Quercus suber Roots Activate Antioxidant and Membrane Protective Processes in Response to High Salinity
Abstract
:1. Introduction
2. Results
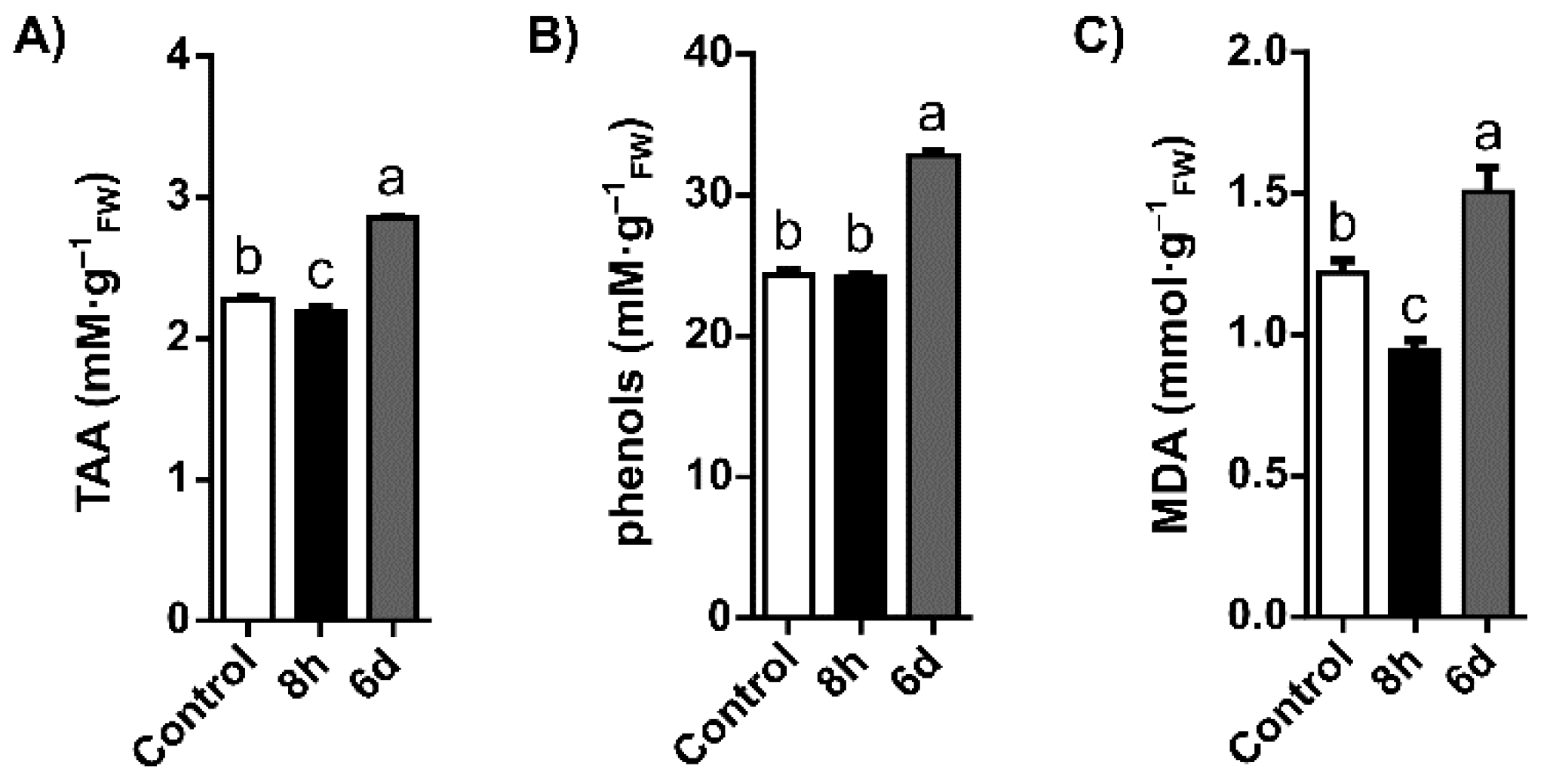
2.1. Oxidative Stress Status
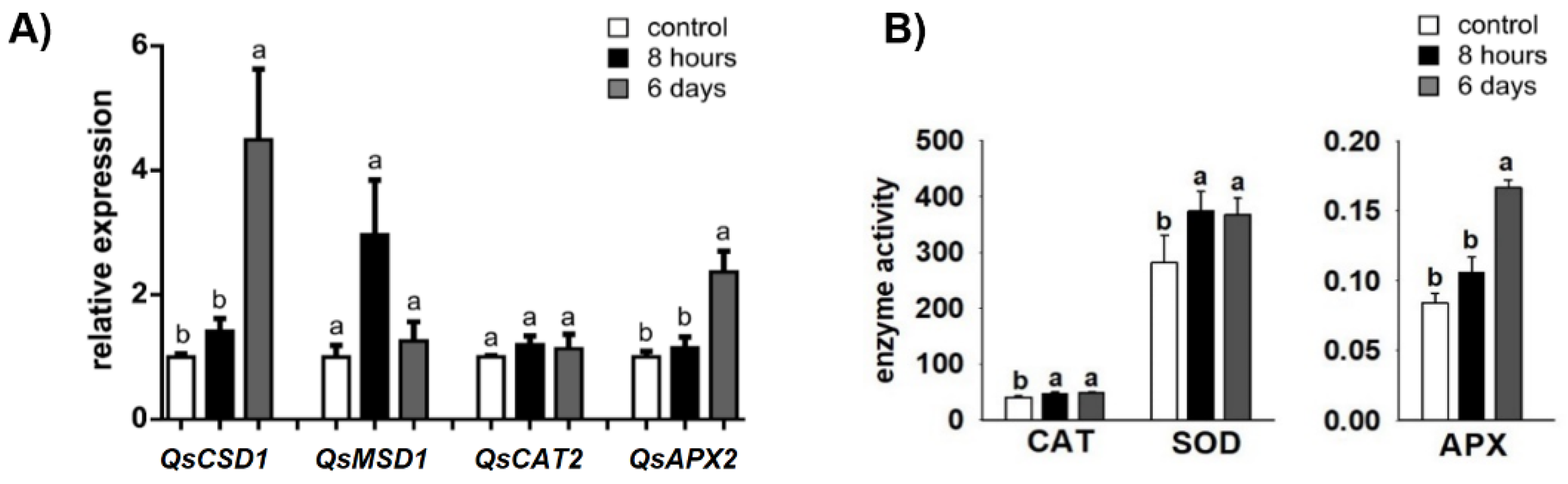
2.2. Antioxidant Enzyme-Encoding Genes and -Activities
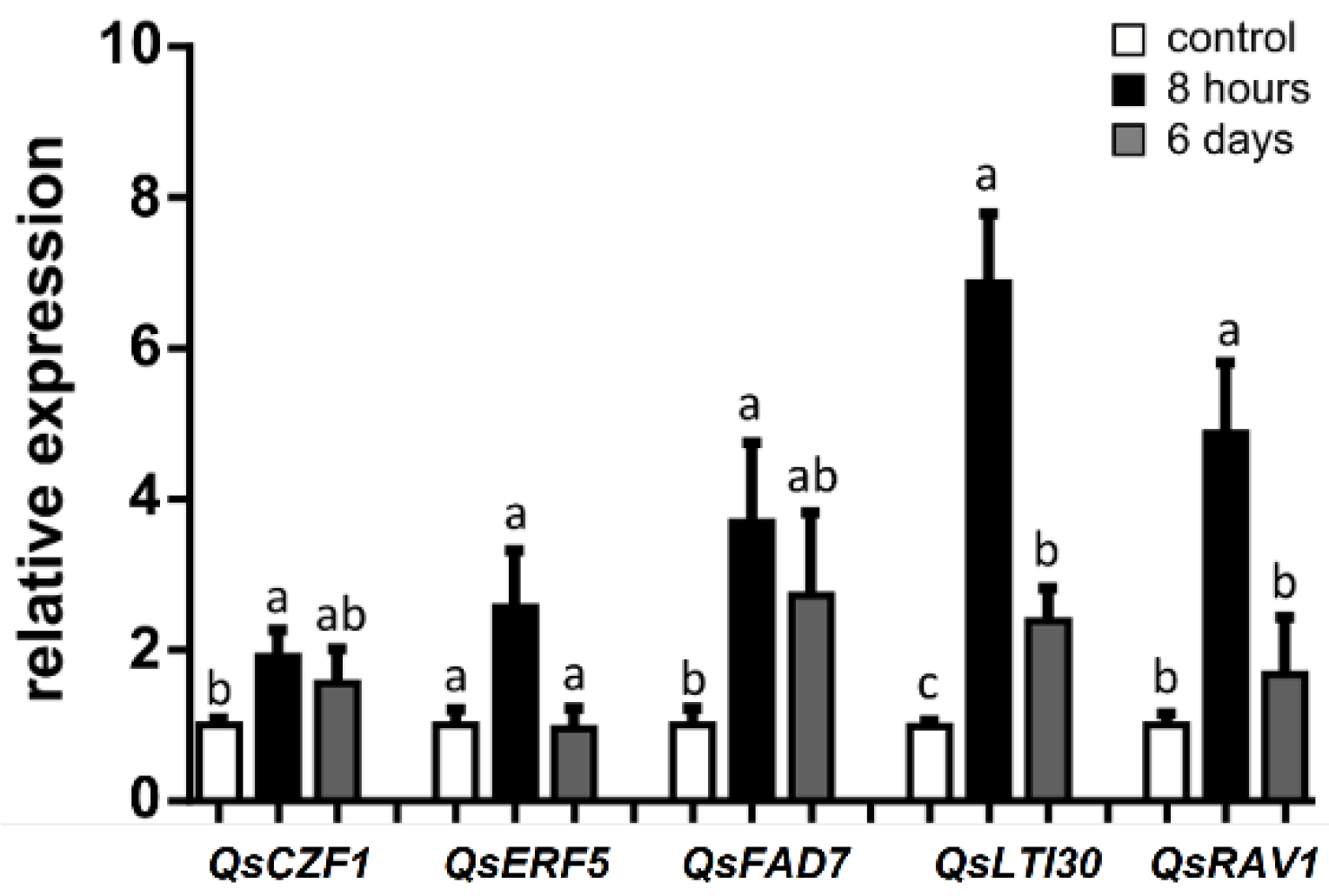
2.3. Relative Expression of Genes Related to Stress Response
2.4. Correlations and Principal Component Analysis
3. Discussion
4. Materials and Methods
4.1. Reagents and Standards
4.2. Plant Material and Experimental Design
4.3. Total Antioxidant Activity (TAA) and Phenol
4.4. Lipid Peroxidation
4.5. RNA Extraction, Primer Design and qPCR
4.6. Antioxidant Enzyme Activities
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Aragüés, R.; Urdanoz, V.; Çetin, M.; Kirda, C.; Daghari, H.; Ltifi, W.; Lahlou, M.; Douaik, A. Soil salinity related to physical soil characteristics and irrigation management in four Mediterranean irrigation districts. Agric. Water Manag. 2011, 98, 959–966. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0378377411000072 (accessed on 6 January 2022). [CrossRef] [Green Version]
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dias, M.C.; Freitas, H. Drought and Salinity Stress Responses and Microbe-Induced Tolerance in Plants. Front. Plant Sci. 2020, 11, 591911. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, Q.; Li, D.; Yang, X.; Li, D. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018. Available online: http://journal.frontiersin.org/article/10.3389/fpls.2017.01018/full (accessed on 6 January 2022). [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Smith, J.A.C.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane lipid remodeling in response to salinity. Int. J. Mol. Sci. 2019, 20, 4264. Available online: https://www.mdpi.com/1422-0067/20/17/4264 (accessed on 6 January 2022). [CrossRef] [Green Version]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Su, W.; Bao, Y.; Wang, S.; He, F.; Wang, D.; Yu, X.; Yin, W.; Liu, C.; Xia, X. Poplar PdPTP1 gene negatively regulates salt tolerance by affecting ion and ROS homeostasis in Populus. Int. J. Mol. Sci. 2020, 21, 1065. Available online: https://www.mdpi.com/1422-0067/21/3/1065 (accessed on 6 January 2022). [CrossRef] [Green Version]
- Kimotho, R.N.; Baillo, E.H.; Zhang, Z. Transcription factors involved in abiotic stress responses in Maize (Zea mays L.) and their roles in enhanced productivity in the post genomics era. PeerJ 2019, 2019, e7211. Available online: https://peerj.com/articles/7211 (accessed on 6 January 2022). [CrossRef] [Green Version]
- Gahlaut, V.; Jaiswal, V.; Kumar, A.; Gupta, P.K. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 2019–2042. [Google Scholar] [CrossRef]
- Rashid, M.; Guangyuan, H.; Guangxiao, Y.; Hussain, J.; Xu, Y. AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol. Bioinforma 2012, 8, 321–355. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Al-Baidhani, H.H.J.; Harris, J.; Riboni, M.; Li, Y.; Mazonka, I.; Bazanova, N.; Chirkova, L.; Sarfraz Hussain, S.; Hrmova, M.; et al. DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: Impact on plant development, stress tolerance and yield. Plant Biotechnol. J. 2020, 18, 829–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.N.; Wang, F.Z.; Yang, C.H.; Yuan, J.Z.; Guo, H.; Zhang, J.L.; Wang, S.M.; Ma, Q. Transcriptomic profiling identifies candidate genes involved in the salt tolerance of the xerophyte Pugionium cornutum. Genes 2019, 10, 1039. Available online: https://www.mdpi.com/2073-4425/10/12/1039 (accessed on 6 January 2022). [CrossRef] [PubMed] [Green Version]
- Sun, J.; Jiang, H.; Xu, Y.; Li, H.; Wu, X.; Xie, Q.; Li, C. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007, 48, 1148–1158. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Pereira-Leal, J.B.; Abreu, I.A.; Alabaça, C.S.; Almeida, M.H.; Almeida, P.; Almeida, T.; Amorim, M.I.; Araújo, S.; Azevedo, H.; Badia, A.; et al. A comprehensive assessment of the transcriptome of Cork oak (Quercus suber) through EST sequencing. BMC Genom. 2014, 15, 371. Available online: http://www.fagaceae (accessed on 6 January 2022). [CrossRef] [Green Version]
- Kim, H.N.; Jin, H.Y.; Kwak, M.J.; Khaine, I.; You, H.N.; Lee, T.Y.; Ahn, T.H.; Woo, S.Y. Why does Quercus suber species decline in Mediterranean areas? J. Asia-Pac. Biodivers. 2017, 10, 337–341. [Google Scholar] [CrossRef]
- Faivre Rampant, P.; Lesur, I.; Boussardon, C.; Bitton, F.; Martin-Magniette, M.L.; Bodénès, C.; Le Provost, G.; Bergès, H.; Fluch, S.; Kremer, A.; et al. Analysis of BAC end sequences in oak, a keystone forest tree species, providing insight into the composition of its genome. BMC Genom. 2011, 12, 292. [Google Scholar] [CrossRef] [Green Version]
- Miguel, A.; de Vega-Bartol, J.; Marum, L.; Chaves, I.; Santo, T.; Leitão, J.; Varela, M.C.; Miguel, C.M. Characterization of the cork oak transcriptome dynamics during acorn development. BMC Plant Biol. 2015, 15, 158. [Google Scholar] [CrossRef] [Green Version]
- Cha-um, S.; Kirdmanee, C. Effects of water stress induced by sodium chloride and mannitol on proline accumulation, photosynthetic abilities and growth characters of eucalyptus (Eucalyptus camaldulensis Dehnh.). New For. 2010, 40, 349–360. [Google Scholar] [CrossRef]
- Hernández, J.A. Salinity tolerance in plants: Trends and perspectives. Int. J. Mol. Sci. 2019, 20, 2408. [Google Scholar] [CrossRef] [Green Version]
- Sedas, A.; Gonzalez, Y.; Winter, K.; Lopez, O.R. Seedling responses to salinity of 26 Neotropical tree species. AoB Plants 2019, 11, plz062. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhou, Z.; Chen, P.; Tang, X.; Shao, H.; Wang, H. Comparative ecophysiological study of salt stress for wild and cultivated soybean species from the yellow river delta, China. Sci. World J. 2014, 2014, 651745. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Rahman, A.; Anee, T.I.; Alam, M.U.; Bhuiyan, T.F.; Oku, H.; Fujita, M. Approaches to enhance salt stress tolerance in wheat. In Wheat Improvement, Management and Utilization; Wanyera, R., Owuoche, J., Eds.; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Rahman, A.; Nahar, K.; Mahmud, J.A.; Hasanuzzaman, M.; Hossain, M.S.; Fujita, M. Salt stress tolerance in rice: Emerging role of exogenous phytoprotectants. In Advances in International Rice Research; Jinquan, L., Ed.; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Ferreira de Oliveira, J.M.P.; Santos, C.; Araújo, M.; Oliveira, M.M.; Dias, M.C. High-salinity activates photoprotective mechanisms in Quercus suber via accumulation of carbohydrates and involvement of non-enzymatic and enzymatic antioxidant pathways. New For. 2021. [Google Scholar] [CrossRef]
- Eriksson, S.K.; Kutzer, M.; Procek, J.; Gröbner, G.; Harryson, P. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 2011, 23, 2391–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puhakainen, T.; Hess, M.W.; Mäkelä, P.; Svensson, J.; Heino, P.; Palva, E.T. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 2004, 54, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chen, Y.; Qian, Y.; Chan, Z. Low Temperature-Induced 30 (LTI30) positively regulates drought stress resistance in Arabidopsis: Effect on abscisic acid sensitivity and hydrogen peroxide accumulation. Front. Plant Sci. 2015, 6, 893. [Google Scholar] [CrossRef] [Green Version]
- Nishiuchi, T.; Hamada, T.; Kodama, H.; Iba, K. Wounding changes the spatial expression pattern of the Arabidopsis plastid ω-3 fatty acid desaturase gene (FAD7) through different signal transduction pathways. Plant Cell 1997, 9, 1701–1712. [Google Scholar] [CrossRef]
- Teixeira, M.C.; Carvalho, I.S.; Brodelius, M. ω-3 Fatty acid desaturase genes isolated from purslane (Portulaca oleracea L.): Expression in different tissues and response to cold and wound stress. J. Agric. Food Chem. 2010, 58, 1870–1877. [Google Scholar] [CrossRef]
- Feng, J.; Dong, Y.; Liu, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Genome-wide identification of membrane-bound fatty acid desaturase genes in Gossypium hirsutum and their expressions during abiotic stress. Sci. Rep. 2017, 7, 45711. [Google Scholar] [CrossRef] [Green Version]
- Wickramanayake, J.S.; Goss, J.A.; Zou, M.; Goggin, F.L. Loss of Function of Fatty Acid Desaturase 7 in Tomato Enhances Photosynthetic Carbon Fixation Efficiency. Front. Plant Sci. 2020, 11, 932. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magalhães, A.P.; Verde, N.; Reis, F.; Martins, I.; Costa, D.; Lino-Neto, T.; Castro, P.H.; Tavares, R.M.; Azevedo, H. RNA-Seq and gene network analysis uncover activation of an ABA-dependent signalosome during the cork oak root response to drought. Front. Plant Sci. 2016, 6, 1195. Available online: http://journal.frontiersin.org/Article/10.3389/fpls.2015.01195/abstract (accessed on 6 January 2022). [CrossRef] [PubMed] [Green Version]
- Min, Z.; Yanming, F.; Yonghua, J.; Zeping, J.; Lei, W. Effects of salt stress on ion content, antioxidant enzymes and protein profile in different tissues of Broussonetia papyrifera. S. Afr. J. Bot. 2013, 85, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Mittova, V.; Guy, M.; Tal, M.; Volokita, M. Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J. Exp. Bot. 2004, 55, 1105–1113. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lim, J.-H.; Park, M.R.; Kim, Y.J.; Park, T.I.; Seo, Y.W.; Choi, K.G.; Yun, S.J. Enhanced antioxidant enzymes are associated with reduced hydrogen peroxide in barley roots under saline stress. J. Biol. Mol. Biol. 2005, 38, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, E.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity stress alters the secondary metabolic profile of M. sativa, M. arborea and their hybrid (Alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef]
- Petridis, A.; Therios, I.; Samouris, G.; Tananaki, C. Salinity-induced changes in phenolic compounds in leaves and roots of four olive cultivars (Olea europaea L.) and their relationship to antioxidant activity. Env. Exp. Bot. 2012, 79, 37–43. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0891584998003153 (accessed on 6 January 2022). [CrossRef]
- Hodges, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Wan, C.Y.; Wilkins, T.A. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem. 1994, 223, 7–12. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0003269784715387 (accessed on 6 January 2022). [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Methods in Molecular Biology; Clifton, N.J., Ed.; Humana Press: Totova, NJ, USA, 2000; pp. 365–386. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, S.; Sairam, R.; Srivastava, G.; Meena, R. Changes in antioxidant enzymes activity and oxidative stress by abscisic acid and salicylic acid in wheat genotypes. Biol. Plant 2005, 49, 541–550. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Beers, R.; Sizer, I. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 1952, 195, 133–140. [Google Scholar] [CrossRef]

| Primers (5′—3′) | Target RNA Q. suber (NCBI ID) | A. thaliana Gene/Locus | Tblastx E-Value (Coverage) |
|---|---|---|---|
| F_CTGAGCGGGAAATTGTTCGTG; R_GCAGTCTCCAACTCCTGCTC | actin (XM_024037498.1) | ACT7 AT5G09810 | 0.0 (70%) |
| F_TACTGTCCCAGAGCTCACCC; R_CACGGAACATAGCTGAGGCA | tubulin (KJ563262.1) | TUB2 AT5G62690 | 0.0 (96%) |
| F_CGCTCTTGGAGACACAACAA; R_CCATCAGCACCAACATTGAC | superoxide dismutase [Cu-Zn] 4A (XM_024050079.1) | CSD1 AT1G08830 | 3 × 10−87 (59%) |
| F_CCTTCTCTCTTCCCGATCTCTC; R_GGAGTCGCCTTTAGCGATG | superoxide dismutase [Mn] (XM_024016741.1) | MSD1 AT3G10920 | 2 × 10−124 (54%) |
| F_GCTAGGGGAGCTAGTGCAAAG; R_CAGGGTTTCAGGGCTACCA | catalase isozyme 3-like (XM_024064618.1) | CAT2 AT4G35090 | 0.0 (81%) |
| F_CGGATCATCTGAGGGATGTATT; R_CAAAAATAAGAGGGTTGCTGGTC | L-ascorbate peroxidase, cytosolic (XM_024061016.1) | APX2 AT3G09640 | 2 × 10−138 (61%) |
| F_GCTCCTTGAGGCATCACACT; R_AGGCGATGGAGTTGGTTCTG | Zn finger CCCH domain-containing protein 29-like (XM_024044287.1) | CZF1 AT2G40140 | 0.0 (58%) |
| F_GCCGTTATCTCCGCATCCTT; R_CCACCAGCACCATTAGCAGA | E-responsive TF ERF105-like (XM_024066622.1) | ERF5 AT5G47230 | 4 × 10−26 (24%) |
| F_GACCCACCTCATACAAGCCC; R_TTAGCCCCCAGTTCCTGTCT | omega-3 fatty acid desaturase (XM_024037902.1) | FAD7 AT3G11170 | 2 × 10−135 (38%) |
| F_ATATGGCAACCCAACCCACC; R_CATACCTTGGAGAGTGGCGG | dehydrin Xero 1-like (XM_024052398.1) | LTI30/XERO2 AT3G50970 | 2 × 10−13 (46%) |
| F_AAGCCGGTTCAACTTTCCCA; R_CGCGTGAACAGCTTTTTGAGA | AP2/ERF and B3 domain-containing TF RAV1-like (XM_024016479.1) | RAV1 AT1G13260 | 2 × 10−102 (66%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias, M.C.; Santos, C.; Araújo, M.; Barros, P.M.; Oliveira, M.; de Oliveira, J.M.P.F. Quercus suber Roots Activate Antioxidant and Membrane Protective Processes in Response to High Salinity. Plants 2022, 11, 557. https://doi.org/10.3390/plants11040557
Dias MC, Santos C, Araújo M, Barros PM, Oliveira M, de Oliveira JMPF. Quercus suber Roots Activate Antioxidant and Membrane Protective Processes in Response to High Salinity. Plants. 2022; 11(4):557. https://doi.org/10.3390/plants11040557
Chicago/Turabian StyleDias, Maria Celeste, Conceição Santos, Márcia Araújo, Pedro M. Barros, Margarida Oliveira, and José Miguel P. Ferreira de Oliveira. 2022. "Quercus suber Roots Activate Antioxidant and Membrane Protective Processes in Response to High Salinity" Plants 11, no. 4: 557. https://doi.org/10.3390/plants11040557
APA StyleDias, M. C., Santos, C., Araújo, M., Barros, P. M., Oliveira, M., & de Oliveira, J. M. P. F. (2022). Quercus suber Roots Activate Antioxidant and Membrane Protective Processes in Response to High Salinity. Plants, 11(4), 557. https://doi.org/10.3390/plants11040557









