,2′:5′,2′′)-terthiophene (81) | Cytotoxicity | MTT/Hec1A | 1.38 µM (IC50) | Cisplatin 120.42 µM (IC50) | [47] |
| Cytotoxicity | MTT/Ishikawa | 7.12 µM (IC50) | Cisplatin 10.11 µM (IC50) | [47] | |
| 3′-Hydroxy-2,2′:5′,2′′-terthiophene-3′-O-β-D-glucopyranoside (82) | Cytotoxicity | MTT/SKOV3 | 58.20 µM (IC50) | Cisplatin 11.25 µM (IC50) | [39] |
| Thiotagetin A (83) | Cytotoxicity | MTT/KB | 2.03 μg/mL (ED50) | Adriamycin 0.26 μg/mL (ED50) | [48] |
| Cytotoxicity | MTT/MCF-7 | 3.88 μg/mL (ED50) | Adriamycin 0.07 μg/mL (ED50) | [48] | |
| Rupestriene A (86) | In vitro anti-inflammatory/NO | LPS-stimulated production in BV-2 microglial cells | 20.3 µM (IC50) | Quercetin 4.3 µM (IC50) | [36] |
| Neuraminidase inhibitory activity | Fluorescence-based assay | 351.15 µM (IC50) | Oseltamivir acid 77.91 µM (IC50) | [15] | |
| 7-[1-(Thiophene-5-yl)-1-formamido]-3-propylenyl-3-cephem-4-carboxylic acid (CAx1) (87) | Antibacterial | Broth microdilution/S. aureus MTCC 740 | 0.2 µg/mL (MIC) 2.0 µg/mL (MBC) | Penicillin 32.0 µg/mL (MIC) 64.0 µg/mL (MBC) | [40] |
| Antibacterial | Broth microdilution/B. subtilis MTCC 736 | 0.25 µg/mL (MIC) 0.5 µg/mL (MBC) | Penicillin 0.5 µg/mL (MIC) 4.0 µg/mL (MBC) | [50] | |
| Antibacterial | Broth microdilution/E. coli MTCC 739 | 4.0 µg/mL (MIC) 8.0 µg/mL (MBC) | Penicillin 4.0 µg/mL (MIC) 16.0 µg/mL (MBC) | [50] | |
| Antibacterial | Broth microdilution/K. pneumonia MTCC 661 | 4.0 µg/mL (MIC) 16.0 µg/mL (MBC) | Penicillin 16.0 µg/mL (MIC) 64.0 µg/mL (MBC) | [50] | |
| 2,5-Bis(5-tert-butyl-2-benzoxazolyl)thiophene (88) | Antimicrobial | Broth microdilution/E. faecalis ATCC29212 | 256.0 µg/mL (MIC) | Streptomycin 256.0 μg/mL (MIC) | [51] |
| Thiocarboxylic A (89) | Antimicrobial | Broth microdilution/E. coli ATCC35218 | 1.7 µg/mL (MIC) | Streptomycin 2.3 µg/mL (MIC) | [16] |
| Broth microdilution/S. aureus ATCC25923 | 1.7 µg/mL (MIC) | Streptomycin 0.1 µg/mL (MIC) | [16] | ||
| Broth microdilution/C. albicans ATCC10231 | 3.3 µg/mL (MIC) | Amphotericin B 0.1 µg/mL (MIC) | [16] | ||
| Thiocarboxylic B (90) | Antimicrobial | Broth microdilution/E. coli ATCC35218 | 0.9 µg/mL (MIC) | Streptomycin 2.3 µg/mL (MIC) | [16] |
| Broth microdilution/S. aureus ATCC25923 | 1.9 µg/mL (MIC) | Streptomycin 0.1 µg/mL (MIC) | [16] | ||
| Broth microdilution/C. albicans ATCC10231 | 3.9 µg/mL (MIC) | Amphotericin B 0.1 µg/mL (MIC) | [16] | ||
| Thiocarboxylic C1 (91) | Antimicrobial | Broth microdilution/E. coli ATCC35218 | 7.0 µg/mL (MIC) | Streptomycin 2.3 µg/mL (MIC) | [16] |
| Broth microdilution/S. aureus ATCC25923 | 3.5 µg/mL (MIC) | Streptomycin 0.1 µg/mL (MIC) | [16] | ||
| Broth microdilution/C. albicans ATCC10231 | 7.0 µg/mL (MIC) | Amphotericin B 0.1 µg/mL (MIC) | [16] | ||
| Thiocarboxylic C2 (92) | Antimicrobial | Broth microdilution/E. coli ATCC35218 | 7.0 µg/mL (MIC) | Streptomycin 2.3 µg/mL (MIC) | [16] |
| Broth microdilution/S. aureus ATCC25923 | 3.5 µg/mL (MIC) | Streptomycin 0.1 µg/mL (MIC) | [16] | ||
| Broth microdilution/C. albicans ATCC10231 | 7.0 µg/mL (MIC) | Amphotericin B 0.1 µg/mL (MIC) | [16] | ||
| Thiocarboxylic D1 (93) | Antimicrobial | Broth microdilution/E. coli ATCC35218 | 3.5 µg/mL (MIC) | Streptomycin 2.3 µg/mL (MIC) | [16] |
| Broth microdilution/S. aureus ATCC25923 | 3.5 µg/mL (MIC) | Streptomycin 0.1 µg/mL (MIC) | [16] | ||
| Broth microdilution/C. albicans ATCC10231 | 7.0 µg/mL (MIC) | Amphotericin B 0.1 µg/mL (MIC) | [16] | ||
| Thiocarboxylic D2 (94) | Antimicrobial | Broth microdilution/E. coli (ATCC35218) | 3.5 µg/mL (MIC) | Streptomycin 2.3 µg/mL (MIC) | [16] |
| Broth microdilution/S. aureus ATCC25923 | 3.5 µg/mL (MIC) | Streptomycin 0.1 µg/mL (MIC) | [16] | ||
| Broth microdilution/C. albicans ATCC10231 | 7.0 µg/mL (MIC) | Amphotericin B 0.1 µg/mL (MIC) | [16] | ||
| Rupestriene D (95) | Neuraminidase inhibitory activity | Fluorescence-based assay | 986.54 µM (IC50) | Oseltamivir acid 77.91 µM (IC50) | [15] |
| Rupestriene E (96) | Neuraminidase inhibitory activity | Fluorescence-based assay | 365.40 µM (IC50) | Oseltamivir acid 77.91 µM (IC50) | [15] |
4.2. Cytotoxic Activity
4.3. Antimicrobial Activity
4.4. Antimalarial Activity
4.5. Larvicidal Activity
4.6. Nematicidal Activity
4.7. Antioxidant and Anti-Influenza Activities
5. AI Target-Based Prediction vs. (Virtual Screening), and MD (Molecular Dynamics) for Thiophene Derivatives
5.1. In Silico ADMET Properties of Selected Ligands
5.2. Ligands and Proteins Preparations
5.3. Molecular Docking Studies
5.4. Molecular Dynamics Simulation
5.5. Materials and Methods
5.5.1. Preparation of PDB Structures
5.5.2. ADME Properties Prediction
5.5.3. Receptor Grids Generation and Docking
5.5.4. MD Simulations of Compound 30 in Complex with 4OD9
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Martins, P.; Jesus, J.; Santos, S.; Raposo, L.R.; Roma-Rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Heterocyclic anticancer compounds: Recent advances and the paradigm shift towards the use of nanomedicine’s toolbox. Molecules 2015, 20, 16852–16891. [Google Scholar] [CrossRef] [PubMed]
- Tilby, M.J.; Willis, M.C. How do we address neglected sulfur pharmacophores in drug discovery? Expert Opin. Drug Discov. 2021, 16, 1227–1231. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.P.; DeNicola, G.M. Sulfur metabolism and its contribution to malignancy. Cell. Nutr. Util. Cancer 2019, 347, 39–103. [Google Scholar]
- Francioso, A.; Baseggio Conrado, A.; Mosca, L.; Fontana, M. Chemistry and biochemistry of sulfur natural compounds: Key intermediates of metabolism and redox biology. Oxid. Med. Cell Longev. 2020, 2020, 8294158. [Google Scholar] [CrossRef]
- Rakesh, K.P.; Wang, S.M.; Leng, J.; Ravindar, L.; Asiri, A.M.; Marwani, H.M.; Qin, H.L. Recent development of sulfonyl or sulfonamide hybrids as potential anticancer agents: A key review. Anti-Cancer Agents. Med. Chem. 2018, 18, 488–505. [Google Scholar] [CrossRef] [PubMed]
- McGrath, N.A.; Brichacek, M.; Njardarson, J.T. A graphical journey of innovative organic architectures that have improved our lives. J. Chem. Educ. 2010, 87, 1348–1349. [Google Scholar] [CrossRef]
- Hai, Y.; Wei, M.Y.; Wang, C.Y.; Gu, Y.C.; Shao, C.L. The intriguing chemistry and biology of sulfur-containing natural products from marine microorganisms (1987–2020). Mar. Life Sci. Technol. 2021, 3, 488–518. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Abdallah, H.M.; El-Halawany, A.M.; Mohamed, G.A. Naturally occurring thiophenes: Isolation, purification, structural elucidation, and bioactivities. Phytochem. Rev. 2016, 15, 197–220. [Google Scholar] [CrossRef]
- Shah, R.; Verma, P.K. Therapeutic importance of synthetic thiophene. Chem. Cent. J. 2018, 12, 137. [Google Scholar] [CrossRef] [Green Version]
- Keri, R.S.; Chand, K.; Budagumpi, S.; Balappa Somappa, S.; Patil, S.A.; Nagaraja, B.M. An overview of benzo[b]thiophene-based medicinal chemistry. Eur. J. Med. Chem. 2017, 138, 1002–1033. [Google Scholar] [CrossRef]
- Gramec, D.; Masic, L.P.; Sollner Dolenc, M. Bioactivation potential of thiophene-containing drugs. Chem. Res. Toxicol. 2014, 27, 1344–1358. [Google Scholar] [CrossRef]
- Joshi, E.M.; Heasley, B.H.; Chordia, M.D.; Macdonald, T.L. In vitro metabolism of 2-acetylbenzothiophene: Relevance to Zileuton Hepatotoxicity. Chem. Res. Toxicol. 2004, 17, 137–143. [Google Scholar] [CrossRef]
- Gil, A.; Ghersa, C.M.; Perelman, S. Root thiophenes in Tagetes minuta L. accessions from Argentina: Genetic and environmental contribution to changes in concentration and composition. Biochem. Syst. Ecol. 2002, 30, 1–13. [Google Scholar] [CrossRef]
- Champagne, D.E.; Arnason, J.T.; Philogene, B.J.R.; Campbell, G.; McLachlan, D.G. Photosensitization and feeding deterrence of Euxoa messoria (Lepidoptera: Noctuiidae) by α-terthienyl, a naturally occurring thiophene from the Asteraceae. Experientia 1984, 40, 577–578. [Google Scholar] [CrossRef]
- Cao, Y.; Zang, Y.; Huang, X.; Cheng, Z. Chemical constituents from Artemisia rupestris and their neuraminidase inhibitory activity. Nat. Prod. Res. 2021, 35, 1775–1782. [Google Scholar] [CrossRef]
- Chang, J.L.; Xu, H.Z.; Zhou, J.; Zhou, M.; Zhang, X.; Guo, Y.; Ruan, H.L. Antimicrobial furancarboxylic acids from a Penicillium sp. J. Nat. Prod. 2020, 83, 3606–3613. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.B.; Wu, H.B.; Kuang, M.S.; Lan, H.P.; Wen, Y.X.; Liu, T.T. Novel bithiophene dimers from Echinops latifolius as potential antifungal and nematicidal agents. J. Agric. Food Chem. 2020, 68, 11939–11945. [Google Scholar] [CrossRef]
- Yu, S.J.; Yu, J.H.; He, F.; Bao, J.; Zhang, J.S.; Wang, Y.Y.; Zhang, H. New antibacterial thiophenes from Eclipta prostrata. Fitoterapia 2020, 142, 104471. [Google Scholar] [CrossRef]
- Liu, T.; Wu, H.; Jiang, H.; Zhang, L.; Zhang, Y.; Mao, L. Thiophenes from Echinops grijsii as a preliminary approach to control disease complex of root-knot nematodes and soil-borne fungi: Isolation, activities, and structure-nonphototoxic activity relationship analysis. J. Agric. Food Chem. 2019, 67, 6160–6168. [Google Scholar] [CrossRef] [PubMed]
- Preya, U.H.; Lee, K.T.; Kim, N.J.; Lee, J.Y.; Jang, D.S.; Choi, J.H. The natural terthiophene α-terthienylmethanol induces S phase cell cycle arrest of human ovarian cancer cells via the generation of ROS stress. Chem. Biol. Interact. 2017, 272, 72–79. [Google Scholar] [CrossRef]
- Da Cruz, R.M.D.; Mendonça-Junior, F.J.B.; de Mélo, N.B.; Scotti, L.; de Araújo, R.S.A.; de Almeida, R.N.; de Moura, R.O. Thiophene-based compounds with potential anti-inflammatory activity. Pharmaceuticals 2021, 14, 692. [Google Scholar] [CrossRef]
- Caballero, R.; Cohen, B.; Gutiérrez, M. Thiophene-based covalent organic frameworks: Synthesis, photophysics and light-driven applications. Molecules 2021, 26, 7666. [Google Scholar] [CrossRef] [PubMed]
- Bhilare, N.V.; Auti, P.B.; Marulkar, V.S.; Pise, V.J. Diverse thiophenes as scaffolds in anti-cancer drug development: A concise review. Mini-Rev. Med. Chem. 2021, 2, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Abedinifar, F.; Babazadeh Rezaei, E.; Biglar, M.; Larijani, B.; Hamedifar, H.; Ansari, S.; Mahdavi, M. Recent strategies in the synthesis of thiophene derivatives: Highlights from the 2012–2020 literature. Mol. Divers. 2021, 25, 2571–2604. [Google Scholar] [CrossRef] [PubMed]
- Chitsazian-Yazdi, M.; Agnolet, S.; Lorenz, S.; Schneider, B.; Es’haghi, Z.; Kasaian, J.; Khameneh, B.; Iranshahi, M. Foetithiophenes C-F, thiophene derivatives from the roots of Ferula foetida. Pharm. Biol. 2015, 53, 710–714. [Google Scholar] [CrossRef]
- Zhou, X.D.; Zhang, C.; He, S.; Zheng, B.; Zeng, K.W.; Zhao, M.B.; Jiang, Y.; Tu, P.F. New terpenoids and thiophene derivatives from the aerial parts of Artemisia sieversiana. Bioorg. Med. Chem. Lett. 2017, 27, 5441–5445. [Google Scholar] [CrossRef]
- Li, L.B.; Xiao, G.D.; Xiang, W.; Yang, X.; Cao, K.X.; Huang, R.S. Novel substituted thiophenes and sulf-polyacetylene ester from Echinops ritro L. Molecules 2019, 24, 805. [Google Scholar] [CrossRef] [Green Version]
- Chang, F.P.; Chen, C.C.; Huang, H.C.; Wang, S.Y.; Chen, J.J.; Yang, C.S.; Ou, C.Y.; Wu, J.B.; Huang, G.J.; Kuo, Y.H. A new bithiophene from the root of Echinops grijsii. Nat. Prod. Commun. 2015, 10, 2147–2149. [Google Scholar] [CrossRef]
- Mbaveng, A.T.; Sandjo, L.P.; Tankeo, S.B.; Ndifor, A.R.; Pantaleon, A.; Nagdjui, B.T.; Kuete, V. Antibacterial activity of nineteen selected natural products against multi-drug resistant Gram-negative phenotypes. Springerplus 2015, 4, 823. [Google Scholar] [CrossRef] [Green Version]
- Sandjo, L.P.; Kuete, V.; Siwe, X.N.; Poumale, H.M.P.; Efferth, T. Cytotoxicity of an unprecedented brominated oleanolide and a new furoceramide from the Cameroonian spice, Echinops giganteus. Nat. Prod. Res. 2016, 30, 2529–2537. [Google Scholar] [CrossRef]
- Boonruang, S.; Prakobsri, K.; Pouyfung, P.; Srisook, E.; Prasopthum, A.; Rongnoparut, P.; Sarapusit, S. Inhibition of human cytochromes P450 2A6 and 2A13 by flavonoids, acetylenic thiophenes and sesquiterpene lactones from Pluchea indica and Vernonia cinerea. J. Enzyme Inhib. Med. Chem. 2017, 32, 1136–1142. [Google Scholar] [CrossRef] [Green Version]
- Bitew, H.; Mammo, W.; Hymete, A.; Yeshak, M.Y. Antimalarial activity of acetylenic thiophenes from Echinops hoehnelii Schweinf. Molecules 2017, 22, 1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.G.; Lv, W.; Dai, C.Y.; Zhu, F.F.; Xu, G.H.; Ma, Z.J.; Chen, Z. 2-(Pro-1-ynyl)-5-(5,6-dihydroxypenta-1,3-diynyl) thiophene induces apoptosis through reactive oxygen species-mediated JNK activation in human colon cancer SW620 cells. Anat. Rec. 2015, 298, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Lee, J.W.; Jang, H.; Choi, J.E.; Kim, H.S.; Lee, D.; Hong, J.T.; Lee, M.K.; Hwang, B.Y. Dimeric sesquiterpene and thiophenes from the roots of Echinops latifolius. Bioorg. Med. Chem. Lett. 2016, 26, 5995–5998. [Google Scholar] [CrossRef]
- Ruan, J.; Li, Z.; Yan, J.; Huang, P.; Yu, H.; Han, L.; Zhang, Y.; Wang, T. Bioactive constituents from the aerial parts of Pluchea indica Less. Molecules 2018, 23, 2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Liu, B.Y.; Zeng, K.W.; Guo, X.Y.; Jiang, Y.; Tu, P.F. New sesquiterpene and thiophene derivatives from Artemisia rupestris. J. Asian Nat. Prod. Res. 2015, 17, 1129–1136. [Google Scholar] [CrossRef]
- Feng, Z.M.; Xu, K.; Wang, W.; Du, N.; Zhang, J.H.; Yang, Y.N.; Jiang, J.S.; Zhang, P.C. Two new thiophene polyacetylene glycosides from Atractylodes lancea. J. Asian Nat. Prod. Res. 2018, 20, 531–537. [Google Scholar] [CrossRef]
- Yu, S.J.; Zhang, J.S.; He, H.; Yu, J.H.; Bao, J.; Zhang, H. Thiophene enantiomers from the aerial parts of Eclipta prostrata. J. Asian Nat. Prod. Res. 2021, 23, 745–753. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kim, H.M.; Ryu, B.; Lee, J.S.; Choi, J.H.; Jang, D.S. Constituents of the aerial parts of Eclipta prostrata and their cytotoxicity on human ovarian cancer cells in vitro. Arch. Pharm. Res. 2015, 38, 1963–1969. [Google Scholar] [CrossRef]
- Postigo, A.; Funes, M.; Petenatti, E.; Bottai, H.; Pacciaroni, A.; Sortino, M. Antifungal photosensitive activity of Porophyllum obscurum (Spreng.) DC.: Correlation of the chemical composition of the hexane extract with the bioactivity. Photodiagn. Photodyn. Ther. 2017, 20, 263–272. [Google Scholar] [CrossRef]
- Zhao, M.P.; Liu, Q.Z.; Liu, Q.; Liu, Z.L. Identification of larvicidal constituents of the essential oil of Echinops grijsii roots against the three species of mosquitoes. Molecules 2017, 22, 205. [Google Scholar] [CrossRef] [Green Version]
- Kiyekbayeva, L.; Mohamed, N.M.; Yerkebulan, O.; Mohamed, E.I.; Ubaidilla, D.; Nursulu, A.; Assem, M.; Srivedavyasasri, R.; Ross, S.A. Phytochemical constituents and antioxidant activity of Echinops albicaulis. Nat. Prod. Res. 2018, 32, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.R.M.; Mohamed, G.A.; Al Haidari, R.A.; El-Kholy, A.A.; Zayed, M.F.; Khayat, M.T. Tagetnoic acid, a new lipoxygenase inhibitor peroxy fatty acid from Tagetes minuta growing in Saudi Arabia. Nat. Prod. Res. 2020, 34, 474–481. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Abdallah, H.M.; El-Halawany, A.M.; Esmat, A.; Mohamed, G.A. Thiotagetin B and tagetannins A and B, new acetylenic thiophene and digalloyl glucose derivatives from Tagetes minuta and evaluation of their in vitro antioxidative and anti-inflammatory activity. Fitoterapia 2018, 125, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, S.; Zhang, Z.; Sauriol, F.; Shi, Q.; Yang, J. New thiophene acetylene from Echinops spinosissimus subsp. Spinosus. Chem. Nat. Compd. 2017, 53, 933–934. [Google Scholar] [CrossRef]
- Politi, F.A.S.; Bueno, R.V.; Zeoly, L.A.; Fantatto, R.R.; Eloy, J.O.; Chorilli, M.; Coelho, F.; Guido, R.V.C.; Chagas, A.C.S.; Furlan, M. Anthelmintic activity of a nanoformulation based on thiophenes identified in Tagetes patula L. (Asteraceae) against the small ruminant nematode Haemonchus contortus. Acta Trop. 2021, 219, 105920. [Google Scholar] [CrossRef]
- Lee, J.S.; Ahn, J.H.; Cho, Y.J.; Kim, H.Y.; Yang, Y.I.; Lee, K.T.; Jang, D.S.; Choi, J.H. α-Terthienylmethanol, isolated from Eclipta prostrata, induces apoptosis by generating reactive oxygen species via NADPH oxidase in human endometrial cancer cells. J Ethnopharmacol. 2015, 169, 426–434. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A. Thiotagetin A, new cytotoxic thiophene from Tagetes minuta. Nat. Prod. Res. 2017, 31, 543–547. [Google Scholar] [CrossRef]
- Shi, Y.S.; Li, L.; Liu, Y.B.; Ma, S.G.; Li, Y.; Qu, J.; Liu, Q.; Shen, Z.F.; Chen, X.G.; Yu, S.S. A new thiophene and two new monoterpenoids from Xanthium sibiricum. J. Asian Nat. Prod. Res. 2015, 17, 1039–1047. [Google Scholar] [CrossRef]
- Bhattacharjee, K.; Palepu, N.R.; Rao, K.M.; Joshi, S.R. Precursor-directed combinatorial biosynthesis of cephalosporin analogue by endolithic actinobacterium Streptomyces sp. AL51 by utilizing thiophene derivative. 3 Biotech 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.T.; Tran, V.H.; Vu, V.N.; Mai, H.D.T.; Le, T.H.M.; Vu, T.Q.; Nguyen, H.H.; Chau, V.M.; Pham, V.C. Antimicrobial metabolites from a marine-derived actinomycete Streptomyces sp. G278. Nat. Prod. Res. 2019, 33, 3223–3230. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, C.-J.; Chen, J.; Zhao, Q.-Q.; Li, Y.; Gao, K. Thiophene acetylenes and furanosesquiterpenes from Xanthopappus subacaulis and their antibacterial activities. Phytochemistry 2014, 106, 134–140. [Google Scholar] [CrossRef]
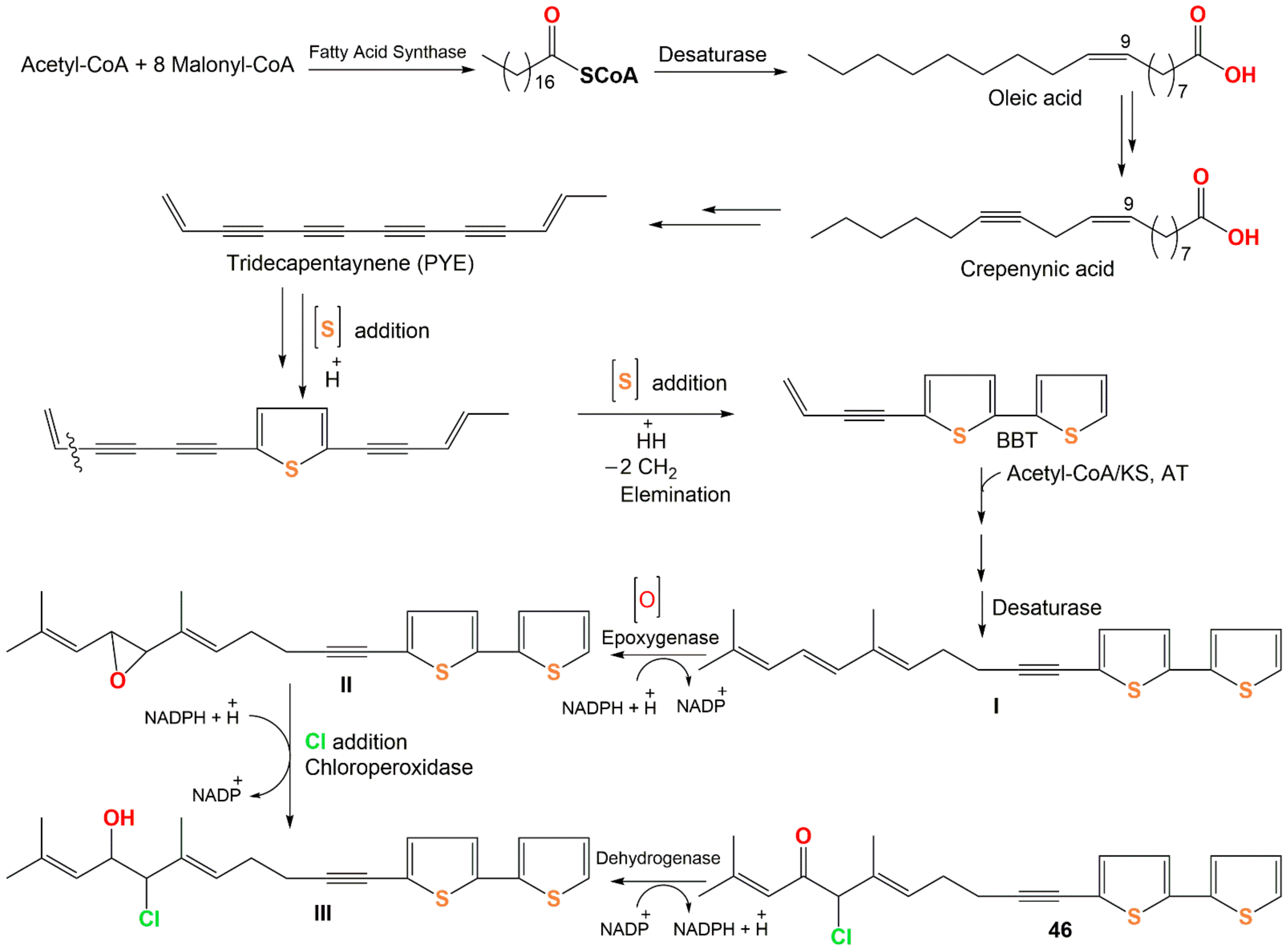
- Velíšek, J.; Cejpek, K. Biosynthesis of food constituents: Lipids. 1. Fatty acids and derived compounds—A review. Czech J. Food Sci. 2006, 24, 193–216. [Google Scholar] [CrossRef] [Green Version]
- Field, J.A. Natural Production of Organohalide Compounds in the Environment. In Organohalide-Respiring Bacteria, 1st ed.; Adrian, L., Löffler, F.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 7–29. [Google Scholar]
- Medzhitov, R. Inflammation 2010: New adventures of an old flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef] [Green Version]
- Yatoo, M.I.; Gopalakrishnan, A.; Saxena, A.; Parray, O.R.; Tufani, N.A.; Chakraborty, S.; Tiwari, R.; Dhama, K.; Iqbal, H. Anti-Inflammatory Drugs and Herbs with Special Emphasis on Herbal Medicines for Countering Inflammatory Diseases and Disorders—A Review. Recent Pat. Inflamm. Allergy Drug Discov. 2018, 12, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Fizeșan, I.; Rusu, M.E.; Georgiu, C.; Pop, A.; Ștefan, M.G.; Muntean, D.M.; Mirel, S.; Vostinaru, O.; Kiss, B.; Popa, D.S. Antitussive, antioxidant, and anti-inflammatory effects of a walnut (Juglans regia L.) septum extract rich in bioactive compounds. Antioxidants 2021, 10, 119. [Google Scholar] [CrossRef]
- Ranaweera, S.S.; Dissanayake, C.Y.; Natraj, P.; Lee, Y.J.; Han, C.H. Anti-inflammatory effect of sulforaphane on LPS-stimulated RAW 264.7 cells and ob/ob mice. J. Vet. Sci. 2020, 21, e91. [Google Scholar] [CrossRef] [PubMed]
- Postigo, A.; Schiavi, P.C.; Funes, M.; Sortino, M. Mechanistic studies of Candida albicans photodynamic inactivation with Porophyllum obscurum hexanic extract and its isolated thiophenic compounds. Photodiagn. Photodyn. Ther. 2019, 26, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 25 December 2021).
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [Green Version]
- Gay, F.; Zougbédé, S.; N’dilimabaka, N.; Rebollo, A.; Mazier, D.; Moreno, A. Cerebral malaria: What is known and what is on research. Rev. Neurol. 2012, 68, 239–256. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). World Malaria Report. 2021. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021 (accessed on 25 December 2021).
- Ibrahim, S.; Mohamed, G.A.; Al Haidari, R.A.; El-Kholy, A.A.; Zayed, M.F. Potential anti-malarial agents from endophytic fungi: A Review. Mini-Rev. Med. Chem. 2018, 18, 1110–1132. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Gao, C.; Yu, Z. Rhabdopeptides from Xenorhabdus budapestensis SN84 and their nematicidal activities against Meloidogyne incognita. J. Agric. Food Chem. 2018, 66, 3833–3839. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, Y.; Gan, X.; Song, B.; Hu, D.; Song, B. Synthesis, nematicidal evaluation, and 3D-QSAR analysis of novel 1,3,4-oxadiazole-cinnamic acid hybrids. J. Agric. Food Chem. 2018, 66, 9616–9623. [Google Scholar] [CrossRef] [PubMed]
- Tocco, G.; Eloh, K.; Onnis, V.; Sasanelli, N.; Caboni, P. Haloacetophenones as newly potent nematicides against Meloidogyne incognita. Ind. Crops Prod. 2017, 110, 94–102. [Google Scholar] [CrossRef]
- Castino, R.; Davies, J.; Beaucourt, S.; Isidoro, C.; Murphy, D. Autophagy is a prosurvival mechanism in cells expressing an autosomal dominant familial neurohypophyseal diabetes insipidus mutant vasopressin transgene. FASEB J. 2005, 19, 1021–1023. [Google Scholar] [CrossRef] [Green Version]
- Houštecká, R.; Hadzima, M.; Fanfrlík, J.; Brynda, J.; Pallová, L.; Hánová, I.; Mertlíková-Kaiserová, H.; Lepšík, M.; Horn, M.; Smrčina, M.; et al. Biomimetic macrocyclic inhibitors of human cathepsin D: Structure-activity relationship and binding mode analysis. J. Med. Chem. 2020, 63, 1576–1596. [Google Scholar] [CrossRef]
- Pranjol, M.Z.I.; Gutowski, N.; Hannemann, M.; Whatmore, J. The potential role of the proteases cathepsin D and cathepsin L in the progression and metastasis of epithelial ovarian cancer. Biomolecules 2015, 5, 3260–3279. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Roth, J.M.; Brooks, P.; Luty, J.; Karpatkin, S. Thrombin up-regulates cathepsin D which enhances angiogenesis, growth, and metastasis. Cancer Res. 2008, 68, 4666–4673. [Google Scholar] [CrossRef] [Green Version]
- Lowry, J.R.; Klegeris, A. Emerging roles of microglial cathepsins in neurodegenerative disease. Brain Res. Bull. 2018, 139, 144–156. [Google Scholar] [CrossRef]
- Yang, S.; Ye, Q.; Ding, J.; Yin, M.; Lu, A.; Chen, X.; Hou, T.; Cao, D. Current advances in ligand-based target prediction. WIREs Comput. Mol. Sci. 2021, 11, e1504. [Google Scholar] [CrossRef]
- Nickel, J.; Gohlke, B.; Erehman, J.; Banerjee, P.; Rong, W.W.; Goede, A.; Dunkel, M.; Preissner, R. SuperPred: Update on drug classification and target prediction. Nucleic Acids Res. 2014, 42, 26. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, LLC. Schrödinger Release 2021-4: LigPrep; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Schrödinger, LLC. Schrödinger Release 2021-4: QikProp; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Schrödinger, LLC. Schrödinger Release 2021-4: Glide; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Schrödinger, LLC. Schrödinger Release 2021-4: Desmond Molecular Dynamics System; D. E. Shaw Research: New York, NY, USA, 2021. [Google Scholar]
- Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2021.
- RCSB PDB: Homepage. Available online: https://www.rcsb.org/ (accessed on 5 January 2022).
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Schrödinger, LLC. Schrödinger Release 2021-4: Protein Preparation Wizard; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Schrödinger, LLC. Schrödinger Release 2021-4: Epik; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Schrödinger, LLC. Schrödinger Release 2021-4: Impact; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Schrödinger, LLC. Schrödinger Release 2021-4: Prime; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Sun, H.; Liu, Z.; Zhao, H.; Ang, E.L. Recent advances in combinatorial biosynthesis for drug discovery. Drug Des. Dev. Ther. 2015, 9, 823–833. [Google Scholar]



















| Compound Name | Source | Extract/Fraction | Mol. Wt. | Mol. Formula | Location | Ref. |
|---|---|---|---|---|---|---|
| I. Monothiophenes | ||||||
| Foetithiophene A (1) | Ferula foetida, roots (Apiaceae) | MeOH/EtOAc-soluble fraction | 170 | C8H10O2S | Dorouneh village mountains, Khorasan Razavi, Iran | [25] |
| Foetithiophene B (2) | Ferula foetida, roots (Apiaceae) | MeOH/EtOAc-soluble fraction | 218 | C9H14O2S2 | Dorouneh village mountains, Khorasan Razavi, Iran | [25] |
| Foetithiophene C (3) | Ferula foetida, roots (Apiaceae) | MeOH/EtOAc-soluble fraction | 186 | C9H14S2 | Dorouneh village mountains, Khorasan Razavi, Iran | [25] |
| Foetithiophene D (4) | Ferula foetida, roots (Apiaceae) | MeOH/EtOAc-soluble fraction | 310 | C16H22S3 | Dorouneh village mountains, Khorasan Razavi, Iran | [25] |
| Foetithiophene E (5) | Ferula foetida, roots (Apiaceae) | MeOH/EtOAc-soluble fraction | 202 | C9H14OS2 | Dorouneh village mountains, Khorasan Razavi, Iran | [25] |
| Foetithiophene F (6) | Ferula foetida, roots (Apiaceae) | MeOH/EtOAc-soluble fraction | 188 | C9H16S2 | Dorouneh village mountains, Khorasan Razavi, Iran | [25] |
| 5-Propinyl-thiophene-2-Carboxylic acid = Junipic acid (7) | Artemisia sieversiana, aerial parts (Asteraceae) | EtOH/CH3Cl-soluble fraction | 166 | C8H6O2S | Qinghai, China | [26] |
| Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | - | - | Qinghe County, Xinjiang, China | [27] | |
| 3-Hydroxy-5-propinyl-2-acetyl-thiophene (8) | Artemisia sieversiana, aerial parts (Asteraceae) | EtOH/CH3Cl-soluble fraction | 180 | C9H8O2S | Qinghai, China | [26] |
| 2-(3,4-Dihydroxybut-1-ynyl)-5-(penta-1,3-diynyl)thiophene = 2-(Penta-1,3-diynyl)-5-(3,4-dihydroxybut-1-ynyl)-thiophene = 4-(5-(Penta-1,3-diyn-1-yl)thiophen-2-yl)but-3-yne-1,2-diol = 2-(Penta-1,3-diynyl)-5-(3,4-dihydroxybut-1-ynyl)-thiophene = 5-(Penta-1,3-diynyl)-2-(3,4-dihydroxybut-1-ynyl)-thiophene (9) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 230 | C13H10O2S | Nantou, Taiwan | [28] |
| Echinops giganteus, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | - | - | Dschang, Western Region of Cameroon | [29] | |
| Echinops giganteus, rhizomes and aerial parts (Asteraceae) | MeOH/n-hexane-soluble fraction | - | - | Dschang, Western Region of Cameroon | [30] | |
| Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | - | - | Qinghe County, Xinjiang, China | [27] | |
| 2-(4-Hydroxybut-1-ynyl)-5-(penta-1,3-diynyl)thiophene (10) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 214 | C13H10OS | Nantou, Taiwan | [28] |
| 2-(Penta-1,3-diyn-1-yl)-5–(4-acetoxy-3-hydroxybuta-1-yn-1-yl) thiophene (11) | Pluchea indica, aerial parts (Asteraceae) | EtOH/n-hexane-soluble fraction | 256 | C15H12O2S | Bon subdistrict, Khlung, Chantaburi, Thailand | [31] |
| 5-(Penta-1,3-diynyl)-2-(3-hydroxy-4-acetoxybut-1-ynyl)-thiophene (12) | Echinops hoehnelii, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 272 | C15H12O3S | Bale Mountains National Park, Ethiopia | [32] |
| 5-(Penta-1,3-diynyl)-2-(3-methoxy-4-acetoxy-but-1-yn)-thiophene (13) | Echinops hoehnelii, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 286 | C16H14O3S | Bale Mountains National Park, Ethiopia | [32] |
| 5-(Penta-1,3-diynyl)-2-(3-chloro-4-acetoxy-but-1-yn)-thiophene (14) | Echinops hoehnelii, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 290 | C15H11ClO2S | Bale Mountains National Park, Ethiopia | [32] |
| Echinothiophene A (15) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 248 | C13H9ClOS | Zhangjiakou Hebei, China | [19] |
| Echinothiophene B (16) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 290 | C15H11ClO2S | Zhangjiakou Hebei, China | [19] |
| Echinothiophene C (17) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 398 | C23H26O4S | Zhangjiakou Hebei, China | [19] |
| 2-(Pro-1-ynyl)-5-(5,6-dihydroxypenta-1,3-diynyl) thiophene (PYDDT) = 2-(Prop-1-ynyl)-5(5,6-dihydroxyhexa-1, 3-diynyl)-thiophene = PITC-2 = R/J/3 (18) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 230 | C13H10O2S | Bozhou north of Anhui, China | [33] |
| Pluchea indica, aerial parts (Asteraceae) | EtOH/n-hexane-soluble fraction | - | - | Bon subdistrict, Khlung, Chantaburi, Thailand | [31] | |
| 5-(1,2-Dihydroxyethyl)-2-(E)-hept-5-ene-1,3-diynylthiophene (19) | Echinops latifolius, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 232 | C13H12O2S | Seoul, Korea | [34] |
| 5-(1,2-Dihydroxy-ethyl)-2-(Z)-hept-5-ene-1,3-diynylthiophene (20) | Echinops latifolius, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 232 | C13H12O2S | Seoul, Korea | [34] |
| 5-(Penta-1,3-diynyl)-2-(3-methoxy-4-hydroxybut-1-ynyl)-thiophen (21) | Echinops hoehnelii, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 244 | C14H12O2S | Bale Mountains National Park, Ethiopia | [32] |
| 2-(Prop-1-inyl)-5-(6-acetoxy-5-hydroxyhexa-1,3-diinyl) thiophene (22) | Pluchea indica, aerial parts (Asteraceae) | EtOH/n-hexane-soluble fraction | 256 | C15H12O2S | Bon subdistrict, Khlung, Chantaburi, Thailand | [31] |
| 53′′R-Pluthiophenol (23) | Pluchea indica, aerial parts (Asteraceae) | EtOH/n-hexane-soluble fraction | 230 | C13H10O2S | Hepu, Guangxi, China | [35] |
| 3′′R-Pluthiophenol-4′′-acetate (24) | Pluchea indica, aerial parts (Asteraceae) | EtOH/n-hexane-soluble fraction | 272 | C15H12O3S | Hepu, Guangxi, China | [35] |
| 3′′-Ethoxy-3′′S-pluthiophenol (25) | Pluchea indica, aerial parts (Asteraceae) | EtOH/n-hexane-soluble fraction | 258 | C15H14O2S | Hepu, Guangxi, China | [3] |
| 3′′-Ethoxy-3′′S-pluthiophenol-4′′-acetate (26) | Pluchea indica, aerial parts (Asteraceae) | EtOH/n-hexane-soluble fraction | 300 | C17H16O3S | Hepu, Guangxi, China | [35] |
| Rupestriene B (27) | Artemisia rupestris, whole plant (Asteraceae) | EtOH/CH3Cl-soluble fraction | 234 | C13H14O2S | Xinjiang Uygur Autonomous, China | [36] |
| Rupestriene C (28) | Artemisia rupestris, whole plant (Asteraceae) | EtOH/CH3Cl-soluble fraction | 234 | C13H14O2S | Xinjiang Uygur Autonomous, China | [36] |
| Atracthioenyneside A (29) | Atractylodes lancea, rhizomes (Asteraceae) | EtOH/n-BuOH extract | 414 | C19H26O8S | Huanggang, Hubei, China | [37] |
| Atracthioenyneside B (30) | Atractylodes lancea, rhizomes (Asteraceae) | EtOH/n-BuOH extract | 386 | C17H22O8S | Huanggang, Hubei, China | [37] |
| (Z)-6-(5-(Prop-1-yn-1-yl)thiophen-2-yl)hex-3-en-5-yne-1,2-diol (31) | Eclipta prostrata, aerial parts (Asteraceae) | EtOH/PE-soluble fraction | 232 | C13H12O2S | Mount Kunyu area, Shandong, China | [38] |
| II. Bithiophenes | ||||||
| 5-(4-Hydroxy-3-methoxy-1-butyny)-2,2′-bithiophene (32) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 264 | C13H12O2S2 | Nantou, Taiwan | [28] |
| 5-(3,4-Dihydroxybut-1-ynyl)-2,2′-bithiophene = 4-([2,2′-bithiophen]-5-yl) but-3-yne-1,2-diol (33) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 250 | C12H10O2S2 | Nantou, Taiwan | [28] |
| Eclipta prostrata, aerial parts (Asteraceae) | MeOH/EtOAc-soluble fraction | - | - | Seoul, Korea | [39] | |
| Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | - | - | Qinghe County, Xinjiang, China | [27] | |
| 5-Acetyl-2,2′-bithiophene = Ethanone (34) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 208 | C10H8OS2 | Nantou, Taiwan | [28] |
| Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | - | - | Qinghe County, Xinjiang, China | [27] | |
| 5-Formyl-2,2′-bithiophene (35) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 194 | C9H6OS2 | Nantou, Taiwan | [28] |
| Methyl 2,2′-bithiophene-5-carboxylate (36) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 224 | C10H8O2S2 | Nantou, Taiwan | [28] |
| 5-(But-3-en-1-ynyl)-2,2′-bithiophene (5-BBT) (37) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 216 | C12H8S2 | Nantou, Taiwan | [28] |
| Porophyllum obscurum, aerial parts (Asteraceae) | n-Hexane extract | - | - | Las Chacras, province of San Luis, Argentina | [40] | |
| Echinops grijisii, roots (Asteraceae) | Essential oil, hydrodistillation | - | - | Nanjing, Jiangsu, China | [41] | |
| Echinops albicaulis, aerial parts (Asteraceae) | MeOH/CH2Cl2 soluble fraction | - | - | Malaysary gorge, Kazakhstan | [42] | |
| 5-(4-Isovaleroyloxybut-1-ynyl)-2,2′-bithiophene (5-IBT) (38) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 318 | C17H18O2S2 | Nantou, Taiwan | [28] |
| Echinops grijisii, roots (Asteraceae) | Essential oil, hydrodistillation | - | - | Nanjing, Jiangsu, China | [41] | |
| Cardopatine (39) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 432 | C24H16S4 | Nantou, Taiwan | [28] |
| Isocardopatine (40) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 432 | C24H16S4 | Nantou, Taiwan | [28] |
| 5-(3-Hydroxy-4-isovaleroyloxybut-1-ynyl)-2,2′-bithiophene (41) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 334 | C17H18O3S2 | Nantou, Taiwan | [28] |
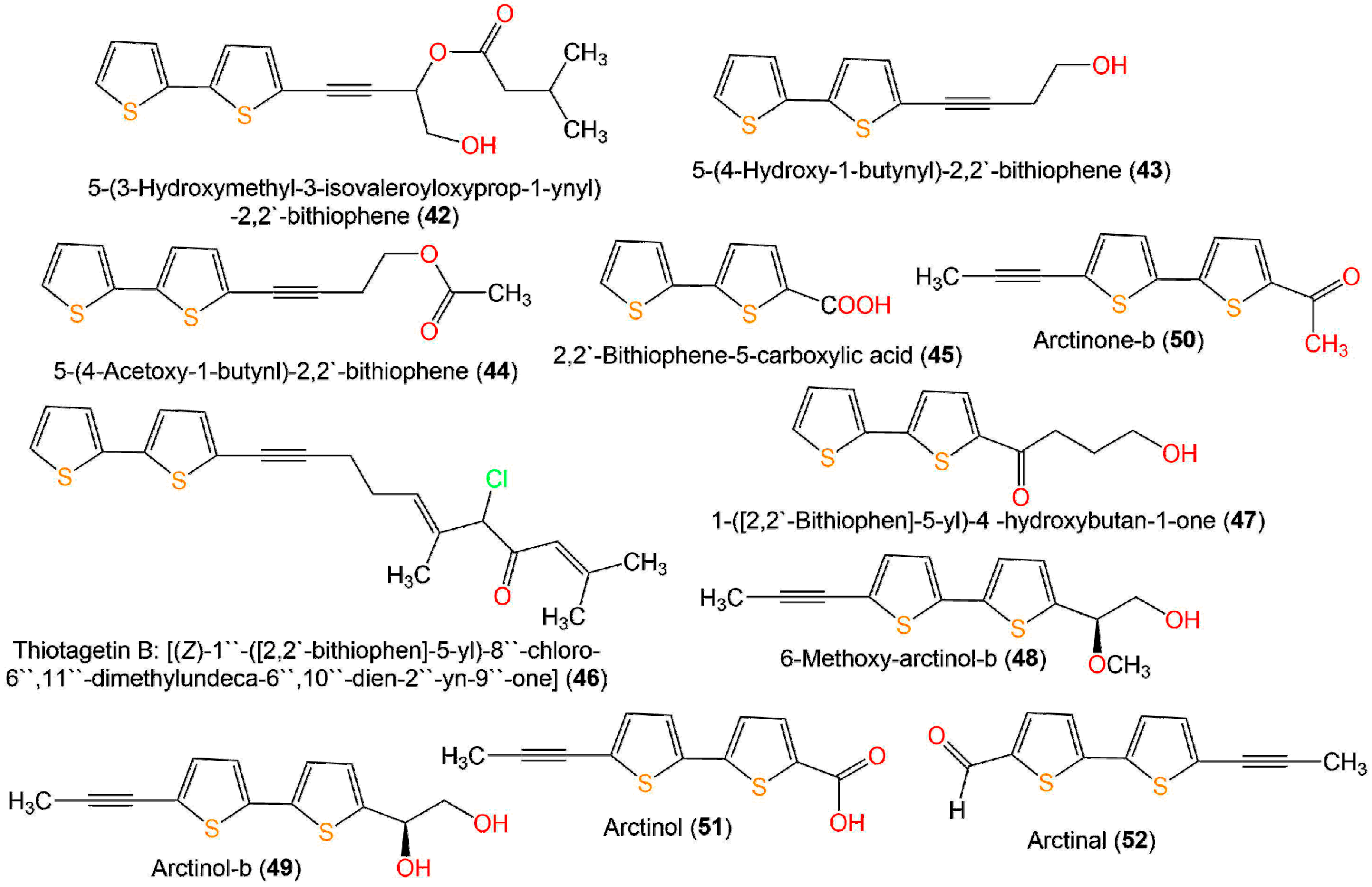
| 5-(3-Hydroxymethyl-3-isovaleroyloxyprop-1-ynyl)-2,2′-bithiophene (42) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 334 | C17H18O3S2 | Nantou, Taiwan | [28] |
| 5-(4-Hydroxy-1-butynyl)-2,2′-bithiophene = 4-([2,2′-bithiophen]-5-yl)but-3-yn-1-ol = 4-(5′-(hydroxymethyl)-[2,2′-bithiophene]-5-yl)but-3-yn-1-ol) (43) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 234 | C12H10OS2 | Nantou, Taiwan | [28] |
| Porophyllum obscurum, aerial parts (Asteraceae) | n-Hexane extract | - | - | Las Chacras, province of San Luis, Argentina | [40] | |
| Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | - | - | Qinghe County, Xinjiang, China | [27] | |
| Tagetes minuta, aerial parts (Asteraceae) | MeOH/n-hexane-soluble fraction | - | - | Al-Baha city, Saudi Arabia | [43] | |
| 5-(4-Acetoxy-1-butynl)-2,2′-bithiophene (44) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | Nantou, Taiwan | [28] | ||
| Porophyllum obscurum, aerial parts (Asteraceae) | n-Hexane extract | - | - | Las Chacras, province of San Luis, Argentina | [40] | |
| 2,2′-Bithiophene-5-carboxylic acid (45) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 209 | C9H6O2S2 | Nantou, Taiwan | [28] |
| Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | - | - | Qinghe County, Xinjiang, China | [27] | |
| Thiotagetin B: [(Z)-1′′-([2,2′-bithiophen]-5-yl)-8′′-chloro-6′′,11′′-dimethylundeca-6′′,10′′-dien-2′′-yn-9′′-one] (46) | Tagetes minuta, aerial parts (Asteraceae) | MeOH/n-hexane-soluble fraction | 388 | C21H21ClOS5 | Al-Baha city, Saudi Arabia | [44] |
| 1-([2,2′-Bithiophen]-5-yl)-4 -hydroxybutan-1-one (47) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 252 | C12H12O2S2 | Qinghe County, Xinjiang, China | [27] |
| 6-Methoxy-arctinol-b (48) | Echinops latifolius, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 278 | C14H14O2S2 | Seoul, Korea | [34] |
| Arctinol-b (49) | Echinops latifolius, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 264 | C13H12O2S2 | Seoul, Korea | [34] |
| Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | - | - | Qinghe County, Xinjiang, China | [27] | |
| Arctinone-b (50) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 246 | C13H10OS2 | Zhangjiakou Hebei, China | [19] |
| Arctinol (51) | Echinops latifolius, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 248 | C12H8O2S2 | Seoul, Korea | [34] |
| Arctinal (52) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 232 | C12H8OS2 | Qinghe County, Xinjiang, China | [27] |
| Arctinol A (53) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 234 | C12H10OS2 | Qinghe County, Xinjiang, China | [27] |
| Arctic acid (54) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 248 | C12H8O2S2 | Qinghe County, Xinjiang, China | [27] |
| Methyl [5′-(1-propynyf)-2,2′-bithienyl-5-yl] carboxylate (55) | Echinops latifolius, roots (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 262 | C13H10O2S2 | Seoul, Korea | [34] |
| 2,2-Dimethyl-4-[5′-(prop-1-ynyl)-2,2′-bithiophen-5-yl]-1,3-dioxolane (56) | Echinops spinosissimus subsp. Spinosus, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 304 | C16H16O2S2 | Morocco | [45] |
| 5′-(3,4-Dihydroxybut-1-yn-1-yl)-[2,2′-bithiophene]-5-carbaldehyde = 5-[l-(4-hydroxybut-l-ynyl)]-2,2′-bithiophene-5′-carbaldehyde (57) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 278 | C13H10O3S2 | Qinghe County, Xinjiang, China | [27] |
| 4-Hydroxy-1-(5′-methyl-[2,2′-bithiophen]-5-yl)butan-1-one (58) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 266 | C13H14O2S2 | Qinghe County, Xinjiang, China | [27] |
| 5′-(3,4-Dihydroxybut-1-yn-1-yl)-[2,2′-bithiophene]-5-carboxylic acid (59) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 294 | C13H10O4S2 | Qinghe County, Xinjiang, China | [27] |
| 4-(5′-Methyl-[2,2′-bithiophen]-5-yl)but-3-yn-1-ol (60) | Echinops ritro, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 248 | C13H12OS2 | Qinghe County, Xinjiang, China | [27] |
| Echinothiophene D (61) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 298 | C13H14O4S2 | Zhangjiakou Hebei, China | [19] |
| Echinothiophene E (62) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 366 | C18H19ClO2S2 | Zhangjiakou Hebei, China | [19] |
| Echinothiophene F (63) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 432 | C23H28O4S2 | Zhangjiakou Hebei, China | [19] |
| 2-Prop-1-inyl-5′-(2-hydroxy-3-chloropropyl) dithiophene (64) | Echinops grijisii, roots (Asteraceae) | EtOH/CH2Cl2-soluble fraction | 282 | C13H11ClOS2 | Zhangjiakou Hebei, China | [19] |
| Ecliprostin A (65) | Eclipta prostrata, aerial parts (Asteraceae) | EtOH/EtOAc-soluble fraction | 348 | C18H20O3S2 | Mount Kunyu area, Shandong, China | [18] |
| Ecliprostin B (66) | Eclipta prostrata, aerial parts (Asteraceae) | EtOH/EtOAc-soluble fraction | 348 | C18H20O3S2 | Mount Kunyu area, Shandong, China, | [18] |
| Ecliprostin C (67) | Eclipta prostrata, aerial parts (Asteraceae) | EtOH/EtOAc-soluble fraction | 678 | C36H38O5S4 | Mount Kunyu area, Shandong, China | [18] |
| Echinbithiophenedimer A (68) | Echinops latifolius, roots (Asteraceae) | EtOH/n-hexane:acetone (8:1) soluble fraction | 492 | C26H20O2S4 | Mentougou, Beijing, China | [17] |
| Echinbithiophenedimer B (69) | Echinops latifolius, roots (Asteraceae) | EtOH/n-hexane:acetone (8:1) soluble fraction | 492 | C26H20O2S4 | Mentougou, Beijing, China | [17] |
| Echinbithiophenedimer C (70) | Echinops latifolius, roots (Asteraceae) | EtOH/n-hexane:acetone (8:1) soluble fraction | 492 | C26H20O2S4 | Mentougou, Beijing, China | [17] |
| (R)-(5′-(3,4-dihydroxybut-1-yn-1-yl)-[2,2′-bithiophen]-5-yl)methyl 3-methylisovalerate (71) | Eclipta prostrata, aerial parts (Asteraceae) | EtOH/PE-soluble fraction | 364 | C18H20O4S2 | Mount Kunyu area, Shandong, China | [38] |
| 5-[l-(4-hydroxybut-l-ynyl)]-2,2′-bithiophene-5′-carbaldehyde (72) | Eclipta prostrata, aerial parts (Asteraceae) | EtOH/PE-soluble fraction | 262 | C13H10O2S2 | Mount Kunyu area, Shandong, China | [38] |
| 5′-Hydroxymethyl-5-(3-butene-1-ynyl)-2,2′-bithiophene (73) | Eclipta prostrata, aerial parts (Asteraceae) | EtOH/PE-soluble fraction | 246 | C13H10OS2 | Mount Kunyu area, Shandong, China | [38] |
| 4-(5′-(hydroxymethyl)-[2,2′-bithiophene]-5-yl)but-3-yn-1-ol) (Thio1) (74) | Tagetes patula, aerial parts (Asteraceae) | Synthesis | 264 | C13H12O2S2 | - | [46] |
| III. Terthiophene | ||||||
| 2,2′:5′,2′′-Terthiophene (α-Terthienyl) (α-T) (75) | Echinops grijisii, roots (Asteraceae) | MeOH/EtOAc-soluble fraction | 248 | C12H8S3 | Nantou, Taiwan | [28] |
| Eclipta prostrata, aerial parts (Asteraceae) | MeOH/EtOAc-soluble fraction | - | - | Seoul, Korea | [39] | |
| Porophyllum obscurum, aerial parts (Asteraceae) | n-Hexane extract | - | - | Las Chacras, province of San Luis, Argentina | [40] | |
| Echinops grijisii, roots (Asteraceae) | Essential oil, hydrodistillation | - | - | Nanjing, Jiangsu, China | [41] | |
| Echinops albicaulis, aerial parts (Asteraceae) | MeOH/CH2Cl2 soluble fraction | - | - | Malaysary gorge, Kazakhstan | [42] | |
| 5-Formyl-2,2′:5′,2′′-terthiophene (Ecliptal) (76) | Eclipta prostrata, aerial parts (Asteraceae) | MeOH/EtOAc-soluble fraction | 276 | C13H8OS3 | Seoul, Korea | [39] |
| Eclipta prostrata, whole plant (Asteraceae) | EtOH/n-hexane-soluble fraction | - | - | Seoul, Korea | [47] | |
| Tagetes minuta, aerial parts (Asteraceae) | MeOH/n-hexane-soluble fraction | - | - | Al-Baha city, Saudi Arabia | [43] | |
| 5-Hydroxymethyl-2,2′:5′,2′′- terthiophene (α-Terthienylmethanol) (77) | Eclipta prostrata, aerial parts (Asteraceae) | MeOH/EtOAc-soluble fraction | 278 | C13H10OS3 | Seoul, Korea | [39] |
| Eclipta prostrata, whole plant (Asteraceae) | EtOH/n-hexane-soluble fraction | - | - | Seoul, Korea | [47] | |
| Eclipta prostrata, whole plant (Asteraceae) | EtOH/n-hexane-soluble fraction | - | - | Seoul, Korea | [20] | |
| 3′-Methoxy-2,2′:5′,2′′-terthiophene (78) | Eclipta prostrata, aerial parts (Asteraceae) | MeOH/EtOAc-soluble fraction | 278 | C13H10OS3 | Seoul, Korea | [39] |
| 5-Hydroxymethyl-(2,2′:5′,2′′)-terthienyl angelate (79) | Eclipta prostrata, whole plant (Asteraceae) | EtOH/n-hexane-soluble fraction | 360 | C18H16O2S3 | Seoul, Korea | [47] |
| 5-Hydroxymethyl-(2,2′:5′,2′′)-terthienyl tiglate (80) | Eclipta prostrata, whole plant (Asteraceae) | EtOH/n-hexane-soluble fraction | 360 | C18H16O2S3 | Seoul, Korea | [47] |
| 5-Methoxy-(2,2′:5′,2′′)-terthiophene (81) | Eclipta prostrata, whole plant (Asteraceae) | EtOH/n-hexane-soluble fraction | 292 | C14H12OS3 | Seoul, Korea | [47] |
| 3′-Hydroxy-2,2′:5′,2′′-terthiophene-3′-O-β-D-glucopyranoside (82) | Eclipta prostrata, aerial parts (Asteraceae) | MeOH/EtOAc-soluble fraction | 426 | C18H18O6S3 | Seoul, Korea | [39] |
| IV. Quinquethiophenes | ||||||
| Thiotagetin A (83) | Tagetes minuta, aerial parts, (Asteraceae) | MeOH/n-hexane-soluble fraction | 540 | C27H24O2S5 | Al-Baha city, Saudi Arabia | [48] |
| V. Miscellaneous | ||||||
| Sibiricumthionol (84) | Xanthium sibiricum, fruits (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 197 | C9H11NO2S | Helen town, Heilongjiang, China | [49] |
| (+)-Xanthienopyran (85) | Xanthium sibiricum, fruits (Asteraceae) | MeOH/CH2Cl2-soluble fraction | 316 | C17H16O4S | Helen town, Heilongjiang, China | [49] |
| Rupestriene A (86) | Artemisia rupestris, whole plant (Asteraceae) | EtOH/CH3Cl-soluble fraction | 280 | C15H20O3S | Xinjiang Uygur Autonomous, China | [36] |
| 7-[1-(Thiophene-5-yl)-1-formamido]-3-propylenyl-3-cephem-4-carboxylic acid (CAx1) (87) | Endolithic Streptomyces sp. AL51 | Spore MeOH extract | 350 | C15H14N2O4S2 | Granite rock, Mylliem, Meghalaya, India | [50] |
| 2,5-Bis(5-tert-butyl-2-benzoxazolyl)thiophene (88) | Marine-derived actinomycete Streptomyces sp. G278 isolated from echinoderm Holothuria edulis | Fermentation broth | 430 | C26H26N2O2S | Cu Lao Cham- Quang Nam, Vietnam | [51] |
| Thiocarboxylic A (89) | Penicillium sp. sb62 was isolated from soil collected near the fibrous roots of Schisandra bicolor var. tuberculata (Schisandraceae) | EtOAc of fermented material | 332 | C18H20O4S | Xinning, Hunan, China | [16] |
| Thiocarboxylic B (90) | Penicillium sp. sb62 was isolated from soil collected near the fibrous roots of Schisandra bicolor var. tuberculata (Schisandraceae) | EtOAc of fermented material | 376 | C19H20O6S | Xinning, Hunan, China | [16] |
| Thiocarboxylic C1 (91) | Penicillium sp. sb62 was isolated from soil collected near the fibrous roots of Schisandra bicolor var. tuberculata (Schisandraceae) | EtOAc of fermented material | 348 | C18H20O5S | Xinning, Hunan, China | [16] |
| Thiocarboxylic C2 (92) | Penicillium sp. sb62 was isolated from soil collected near the fibrous roots of Schisandra bicolor var. tuberculata (Schisandraceae) | EtOAc of fermented material | 348 | C18H20O5S | Xinning, Hunan, China | [16] |
| Thiocarboxylic D1 (93) | Penicillium sp. sb62 was isolated from soil collected near the fibrous roots of Schisandra bicolor var. tuberculata (Schisandraceae) | EtOAc of fermented material | 350 | C18H22O5S | Xinning, Hunan, China | [16] |
| Thiocarboxylic D2 (94) | Penicillium sp. sb62 was isolated from soil collected near the fibrous roots of Schisandra bicolor var. tuberculata (Schisandraceae) | EtOAc of fermented material | 350 | C18H22O5S | Xinning, Hunan, China | [16] |
| Rupestriene D (95) | Artemisia rupestris, whole plant (Asteraceae) | EtOH/PE-soluble fraction | 234 | C13H14O2S | Xinjiang Uygur Autonomous, China | [15] |
| Rupestriene E (96) | Artemisia rupestris, whole plant (Asteraceae) | EtOH/EtOAc-soluble fraction | 294 | C15H18O4S | Xinjiang Uygur Autonomous, China | [15] |
| 9 | Probability * | Model Accuracy ** |
|---|---|---|
| 9 | 96.04% | 96.09% |
| 20 | 93.69% | 96.09% |
| 28 | 82.30% | 98.95% |
| 29 | 85.79% | 98.95% |
| 30 | 82.76% | 98.95% |
| 33 | 87.63% | 98.95% |
| 43 | 82.00% | 98.95% |
| 46 | 94.79% | 98.95% |
| 57 | 89.12% | 98.95% |
| 67 | 91.20% | 98.95% |
| 68 | 77.77% | 90.17% |
| 70 | 88.21% | 98.95% |
| 75 | 79.91% | 98.95% |
| 76 | 87.50% | 98.95% |
| 77 | 85.75% | 98.95% |
| 80 | 90.87% | 98.95% |
| 81 | 90.32% | 98.95% |
| 82 | 88.71% | 98.95% |
| Title | mol MW | # Stars | Dipole | SASA | DonorHB | AccptHB | QPlogPo/w | QPlogS | QPlogKhsa | # Metab | QPlogBB | %HumOral Absorption | QPlogHERG | CNS | # RtvFG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Recommended range | (130–725) | (0.0–5.0) | (1–12.50) | (300–1000) | (0–6) | (2.0–20.0) | (−2–6.5) | (−6.5–0.5) | (−1.5–1.5) | (1–8) | (−3–1.2) | (<25% poor; >80% high) | concern below −5 | (−2 inactive) (+2 active) | (0–2) |
| 1 | 170.23 | 1 | 2.8 | 375.85 | 1 | 2 | 2.15 | −2.47 | −0.34 | 4 | −0.19 | 83.82 | −1.25 | −1 | - |
| 2 | 218.33 | - | 7.35 | 456.52 | - | 4 | 2.1 | −1.26 | −0.4 | 6 | 0.01 | 75.45 | −3.63 | 1 | - |
| 2 | 218.33 | - | 7.35 | 456.52 | - | 4 | 2.1 | −1.26 | −0.4 | 6 | 0.01 | 75.45 | −3.63 | 1 | - |
| 3 | 186.33 | 5 | 3.11 | 416.69 | - | 0.5 | 3.73 | −4.49 | 0.38 | 5 | 0.61 | 100 | −3.22 | 2 | - |
| 4 | 310.53 | 7 | 0.48 | 576.92 | - | 0.5 | 5.35 | −8.24 | 1.34 | 10 | 0.96 | 100 | −3.9 | 2 | - |
| 5 | 202.33 | - | 5.95 | 441.45 | - | 4 | 2.01 | −1.08 | −0.44 | 6 | 0.24 | 78.76 | −3.53 | 1 | - |
| 5 | 202.33 | - | 5.95 | 441.45 | - | 4 | 2.01 | −1.08 | −0.44 | 6 | 0.24 | 78.76 | −3.53 | 1 | - |
| 6 | 188.35 | 5 | 2.31 | 409.89 | - | 1 | 3.11 | −3.82 | 0.3 | 3 | 0.45 | 100 | −2.88 | 1 | - |
| 7 | 166.19 | - | 3.67 | 386.76 | 1 | 2 | 1.96 | −1.92 | −0.42 | 2 | −0.4 | 80.8 | −2.35 | −1 | - |
| 8 | 180.22 | 3 | 6.7 | 417.32 | - | 1.75 | 2.21 | −2.75 | −0.11 | 3 | −0.39 | 95.04 | −4.18 | - | - |
| 8 | 180.22 | 3 | 6.61 | 417.82 | - | 1.75 | 2.22 | −2.76 | −0.11 | 3 | −0.39 | 95.17 | −4.18 | - | - |
| 9 | 230.28 | - | 3.18 | 547.88 | 2 | 3.4 | 2.52 | −4.13 | −0.08 | 4 | −0.8 | 95.43 | −5.93 | −1 | - |
| 9 | 230.28 | - | 3.09 | 547.44 | 2 | 3.4 | 2.52 | −4.12 | −0.08 | 4 | −0.8 | 95.42 | −5.92 | −1 | - |
| 10 | 214.28 | 1 | 2.07 | 536.85 | 1 | 1.7 | 3.7 | −4.76 | 0.3 | 4 | −0.22 | 100 | −5.9 | - | - |
| 11 | 256.32 | - | 3.01 | 596.59 | - | 2.7 | 3.92 | −5.33 | 0.4 | 4 | −0.73 | 100 | −6.01 | −1 | 1 |
| 11 | 256.32 | - | 3.22 | 600.04 | - | 2.7 | 3.91 | −5.41 | 0.4 | 4 | −0.77 | 100 | −6.07 | −1 | 1 |
| 12 | 272.32 | - | 4.71 | 639.64 | 1 | 3.7 | 3.61 | −5.81 | 0.34 | 3 | −1.03 | 100 | −6.48 | −2 | 1 |
| 12 | 272.32 | - | 4.76 | 640.28 | 1 | 3.7 | 3.61 | −5.82 | 0.34 | 3 | −1.03 | 100 | −6.49 | −2 | 1 |
| 13 | 286.35 | - | 4.06 | 664.62 | - | 3.7 | 4.35 | −6.09 | 0.44 | 4 | −0.45 | 100 | −6.48 | - | 1 |
| 13 | 286.35 | - | 4.29 | 664.92 | - | 3.7 | 4.36 | −6.11 | 0.44 | 4 | −0.44 | 100 | −6.46 | - | 1 |
| 14 | 290.76 | 1 | 5.18 | 641.98 | - | 2 | 5.2 | −7.01 | 0.83 | 3 | −0.19 | 100 | −6.36 | - | 2 |
| 14 | 290.76 | 1 | 5.25 | 642.37 | - | 2 | 5.2 | −7.01 | 0.83 | 3 | −0.18 | 100 | −6.36 | - | 2 |
| 15 | 248.73 | 1 | 5.08 | 543.19 | 1 | 1.7 | 4.16 | −5.53 | 0.46 | 3 | 0.24 | 100 | −5.84 | 1 | - |
| 16 | 290.76 | 1 | 3.84 | 623.15 | - | 2 | 5.14 | −6.93 | 0.85 | 3 | 0.1 | 100 | −6.22 | 1 | 1 |
| 17 | 398.52 | 2 | 2.11 | 825.87 | - | 4 | 6.39 | −8.4 | 1.32 | 5 | −0.97 | 100 | −6.6 | −1 | 2 |
| 18 | 230.28 | - | 3.09 | 548.17 | 2 | 3.4 | 2.52 | −4.13 | −0.08 | 4 | −0.8 | 95.44 | −5.93 | −1 | - |
| 18 | 230.28 | - | 3.07 | 548.16 | 2 | 3.4 | 2.52 | −4.13 | −0.08 | 4 | −0.8 | 95.44 | −5.93 | −1 | - |
| 19 | 232.3 | - | 3.57 | 540.4 | 2 | 3.4 | 2.59 | −3.96 | −0.05 | 4 | −0.68 | 100 | −5.63 | - | - |
| 19 | 232.3 | - | 3.49 | 538.9 | 2 | 3.4 | 2.64 | −3.94 | −0.06 | 4 | −0.61 | 100 | −5.6 | - | - |
| 20 | 232.3 | - | 3.7 | 536.62 | 2 | 3.4 | 2.57 | −3.9 | −0.06 | 4 | −0.67 | 100 | −5.55 | - | - |
| 20 | 232.3 | - | 1.64 | 534.91 | 2 | 3.4 | 2.62 | −3.87 | −0.06 | 4 | −0.6 | 100 | −5.5 | - | - |
| 21 | 244.31 | - | 2.9 | 571.71 | 1 | 3.4 | 3.53 | −4.67 | 0.15 | 5 | −0.27 | 100 | −5.9 | - | - |
| 21 | 244.31 | - | 2.85 | 572.08 | 1 | 3.4 | 3.53 | −4.68 | 0.15 | 5 | −0.27 | 100 | −5.91 | - | - |
| 22 | 256.32 | - | 3.42 | 600.19 | - | 2.7 | 3.91 | −5.41 | 0.4 | 4 | −0.77 | 100 | −6.08 | −1 | 1 |
| 22 | 256.32 | - | 3.45 | 600.15 | - | 2.7 | 3.91 | −5.41 | 0.4 | 4 | −0.77 | 100 | −6.08 | −1 | 1 |
| 23 | 230.28 | - | 3.18 | 547.88 | 2 | 3.4 | 2.52 | −4.13 | −0.08 | 4 | −0.8 | 95.43 | −5.93 | −1 | - |
| 24 | 272.32 | - | 4.71 | 639.63 | 1 | 3.7 | 3.61 | −5.81 | 0.34 | 3 | −1.03 | 100 | −6.48 | −2 | 1 |
| 25 | 258.33 | - | 2.66 | 619.03 | 1 | 3.4 | 4 | −5.37 | 0.3 | 5 | −0.35 | 100 | −6.24 | - | - |
| 26 | 300.37 | 2 | 4.36 | 710.36 | - | 3.7 | 4.83 | −6.8 | 0.61 | 4 | −0.55 | 100 | −6.73 | - | 1 |
| 27 | 234.31 | - | 3.3 | 548.83 | - | 2.7 | 3.45 | −4.13 | 0.11 | 5 | −0.82 | 100 | −5.6 | −1 | 1 |
| 28 | 234.31 | 1 | 6.27 | 543.84 | - | 2.7 | 3.44 | −4.03 | 0.09 | 5 | −0.77 | 100 | −5.53 | −1 | 1 |
| 29 | 414.47 | 2 | 4.68 | 754.8 | 6 | 13.6 | -0.11 | −2.92 | −1.06 | 8 | −3.29 | 40.05 | −5.89 | −2 | 1 |
| 29 | 414.47 | 2 | 4.64 | 734.02 | 6 | 13.6 | -0.19 | −2.69 | −1.1 | 8 | −3.13 | 40.7 | −5.7 | −2 | 1 |
| 29 | 414.47 | 2 | 5.17 | 755.35 | 6 | 13.6 | -0.19 | −2.92 | −1.08 | 8 | −3.35 | 39.14 | −5.94 | −2 | 1 |
| 29 | 414.47 | 2 | 5.73 | 728.91 | 6 | 13.6 | -0.23 | −2.61 | −1.08 | 8 | −3.16 | 39.41 | −5.63 | −2 | 1 |
| 30 | 386.42 | - | 2.77 | 688.65 | 6 | 13.6 | -0.66 | −2.47 | −1.18 | 7 | −2.88 | 39.15 | −5.64 | −2 | 1 |
| 30 | 386.42 | - | 3.22 | 682.74 | 6 | 13.6 | -0.75 | −2.36 | −1.15 | 7 | −2.99 | 36.13 | −5.48 | −2 | 1 |
| 30 | 386.42 | 1 | 4.11 | 714.6 | 6 | 13.6 | -0.74 | −2.75 | −1.18 | 7 | −3.27 | 34.82 | −5.96 | −2 | 1 |
| 30 | 386.42 | 1 | 1.2 | 710.72 | 6 | 13.6 | -0.73 | −2.7 | −1.18 | 7 | −3.22 | 35.33 | −5.91 | −2 | 1 |
| 31 | 232.3 | - | 2.7 | 541.33 | 2 | 3.4 | 2.43 | −3.82 | −0.08 | 4 | −0.79 | 95.57 | −5.54 | −1 | - |
| 31 | 232.3 | - | 2.84 | 542.23 | 2 | 3.4 | 2.43 | −3.84 | −0.08 | 4 | −0.8 | 95.44 | −5.56 | −1 | - |
| 32 | 264.36 | - | 1.82 | 533.75 | 1 | 3.4 | 3.52 | −4.3 | 0.12 | 5 | −0.08 | 100 | −5.49 | - | - |
| 32 | 264.36 | - | 1.93 | 538.34 | 1 | 3.4 | 3.54 | −4.4 | 0.13 | 5 | −0.08 | 100 | −5.56 | - | - |
| 33 | 250.33 | - | 2.38 | 509.39 | 2 | 3.4 | 2.66 | −3.76 | −0.09 | 4 | −0.56 | 96.23 | −5.48 | - | - |
| 33 | 250.33 | - | 2.37 | 509.81 | 2 | 3.4 | 2.66 | −3.77 | −0.09 | 4 | −0.56 | 96.24 | −5.49 | - | - |
| 34 | 208.29 | - | 4.09 | 422.53 | - | 2 | 3.01 | −3.34 | 0.01 | 2 | 0.23 | 100 | −4.42 | 1 | - |
| 35 | 194.27 | - | 4.61 | 394.08 | - | 2 | 2.75 | −2.53 | −0.14 | 2 | 0.03 | 100 | −4.24 | 1 | - |
| 36 | 224.29 | - | 3.29 | 451.72 | - | 2 | 3.15 | −3.89 | 0.14 | 2 | 0.14 | 100 | −4.8 | 1 | - |
| 37 | 216.32 | 5 | 0.44 | 474.39 | - | - | 5.8 | −5.65 | 0.72 | 2 | 0.35 | 100 | −5.56 | 1 | - |
| 38 | 318.45 | 1 | 3.17 | 674.19 | - | 2 | 5.85 | −7.32 | 1.08 | 4 | −0.2 | 100 | −6.41 | - | 1 |
| 39 | 432.63 | 9 | 0.65 | 807.6 | - | - | 11.73 | −12.03 | 2.41 | 6 | 0.5 | 100 | −8.14 | 2 | - |
| 39 | 432.63 | 8 | 0.93 | 700.3 | - | - | 9.78 | −11.17 | 2.17 | 6 | 0.52 | 100 | −6.51 | 2 | - |
| 40 | 432.63 | 9 | 0.65 | 807.6 | - | - | 11.73 | −12.03 | 2.41 | 6 | 0.5 | 100 | −8.14 | 2 | - |
| 40 | 432.63 | 8 | 0.93 | 700.3 | - | - | 9.78 | −11.17 | 2.17 | 6 | 0.52 | 100 | −6.51 | 2 | - |
| 41 | 334.45 | - | 4.91 | 630.13 | 1 | 3.7 | 4.38 | −5.51 | 0.58 | 4 | −0.77 | 100 | −5.67 | −1 | 1 |
| 41 | 334.45 | - | 4.93 | 626.38 | 1 | 3.7 | 4.38 | −5.47 | 0.58 | 4 | −0.74 | 100 | −5.58 | −1 | 1 |
| 42 | 334.45 | - | 4.82 | 652.88 | 1 | 3.7 | 4.57 | −5.96 | 0.64 | 5 | −0.76 | 100 | −5.95 | −1 | 1 |
| 42 | 334.45 | - | 4.4 | 673.42 | 1 | 3.7 | 4.8 | −6.38 | 0.68 | 5 | −0.64 | 100 | −6.24 | - | 1 |
| 43 | 234.33 | 1 | 2.01 | 498.72 | 1 | 1.7 | 3.71 | −4.39 | 0.27 | 4 | −0.03 | 100 | −5.45 | - | - |
| 44 | 276.37 | - | 2.76 | 592.07 | - | 2 | 4.28 | −6.08 | 0.67 | 3 | −0.21 | 100 | −6.15 | - | 1 |
| 45 | 210.27 | - | 3.5 | 401.73 | 1 | 2 | 2.72 | −3.04 | −0.23 | 2 | −0.22 | 85.2 | −2.48 | −1 | - |
| 46 | 388.97 | 2 | 4.59 | 617.73 | - | 2 | 6.5 | −6.17 | 1.21 | 8 | 0.19 | 100 | −4.68 | 1 | 2 |
| 46 | 388.97 | 1 | 6.57 | 609.65 | - | 2 | 6.37 | −6.02 | 1.17 | 8 | 0.12 | 100 | −4.58 | 1 | 2 |
| 47 | 252.35 | - | 4.52 | 495.92 | 1 | 3.7 | 2.67 | −3.53 | −0.07 | 4 | −0.5 | 96.52 | −4.88 | - | - |
| 48 | 278.38 | - | 2.94 | 560.83 | 1 | 3.4 | 3.75 | −4.74 | 0.27 | 6 | −0.12 | 100 | −5.36 | - | - |
| 49 | 264.36 | - | 2.49 | 532.94 | 2 | 3.4 | 2.98 | −4.12 | 0.04 | 5 | −0.44 | 100 | −5.28 | - | - |
| 50 | 246.34 | - | 5.04 | 515.59 | - | 2 | 3.88 | −4.92 | 0.41 | 3 | 0.1 | 100 | −5.25 | 1 | - |
| 51 | 248.31 | - | 4.21 | 494.76 | 1 | 2 | 3.56 | −4.39 | 0.13 | 3 | −0.42 | 90.16 | −3.34 | −1 | - |
| 52 | 232.32 | - | 5.54 | 487.09 | - | 2 | 3.35 | −4.29 | 0.26 | 3 | −0.13 | 100 | −5.11 | - | - |
| 53 | 234.33 | 1 | 1.8 | 496.6 | 1 | 1.7 | 3.64 | −4.49 | 0.3 | 4 | 0.05 | 100 | −5.21 | 1 | - |
| 54 | 248.31 | - | 4.21 | 495.1 | 1 | 2 | 3.57 | −4.39 | 0.13 | 3 | −0.42 | 90.17 | −3.35 | −1 | - |
| 55 | 262.34 | - | 4.22 | 544.7 | - | 2 | 4.12 | −5.47 | 0.54 | 3 | 0 | 100 | −5.55 | - | - |
| 56 | 304.42 | 4 | 1.89 | 591.79 | - | 1.5 | 5.59 | −6.89 | 1.06 | 4 | 0.15 | 100 | −5.48 | 1 | 1 |
| 56 | 304.42 | 4 | 1.89 | 591.47 | - | 1.5 | 5.58 | −6.9 | 1.07 | 4 | 0.15 | 100 | −5.46 | 1 | 1 |
| 57 | 278.34 | 1 | 2.8 | 547.2 | 2 | 5.4 | 1.7 | −3.68 | −0.27 | 4 | −1.47 | 77.15 | −5.44 | −2 | - |
| 57 | 278.34 | 1 | 3.43 | 547.55 | 2 | 5.4 | 1.7 | −3.69 | −0.27 | 4 | −1.47 | 77.15 | −5.45 | −2 | - |
| 58 | 266.37 | - | 4.99 | 531.75 | 1 | 3.7 | 2.98 | −4.13 | 0.1 | 5 | −0.56 | 100 | −4.91 | - | - |
| 59 | 294.34 | 1 | 1.92 | 555.27 | 3 | 5.4 | 1.93 | −3.98 | −0.45 | 4 | −1.78 | 62.81 | −3.68 | −2 | - |
| 59 | 294.34 | 1 | 2.76 | 555.53 | 3 | 5.4 | 1.93 | −3.98 | −0.45 | 4 | −1.78 | 62.81 | −3.68 | −2 | - |
| 60 | 248.36 | 1 | 2.19 | 534.06 | 1 | 1.7 | 4.03 | −5.03 | 0.44 | 5 | −0.06 | 100 | −5.44 | - | - |
| 61 | 298.37 | - | 5.85 | 542.89 | 3 | 7.1 | 1.07 | −2.98 | −0.5 | 5 | −1.4 | 74.44 | −4.8 | −2 | - |
| 61 | 298.37 | - | 4.88 | 542.6 | 3 | 7.1 | 1.06 | −2.98 | −0.5 | 5 | −1.4 | 74.36 | −4.79 | −2 | - |
| 62 | 366.92 | 3 | 3.08 | 712.65 | - | 2 | 6.67 | −8.41 | 1.37 | 5 | 0.21 | 100 | −6.17 | 1 | 1 |
| 63 | 432.59 | 4 | 5.23 | 855.53 | - | 4 | 7 | −9.13 | 1.52 | 6 | −0.86 | 100 | −6.63 | −1 | 2 |
| 64 | 282.8 | 1 | 5.1 | 546.49 | 1 | 1.7 | 4.64 | −5.5 | 0.57 | 4 | 0.34 | 100 | −5.39 | 1 | - |
| 65 | 348.47 | - | 4.77 | 685.58 | 1 | 3.7 | 4.9 | −6.42 | 0.76 | 5 | −0.85 | 100 | −5.81 | −1 | 1 |
| 66 | 348.47 | 1 | 5.79 | 721.29 | 1 | 3.7 | 5.08 | −7.07 | 0.82 | 6 | −0.93 | 100 | −6.37 | −1 | 1 |
| 67 | 678.93 | 10 | 5.34 | 1180.96 | - | 5.7 | 10.73 | −13.34 | 2.73 | 11 | −1.63 | 100 | −8.35 | −2 | 2 |
| 68 | 492.68 | 5 | 2.25 | 916.97 | - | 3.4 | 9.48 | −11.13 | 2.07 | 8 | −0.68 | 100 | −8.28 | - | 1 |
| 69 | 492.68 | 5 | 2.32 | 856.86 | - | 3.4 | 8.46 | −10.47 | 1.88 | 8 | −0.68 | 100 | −7.68 | - | 1 |
| 70 | 492.68 | 6 | 1.67 | 848.1 | - | 3.4 | 8.4 | −10.35 | 1.93 | 10 | −0.68 | 100 | −7.54 | - | - |
| 71 | 364.47 | - | 4.17 | 731.03 | 2 | 5.4 | 3.97 | −6.33 | 0.43 | 6 | −1.58 | 95.78 | −6.38 | −2 | 1 |
| 72 | 262.34 | 1 | 4.69 | 537.04 | 1 | 3.7 | 2.72 | −4.24 | 0.05 | 4 | −0.92 | 91.26 | −5.42 | −1 | - |
| 73 | 246.34 | 1 | 2.19 | 521.45 | 1 | 1.7 | 3.88 | −4.78 | 0.38 | 3 | −0.03 | 100 | −5.63 | - | - |
| 74 | 264.36 | - | 2.47 | 545.82 | 2 | 3.4 | 2.92 | −4.18 | 0.02 | 5 | −0.73 | 96.95 | −5.5 | −1 | - |
| 75 | 248.38 | 6 | 0.96 | 464.31 | - | - | 5.71 | −7.4 | 0.83 | 3 | 0.19 | 100 | −5.33 | 1 | - |
| 76 | 276.39 | 1 | 4.61 | 502.07 | - | 2 | 3.95 | −4.98 | 0.46 | 3 | 0.03 | 100 | −5.34 | 1 | - |
| 77 | 278.4 | 1 | 2.58 | 511.2 | 1 | 1.7 | 4.22 | −5.06 | 0.5 | 4 | 0.2 | 100 | −5.44 | 1 | - |
| 78 | 278.4 | 3 | 1.84 | 505.75 | - | 0.75 | 5.62 | −5.96 | 0.83 | 4 | 0.03 | 100 | −5.34 | 1 | - |
| 79 | 360.5 | 1 | 2.73 | 671.68 | - | 2 | 6.27 | −7.84 | 1.29 | 6 | 0.18 | 100 | −6.45 | 1 | 1 |
| 80 | 360.5 | 1 | 3.82 | 678.33 | - | 2 | 6.24 | −7.98 | 1.3 | 6 | 0.08 | 100 | −6.53 | 1 | 1 |
| 81 | 292.43 | 2 | 2.1 | 542.23 | - | 1.7 | 5.44 | −5.81 | 0.72 | 5 | −0.31 | 100 | −5.61 | - | - |
| 82 | 426.52 | - | 3.15 | 644.26 | 4 | 9.25 | 2.04 | −4.08 | −0.27 | 7 | −1.16 | 83.97 | −5.71 | −2 | 1 |
| 82 | 426.52 | - | 3.4 | 642.05 | 4 | 9.25 | 2.03 | −4.08 | −0.26 | 7 | −1.21 | 82.47 | −5.59 | −2 | 1 |
| 82 | 426.52 | - | 4.89 | 649.11 | 4 | 9.25 | 2.05 | −4.16 | −0.26 | 7 | −1.21 | 83.16 | −5.76 | −2 | 1 |
| 82 | 426.52 | - | 2.93 | 642.67 | 4 | 9.25 | 2.16 | −4.08 | −0.27 | 7 | −0.99 | 87.19 | −5.61 | −1 | 1 |
| 83 | 540.79 | 6 | 4.22 | 945.88 | - | 2 | 9.78 | −12.95 | 2.68 | 6 | −0.12 | 100 | −8.05 | - | - |
| 84 | 197.25 | - | 6.72 | 419.53 | 1 | 4.7 | 1.09 | −2.13 | −0.51 | 4 | −0.42 | 86.7 | −3.64 | - | - |
| 85 | 316.37 | 1 | 3.62 | 593.36 | 2 | 5.45 | 2.42 | −4.27 | 0.05 | 6 | −1.29 | 85.52 | −5.02 | −2 | 1 |
| 86 | 280.38 | - | 4.82 | 549.83 | 1 | 5.4 | 2.64 | −3.42 | −0.09 | 5 | −0.73 | 95.86 | −4.44 | −1 | - |
| 86 | 280.38 | - | 6.54 | 568.11 | 1 | 4.45 | 3.19 | −4.05 | 0.12 | 4 | −0.68 | 100 | −4.85 | - | - |
| 86 | 280.38 | - | 6.55 | 555.35 | 1 | 4.45 | 3.06 | −3.82 | 0.08 | 4 | −0.71 | 100 | −4.68 | −1 | - |
| 86 | 280.38 | - | 5.66 | 564.44 | 1 | 4.45 | 3.08 | −4.01 | 0.09 | 4 | −0.75 | 100 | −4.83 | −1 | - |
| 86 | 280.38 | - | 4.24 | 564.71 | 1 | 4.45 | 3.08 | −3.98 | 0.11 | 4 | −0.79 | 100 | −4.78 | −1 | - |
| 87 | 350.41 | - | 9.97 | 600.63 | 1.25 | 6.25 | 3.04 | −4.55 | −0.14 | 3 | −1.21 | 76.31 | −3.41 | −2 | 1 |
| 87 | 350.41 | - | 3.36 | 593.33 | 1.25 | 6.25 | 2.95 | −4.37 | −0.14 | 3 | −1.23 | 75.36 | −3.25 | −2 | 1 |
| 87 | 350.41 | - | 7.76 | 582.94 | 1.25 | 6.25 | 2.99 | −4.17 | −0.17 | 3 | −1.04 | 78.73 | −3.14 | −2 | 1 |
| 87 | 350.41 | - | 8.61 | 596.55 | 1.25 | 6.25 | 3.04 | −4.47 | −0.14 | 3 | −1.18 | 76.64 | −3.33 | −2 | 1 |
| 88 | 432.58 | 2 | 2.52 | 761.06 | 1 | 3.75 | 6.09 | −7.38 | 1.59 | 3 | 0.48 | 100 | −6.96 | 2 | - |
| 88 | 432.58 | 2 | 2.52 | 761.06 | 1 | 3.75 | 6.09 | −7.38 | 1.59 | 3 | 0.48 | 100 | −6.96 | 2 | - |
| 89 | 332.41 | - | 3.73 | 648.07 | - | 4.75 | 4.13 | −5.57 | 0.45 | 3 | −0.34 | 100 | −5.16 | - | - |
| 90 | 376.42 | - | 5.82 | 693.39 | 1 | 6.75 | 3.51 | −5.75 | 0.2 | 3 | −1.6 | 76.93 | −3.34 | −2 | - |
| 91 | 348.41 | - | 2.67 | 673.35 | 1 | 6.45 | 3.34 | −5.55 | 0.28 | 3 | −0.88 | 100 | −5.43 | −1 | - |
| 92 | 348.41 | - | 5.79 | 664.93 | 1 | 6.45 | 3.2 | −5.39 | 0.28 | 3 | −1.03 | 94.16 | −5.22 | −2 | - |
| 93 | 350.43 | - | 2.82 | 679.38 | 1 | 6.45 | 3.41 | −5.5 | 0.28 | 4 | −0.95 | 100 | −5.29 | −1 | - |
| 94 | 350.43 | - | 4.86 | 665.38 | 1 | 6.45 | 3.35 | −5.25 | 0.26 | 4 | −0.91 | 100 | −5.09 | −1 | - |
| 95 | 234.31 | - | 6.4 | 449.04 | - | 3.7 | 2.41 | −2.95 | −0.18 | 5 | 0.28 | 100 | −3.85 | 1 | - |
| 95 | 234.31 | - | 6.3 | 424.55 | - | 2.75 | 2.8 | −3 | 0.03 | 3 | 0.3 | 100 | −3.42 | 1 | - |
| 95 | 234.31 | - | 6.52 | 444.21 | - | 2.75 | 2.96 | −3.37 | 0.1 | 3 | 0.26 | 100 | −3.77 | 1 | - |
| 95 | 234.31 | - | 6.52 | 444.21 | - | 2.75 | 2.96 | −3.37 | 0.1 | 3 | 0.26 | 100 | −3.77 | 1 | - |
| 95 | 234.31 | - | 6.3 | 424.54 | - | 2.75 | 2.8 | −3 | 0.03 | 3 | 0.3 | 100 | −3.42 | 1 | - |
| 96 | 294.37 | - | 6.75 | 555.68 | 1 | 5.7 | 2.37 | −3.57 | −0.1 | 5 | −0.9 | 90.35 | −4.57 | −1 | 1 |
| 96 | 294.37 | - | 4.21 | 564.47 | 1 | 6.2 | 2.14 | −3.54 | −0.17 | 4 | −1.09 | 85.98 | −4.81 | −2 | 1 |
| 96 | 294.37 | - | 8.99 | 556.37 | 1 | 6.2 | 2.17 | −3.43 | −0.18 | 4 | −0.96 | 87.87 | −4.63 | −1 | 1 |
| 96 | 294.37 | - | 4.8 | 551.9 | 1 | 6.2 | 2.08 | −3.35 | −0.2 | 4 | −1.03 | 86.02 | −4.63 | −2 | 1 |
| 96 | 294.37 | - | 7.82 | 549 | 1 | 6.2 | 2.16 | −3.29 | −0.22 | 4 | −0.88 | 89.33 | −4.65 | −1 | 1 |
| Compd | Docking Score | XP GScore | Glide GScore | Glide Emodel |
|---|---|---|---|---|
| 29 | −9.439 | −9.439 | −9.439 | −62.9 |
| 30 | −9.178 | −9.178 | −9.178 | −61.357 |
| 82 | −8.09 | −8.09 | −8.09 | −64.014 |
| 42 | −6.971 | −6.971 | −6.971 | −45.831 |
| Ref_4OD9 | −6.895 | −7.306 | −7.306 | −71.557 |
| 66 | −6.889 | −6.889 | −6.889 | −56.795 |
| 64 | −6.772 | −6.772 | −6.772 | −47.815 |
| 61 | −6.677 | −6.677 | −6.677 | −45.925 |
| 39 | −6.567 | −6.567 | −6.567 | −68.455 |
| 40 | −6.567 | −6.567 | −6.567 | −68.455 |
| 57 | −6.484 | −6.484 | −6.484 | −44.895 |
| 33 | −6.478 | −6.478 | −6.478 | −37.519 |
| 46 | −6.385 | −6.385 | −6.385 | −54.121 |
| 19 | −6.374 | −6.374 | −6.374 | −39.754 |
| 81 | −6.373 | −6.373 | −6.373 | −40.88 |
| 92 | −6.297 | −6.297 | −6.297 | −50.1 |
| 9 | −6.119 | −6.119 | −6.119 | −37.184 |
| 52 | −6.056 | −6.056 | −6.056 | −39.105 |
| 2 | −5.946 | −5.946 | −5.946 | −35.558 |
| 89 | −5.913 | −5.913 | −5.913 | −41.976 |
| 68 | −5.91 | −5.91 | −5.91 | −58.146 |
| 59 | −5.879 | −5.879 | −5.879 | −43.627 |
| 58 | −5.789 | −5.789 | −5.789 | −39.479 |
| 41 | −5.775 | −5.778 | −5.778 | −50.687 |
| 84 | −5.761 | −5.761 | −5.761 | −35.037 |
| 31 | −5.624 | −5.624 | −5.624 | −39.484 |
| 17 | −5.589 | −5.589 | −5.589 | −58.566 |
| 91 | −5.563 | −5.563 | −5.563 | −39.688 |
| 65 | −5.534 | −5.534 | −5.534 | −52.028 |
| 74 | −5.522 | −5.522 | −5.522 | −50.032 |
| 49 | −5.439 | −5.439 | −5.439 | −37.501 |
| 53 | −5.431 | −5.431 | −5.431 | −42.062 |
| 60 | −5.413 | −5.413 | −5.413 | −48.483 |
| 32 | −5.359 | −5.359 | −5.359 | −36.918 |
| 37 | −5.344 | −5.344 | −5.344 | −38.255 |
| 95 | −5.323 | −5.733 | −5.733 | −31.554 |
| 12 | −5.317 | −5.32 | −5.32 | −48.638 |
| 93 | −5.294 | −5.294 | −5.294 | −41.169 |
| 71 | −5.234 | −5.234 | −5.234 | −56.25 |
| 77 | −5.182 | −5.182 | −5.182 | −47.296 |
| 80 | −5.175 | −5.175 | −5.175 | −52.89 |
| 20 | −5.032 | −5.032 | −5.032 | −38.495 |
| 47 | −4.988 | −4.988 | −4.988 | −41.313 |
| 48 | −4.959 | −4.959 | −4.959 | −45.598 |
| 79 | −4.959 | −4.959 | −4.959 | −53.558 |
| 87 | −4.892 | −4.892 | −4.892 | −42.972 |
| 70 | −4.788 | −4.788 | −4.788 | −66.838 |
| 16 | −4.761 | −4.761 | −4.761 | −39.898 |
| 22 | −4.708 | −4.708 | −4.708 | −43.587 |
| 76 | −4.681 | −4.681 | −4.681 | −42.185 |
| 50 | −4.616 | −4.616 | −4.616 | −42.266 |
| 62 | −4.615 | −4.615 | −4.615 | −53.205 |
| 38 | −4.587 | −4.587 | −4.587 | −44.277 |
| 21 | −4.479 | −4.479 | −4.479 | −34.302 |
| 96 | −4.434 | −4.844 | −4.844 | −43.679 |
| 27 | −4.427 | −4.427 | −4.427 | −32.461 |
| 54 | −4.4 | −4.4 | −4.4 | −42.328 |
| 36 | −4.372 | −4.372 | −4.372 | −31.206 |
| 34 | −4.367 | −4.367 | −4.367 | −32.958 |
| 10 | −4.36 | −4.36 | −4.36 | −37.183 |
| 4 | −4.325 | −4.325 | −4.325 | −44.054 |
| 55 | −4.289 | −4.289 | −4.289 | −41.301 |
| 86 | −4.263 | −4.673 | −4.673 | −34.699 |
| 67 | −4.242 | −4.242 | −4.242 | −71.461 |
| 72 | −4.221 | −4.221 | −4.221 | −45.333 |
| 56 | −4.166 | −4.166 | −4.166 | −45.866 |
| 18 | −4.159 | −4.159 | −4.159 | −38.902 |
| 28 | −4.131 | −4.131 | −4.131 | −38.886 |
| 51 | −4.127 | −4.127 | −4.127 | −38.518 |
| 73 | −4.109 | −4.109 | −4.109 | −41.377 |
| 88 | −4.09 | −4.09 | −4.09 | −46.666 |
| 94 | −4.085 | −4.085 | −4.085 | −40.186 |
| 35 | −4.079 | −4.079 | −4.079 | −30.767 |
| 44 | −4.047 | −4.047 | −4.047 | −41.625 |
| 75 | −4.012 | −4.012 | −4.012 | −39.751 |
| 69 | −3.964 | −3.964 | −3.964 | −65.916 |
| 85 | −3.899 | −3.899 | −3.899 | −35.518 |
| 83 | −3.778 | −3.778 | −3.778 | −63.675 |
| 13 | −3.769 | −3.769 | −3.769 | −38.792 |
| 9 | −3.688 | −3.688 | −3.688 | −41.071 |
| 23 | −3.688 | −3.688 | −3.688 | −41.071 |
| 14 | −3.65 | −3.65 | −3.65 | −38.638 |
| 11 | −3.64 | −3.64 | −3.64 | −42.16 |
| 43 | −3.528 | −3.528 | −3.528 | −37.644 |
| 25 | −3.481 | −3.481 | −3.481 | −39.665 |
| 24 | −3.48 | −3.483 | −3.483 | −44.761 |
| 78 | −3.406 | −3.406 | −3.406 | −37.385 |
| 45 | −3.403 | −3.403 | −3.403 | −30.006 |
| 3 | −3.303 | −3.303 | −3.303 | −26.546 |
| 15 | −3.253 | −3.253 | −3.253 | −35.921 |
| 90 | −3.239 | −3.239 | −3.239 | −43.733 |
| 26 | −3.185 | −3.185 | −3.185 | −40.847 |
| 8 | −2.058 | −2.117 | −2.117 | −23.999 |
| 63 | −1.85 | −1.85 | −1.85 | −70.342 |
| 7 | −1.55 | −1.55 | −1.55 | −18.635 |
| 5 | −0.841 | −0.841 | −0.841 | −10.861 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, S.R.M.; Omar, A.M.; Bagalagel, A.A.; Diri, R.M.; Noor, A.O.; Almasri, D.M.; Mohamed, S.G.A.; Mohamed, G.A. Thiophenes—Naturally Occurring Plant Metabolites: Biological Activities and In Silico Evaluation of Their Potential as Cathepsin D Inhibitors. Plants 2022, 11, 539. https://doi.org/10.3390/plants11040539
Ibrahim SRM, Omar AM, Bagalagel AA, Diri RM, Noor AO, Almasri DM, Mohamed SGA, Mohamed GA. Thiophenes—Naturally Occurring Plant Metabolites: Biological Activities and In Silico Evaluation of Their Potential as Cathepsin D Inhibitors. Plants. 2022; 11(4):539. https://doi.org/10.3390/plants11040539
Chicago/Turabian StyleIbrahim, Sabrin R. M., Abdelsattar M. Omar, Alaa A. Bagalagel, Reem M. Diri, Ahmad O. Noor, Diena M. Almasri, Shaimaa G. A. Mohamed, and Gamal A. Mohamed. 2022. "Thiophenes—Naturally Occurring Plant Metabolites: Biological Activities and In Silico Evaluation of Their Potential as Cathepsin D Inhibitors" Plants 11, no. 4: 539. https://doi.org/10.3390/plants11040539
APA StyleIbrahim, S. R. M., Omar, A. M., Bagalagel, A. A., Diri, R. M., Noor, A. O., Almasri, D. M., Mohamed, S. G. A., & Mohamed, G. A. (2022). Thiophenes—Naturally Occurring Plant Metabolites: Biological Activities and In Silico Evaluation of Their Potential as Cathepsin D Inhibitors. Plants, 11(4), 539. https://doi.org/10.3390/plants11040539