Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsis thaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis
Abstract
:1. Introduction
2. Results
2.1. A Novel Proteomic Approach: 2Dd-CNBr
2.2. 2Dd -CNBr of Arabidopsis Seed Soluble Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Preparation of Soluble Proteins Extracts
4.3. In Gel Oxidation of Soluble Proteins and BSA with Hypochlorous Acid
4.4. D-Diagonal Electrophoresis, Gel Staining, and Image Analysis
4.5. Protein Identification by Mass Spectrometry
4.6. Bioinformatic Tools
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suzuki, Y.J.; Forman, H.J.; Sevanian, A. Oxidants as Stimulators of Signal Transduction. Free Radic. Biol. Med. 1997, 22, 269–285. [Google Scholar] [CrossRef]
- Nyström, T. Role of Oxidative Carbonylation in Protein Quality Control and Senescence. EMBO J. 2005, 24, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and Redox Signalling in the Response of Plants to Abiotic Stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Oxidant and Antioxidant Signalling in Plants: A Re-Evaluation of the Concept of Oxidative Stress in a Physiological Context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive Oxygen Gene Network of Plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Lovigny, Y.; Groot, S.P.C.; Belghazi, M.; Job, C.; Job, D. Proteome-Wide Characterization of Seed Aging in Arabidopsis: A Comparison between Artificial and Natural Aging Protocols. Plant Physiol. 2008, 148, 620–641. [Google Scholar] [CrossRef] [Green Version]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed Germination and Vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajjou, L.; Gallardo, K.; Debeaujon, I.; Vandekerckhove, J.; Job, C.; Job, D. The Effect of Alpha-Amanitin on the Arabidopsis Seed Proteome Highlights the Distinct Roles of Stored and Neosynthesized mRNAs during Germination. Plant Physiol. 2004, 134, 1598–1613. [Google Scholar] [CrossRef] [Green Version]
- Galland, M.; Huguet, R.; Arc, E.; Cueff, G.; Job, D.; Rajjou, L. Dynamic Proteomics Emphasizes the Importance of Selective mRNA Translation and Protein Turnover during Arabidopsis Seed Germination. Mol. Cell. Proteom. 2014, 13, 252–268. [Google Scholar] [CrossRef] [Green Version]
- Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. From Intracellular Signaling Networks to Cell Death: The Dual Role of Reactive Oxygen Species in Seed Physiology. Comptes Rendus. Biol. 2008, 331, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. The Signalling Role of ROS in the Regulation of Seed Germination and Dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef]
- Davies, M. The Oxidative Environment and Protein Damage. Biochim. Biophys. Acta 2005, 1703, 93–109. [Google Scholar] [CrossRef]
- Rinalducci, S.; Murgiano, L.; Zolla, L. Redox Proteomics: Basic Principles and Future Perspectives for the Detection of Protein Oxidation in Plants. J. Exp. Bot. 2008, 59, 3781–3801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.; Weiss, S.J.; Levine, R.L. Methionine Oxidation and Reduction in Proteins. Biochim. Biophys. Acta BBA Gen. Subj. 2014, 1840, 901–905. [Google Scholar] [CrossRef] [Green Version]
- Moskovitz, J. Methionine Sulfoxide Reductases: Ubiquitous Enzymes Involved in Antioxidant Defense, Protein Regulation, and Prevention of Aging-Associated Diseases. Biochim. Biophys. Acta 2005, 1703, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Toennies, G.; Kolb, J.J. Methionine studies VI. dl-methionine sulfone. J. Biol. Chem. 1941, 140, 131–134. [Google Scholar] [CrossRef]
- Stadtman, E.R.; Levine, R.L. Free Radical-Mediated Oxidation of Free Amino Acids and Amino Acid Residues in Proteins. Amino Acids 2003, 25, 207–218. [Google Scholar] [CrossRef]
- Drazic, A.; Winter, J. The Physiological Role of Reversible Methionine Oxidation. Biochim. Biophys. Acta 2014, 1844, 1367–1382. [Google Scholar] [CrossRef] [PubMed]
- Holmström, K.M.; Finkel, T. Cellular Mechanisms and Physiological Consequences of Redox-Dependent Signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef]
- Levine, R.L.; Moskovitz, J.; Stadtman, E.R. Oxidation of Methionine in Proteins: Roles in Antioxidant Defense and Cellular Regulation. IUBMB Life 2000, 50, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Van Remmen, H.; Richardson, A.; Wehr, N.B.; Levine, R.L. Methionine Oxidation and Aging. Biochim. Biophys. Acta 2005, 1703, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Ugarte, N.; Petropoulos, I.; Friguet, B. Oxidized Mitochondrial Protein Degradation and Repair in Aging and Oxidative Stress. Antioxid. Redox Signal. 2010, 13, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Radak, Z.; Zhao, Z.; Goto, S.; Koltai, E. Age-Associated Neurodegeneration and Oxidative Damage to Lipids, Proteins and DNA. Mol. Aspects Med. 2011, 32, 305–315. [Google Scholar] [CrossRef]
- Oien, D.B.; Moskovitz, J. Substrates of the Methionine Sulfoxide Reductase System and Their Physiological Relevance. Curr. Top. Dev. Biol. 2008, 80, 93–133. [Google Scholar] [CrossRef]
- Levine, R.L.; Mosoni, L.; Berlett, B.S.; Stadtman, E.R. Methionine Residues as Endogenous Antioxidants in Proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 15036–15040. [Google Scholar] [CrossRef] [Green Version]
- Moskovitz, J.; Oien, D.B. Protein Carbonyl and the Methionine Sulfoxide Reductase System. Antioxid. Redox Signal. 2010, 12, 405–415. [Google Scholar] [CrossRef]
- Rey, P.; Tarrago, L. Physiological Roles of Plant Methionine Sulfoxide Reductases in Redox Homeostasis and Signaling. Antioxidants 2018, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Li, C.-W.; Lee, S.-H.; Chieh, P.-S.; Lin, C.-S.; Wang, Y.-C.; Chan, M.-T. Arabidopsis Root-Abundant Cytosolic Methionine Sulfoxide Reductase B Genes MsrB7 and MsrB8 Are Involved in Tolerance to Oxidative Stress. Plant Cell Physiol. 2012, 53, 1707–1719. [Google Scholar] [CrossRef] [Green Version]
- Châtelain, E.; Satour, P.; Laugier, E.; Ly Vu, B.; Payet, N.; Rey, P.; Montrichard, F. Evidence for Participation of the Methionine Sulfoxide Reductase Repair System in Plant Seed Longevity. Proc. Natl. Acad. Sci. USA 2013, 110, 3633–3638. [Google Scholar] [CrossRef] [Green Version]
- Tarrago, L.; Kieffer-Jaquinod, S.; Lamant, T.; Marcellin, M.N.; Garin, J.R.M.; Rouhier, N.; Rey, P. Affinity Chromatography: A Valuable Strategy to Isolate Substrates of Methionine Sulfoxide Reductases? Antioxid. Redox Signal. 2012, 16, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.L.; Burke, J. Methionyl Sulfoxide Content and Protein-Methionine-S-Oxide Reductase Activity in Response to Water Deficits or High Temperature. Physiol. Plant. 1994, 90, 253–258. [Google Scholar] [CrossRef]
- Romero, H.M.; Berlett, B.S.; Jensen, P.J.; Pell, E.J.; Tien, M. Investigations into the Role of the Plastidial Peptide Methionine Sulfoxide Reductase in Response to Oxidative Stress in Arabidopsis. Plant Physiol. 2004, 136, 3784–3794. [Google Scholar] [CrossRef] [Green Version]
- Laugier, E.; Tarrago, L.; Vieira Dos Santos, C.; Eymery, F.; Havaux, M.; Rey, P. Arabidopsis thaliana Plastidial Methionine Sulfoxide Reductases B, MSRBs, Account for Most Leaf Peptide MSR Activity and Are Essential for Growth under Environmental Constraints through a Role in the Preservation of Photosystem Antennae. Plant J. Cell Mol. Biol. 2010, 61, 271–282. [Google Scholar] [CrossRef]
- Wojciechowska, N.; Alipour, S.; Stolarska, E.; Bilska, K.; Rey, P.; Kalemba, E.M. Involvement of the MetO/Msr System in Two Acer Species That Display Contrasting Characteristics during Germination. Int. J. Mol. Sci. 2020, 21, 9197. [Google Scholar] [CrossRef]
- Bettinger, J.Q.; Welle, K.A.; Hryhorenko, J.R.; Ghaemmaghami, S. Quantitative Analysis of in Vivo Methionine Oxidation of the Human Proteome. J. Proteome Res. 2020, 19, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.J.; Bettinger, J.Q.; Welle, K.A.; Hryhorenko, J.R.; Ghaemmaghami, S. Global Analysis of Methionine Oxidation Provides a Census of Folding Stabilities for the Human Proteome. Proc. Natl. Acad. Sci. USA 2019, 116, 6081–6090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aledo, J.C.; Aledo, P. Susceptibility of Protein Methionine Oxidation in Response to Hydrogen Peroxide Treatment-Ex Vivo Versus In Vitro: A Computational Insight. Antioxidants 2020, 9, 987. [Google Scholar] [CrossRef]
- Veredas, F.J.; Cantón, F.R.; Aledo, J.C. Methionine Residues around Phosphorylation Sites Are Preferentially Oxidized in Vivo under Stress Conditions. Sci. Rep. 2017, 7, 40403. [Google Scholar] [CrossRef]
- Jacques, S.; Ghesquière, B.; De Bock, P.-J.; Demol, H.; Wahni, K.; Willems, P.; Messens, J.; Van Breusegem, F.; Gevaert, K. Protein Methionine Sulfoxide Dynamics in Arabidopsis thaliana under Oxidative Stress. Mol. Cell. Proteomics 2015, 14, 1217–1229. [Google Scholar] [CrossRef] [Green Version]
- Wojciechowska, N.; Bagniewska-Zadworna, A.; Minicka, J.; Michalak, K.M.; Kalemba, E.M. Localization and Dynamics of the Methionine Sulfoxide Reductases MsrB1 and MsrB2 in Beech Seeds. Int. J. Mol. Sci. 2021, 22, 402. [Google Scholar] [CrossRef] [PubMed]
- El-Maarouf-Bouteau, H.; Meimoun, P.; Job, C.; Job, D.; Bailly, C. Role of Protein and mRNA Oxidation in Seed Dormancy and Germination. Front. Plant Sci. 2013, 4, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sano, N.; Rajjou, L.; North, H.M.; Debeaujon, I.; Marion-Poll, A.; Seo, M. Staying Alive: Molecular Aspects of Seed Longevity. Plant Cell Physiol. 2016, 57, 660–674. [Google Scholar] [CrossRef] [Green Version]
- Dirk, L.M.A.; Downie, A.B. An Examination of Job’s Rule: Protection and Repair of the Proteins of the Translational Apparatus in Seeds. Seed Sci. Res. 2018, 28, 168–181. [Google Scholar] [CrossRef] [Green Version]
- Lonsdale-Eccles, J.D.; Lynley, A.M.; Dale, B.A. Cyanogen Bromide Cleavage of Proteins in Sodium Dodecyl Sulphate/Polyacrylamide Gels. Diagonal Peptide Mapping of Proteins from Epidermis. Biochem. J. 1981, 197, 591–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, E.; Witkop, B. Nonenzymatic Cleavage of Peptide Bonds: The Methionine Residues in Bovine Pancreatic Ribonuclease. J. Biol. Chem. 1962, 237, 1856–1860. [Google Scholar] [CrossRef]
- Gross, E. The Cyanogen Bromide Reaction. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1967; Volume 11, pp. 238–255. [Google Scholar]
- Andreev, Y.A.; Kozlov, S.A.; Vassilevski, A.A.; Grishin, E.V. Cyanogen Bromide Cleavage of Proteins in Salt and Buffer Solutions. Anal. Biochem. 2010, 407, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Samyn, B.; Sergeant, K.; Castanheira, P.; Faro, C.; Van Beeumen, J. A New Method for C-Terminal Sequence Analysis in the Proteomic Era. Nat. Methods 2005, 2, 193–200. [Google Scholar] [CrossRef]
- Morla, A.; Poirier, F.; Pons, S.; Beaulieu, C.; Charrier, J.-P.; Ataman-Onal, Y.; Gléhen, O.; Jolivet, M.; Choquet-Kastylevsky, G. Analysis of High Molecular Mass Proteins Larger than 150 kDa Using Cyanogen Bromide Cleavage and Conventional 2-DE. Electrophoresis 2008, 29, 4158–4168. [Google Scholar] [CrossRef]
- Steers, E.; Craven, G.R.; Anfinsen, C.B.; Bethune, J.L. Evidence for Nonidentical Chains in the Beta-Galactosidase of Escherichia coli K12. J. Biol. Chem. 1965, 240, 2478–2484. [Google Scholar] [CrossRef]
- Pattison, D.I.; Davies, M.J. Absolute Rate Constants for the Reaction of Hypochlorous Acid with Protein Side Chains and Peptide Bonds. Chem. Res. Toxicol. 2001, 14, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Ghesquière, B.; Jonckheere, V.; Colaert, N.; Van Durme, J.; Timmerman, E.; Goethals, M.; Schymkowitz, J.; Rousseau, F.; Vandekerckhove, J.; Gevaert, K. Redox Proteomics of Protein-Bound Methionine Oxidation. Mol. Cell. Proteom. MCP 2011, 10, M110.006866. [Google Scholar] [CrossRef] [Green Version]
- Ghesquière, B.; Gevaert, K. Proteomics Methods to Study Methionine Oxidation. Mass Spectrom. Rev. 2014, 33, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Moskovitz, J.; Levine, R.L. Oxidation of Methionine Residues of Proteins: Biological Consequences. Antioxid. Redox Signal. 2003, 5, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Vogt, W. Oxidation of Methionyl Residues in Proteins: Tools, Targets, and Reversal. Free Radic. Biol. Med. 1995, 18, 93–105. [Google Scholar] [CrossRef]
- Job, C.; Rajjou, L.; Lovigny, Y.; Belghazi, M.; Job, D. Patterns of Protein Oxidation in Arabidopsis Seeds and during Germination. Plant Physiol. 2005, 138, 790–802. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Ross, A.R.S.; Yang, J.; Hegedus, D.D.; Kermode, A.R. Phosphorylation of the 12 S Globulin Cruciferin in Wild-Type and Abi1-1 Mutant Arabidopsis thaliana (Thale Cress) Seeds. Biochem. J. 2007, 404, 247–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojciechowska, N.; Alipour, S.; Stolarska, E.; Bilska, K.; Rey, P.; Kalemba, E.M. Peptide-Bound Methionine Sulfoxide (MetO) Levels and MsrB2 Abundance Are Differentially Regulated during the Desiccation Phase in Contrasted Acer Seeds. Antioxidants 2020, 9, 391. [Google Scholar] [CrossRef] [PubMed]
- Tarrago, L.; Grosse, S.; Siponen, M.I.; Lemaire, D.; Alonso, B.; Miotello, G.; Armengaud, J.; Arnoux, P.; Pignol, D.; Sabaty, M. Rhodobacter sphaeroides Methionine Sulfoxide Reductase P Reduces R- and S-Diastereomers of Methionine Sulfoxide from a Broad-Spectrum of Protein Substrates. Biochem. J. 2018, 475, 3779–3795. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Yang, X.; Jia, S.; Weeks, A.M.; Hornsby, M.; Lee, P.S.; Nichiporuk, R.V.; Iavarone, A.T.; Wells, J.A.; Toste, F.D.; et al. Redox-Based Reagents for Chemoselective Methionine Bioconjugation. Science 2017, 355, 597–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elledge, S.K.; Tran, H.L.; Christian, A.H.; Steri, V.; Hann, B.; Toste, F.D.; Chang, C.J.; Wells, J.A. Systematic Identification of Engineered Methionines and Oxaziridines for Efficient, Stable, and Site-Specific Antibody Bioconjugation. Proc. Natl. Acad. Sci. USA 2020, 117, 5733–5740. [Google Scholar] [CrossRef] [PubMed]
- Hollemeyer, K.; Heinzle, E.; Tholey, A. Identification of Oxidized Methionine Residues in Peptides Containing Two Methionine Residues by Derivatization and Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Proteomics 2002, 2, 1524–1531. [Google Scholar] [CrossRef]
- Gustavsson, N.; Härndahl, U.; Sundby, C.; Emanuelsson, A.; Roepstorff, P. Methionine Sulfoxidation of the Chloroplast Small Heat Shock Protein and Conformational Changes in the Oligomer. Protein Sci. 1999, 8, 2506–2512. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, C.; Nickelsen, J. 2D Diagonal Redox SDS-PAGE of Proteins. Bio-Protoc. 2013, 3, e781. [Google Scholar] [CrossRef]
- Shechter, Y.; Burstein, Y.; Patchornik, A. Selective Oxidation of Methionine Residues in Proteins. Biochemistry 1975, 14, 4497–4503. [Google Scholar] [CrossRef]
- Dure, L.; Crouch, M.; Harada, J.; Ho, T.H.; Mundy, J.; Quatrano, R.; Thomas, T.; Sung, Z.R. Common Amino Acid Sequence Domains among the LEA Proteins of Higher Plants. Plant Mol. Biol. 1989, 12, 475–486. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Wise, M.J. The Continuing Conundrum of the LEA Proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [Green Version]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.-H.; Macherel, D. The Ubiquitous Distribution of Late Embryogenesis Abundant Proteins across Cell Compartments in Arabidopsis Offers Tailored Protection against Abiotic Stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [Green Version]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Kaminski Schierle, G.S.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically Disordered Proteins as Molecular Shields. Mol. Biosyst. 2012, 8, 210–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adachi, M.; Kanamori, J.; Masuda, T.; Yagasaki, K.; Kitamura, K.; Mikami, B.; Utsumi, S. Crystal Structure of Soybean 11S Globulin: Glycinin A3B4 Homohexamer. Proc. Natl. Acad. Sci. USA 2003, 100, 7395–7400. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Yang, Y.-S.; Sutter, B.M.; Wang, Y.; McKnight, S.L.; Tu, B.P. Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell 2019, 177, 711–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perera, S.P.; McIntosh, T.C.; Wanasundara, J.P.D. Structural Properties of Cruciferin and Napin of Brassica napus (Canola) Show Distinct Responses to Changes in PH and Temperature. Plants 2016, 5, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutlu, A.; Chen, X.; Reddy, S.M.; Gal, S. The Aspartic Proteinase Is Expressed in Arabidopsis Thaliana Seeds and Localized in the Protein Bodies. Seed Sci. Res. 1999, 9, 75–84. [Google Scholar] [CrossRef]
- Chen, H.; Osuna, D.; Colville, L.; Lorenzo, O.; Graeber, K.; Küster, H.; Leubner-Metzger, G.; Kranner, I. Transcriptome-Wide Mapping of Pea Seed Ageing Reveals a Pivotal Role for Genes Related to Oxidative Stress and Programmed Cell Death. PLoS ONE 2013, 8, e78471. [Google Scholar] [CrossRef] [Green Version]
- Sano, N.; Permana, H.; Kumada, R.; Shinozaki, Y.; Tanabata, T.; Yamada, T.; Hirasawa, T.; Kanekatsu, M. Proteomic Analysis of Embryonic Proteins Synthesized from Long-Lived mRNAs during Germination of Rice Seeds. Plant Cell Physiol. 2012, 53, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, B.; Gilbert, H.F. Protein Disulfide Isomerase. Biochim. Biophys. Acta 2004, 1699, 35–44. [Google Scholar] [CrossRef]
- Fluhr, R.; Lampl, N.; Roberts, T.H. Serpin Protease Inhibitors in Plant Biology. Physiol. Plant. 2012, 145, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Perozich, J.; Nicholas, H.; Wang, B.C.; Lindahl, R.; Hempel, J. Relationships within the Aldehyde Dehydrogenase Extended Family. Protein Sci. Publ. Protein Soc. 1999, 8, 137–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debeaujon, I.; Léon-Kloosterziel, K.M.; Koornneef, M. Influence of the Testa on Seed Dormancy, Germination, and Longevity in Arabidopsis. Plant Physiol. 2000, 122, 403–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and Biochemistry of Seed Flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-H.; Kim, S.-R.; An, G. Rice Aldehyde Dehydrogenase7 Is Needed for Seed Maturation and Viability. Plant Physiol. 2009, 149, 905–915. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Atamna, H.; Kuratsune, H.; Ames, B.N. Delaying Brain Mitochondrial Decay and Aging with Mitochondrial Antioxidants and Metabolites. Ann. N. Y. Acad. Sci. 2002, 959, 133–166. [Google Scholar] [CrossRef] [PubMed]
- Sadowska-Bartosz, I.; Bartosz, G. Effect of Antioxidants Supplementation on Aging and Longevity. BioMed Res. Int. 2014, 2014, 404680. [Google Scholar] [CrossRef]
- Tahara, E.B.; Barros, M.H.; Oliveira, G.A.; Netto, L.E.S.; Kowaltowski, A.J. Dihydrolipoyl Dehydrogenase as a Source of Reactive Oxygen Species Inhibited by Caloric Restriction and Involved in Saccharomyces Cerevisiae Aging. FASEB J. 2007, 21, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Omarov, R.T.; Akaba, S.; Koshiba, T.; Lips, S.H. Aldehyde Oxidase in Roots, Leaves and Seeds of Barley (Hordeum vulgare L.). J. Exp. Bot. 1999, 50, 63–69. [Google Scholar] [CrossRef]
- Seo, M.; Peeters, A.J.; Koiwai, H.; Oritani, T.; Marion-Poll, A.; Zeevaart, J.A.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis Aldehyde Oxidase 3 (AAO3) Gene Product Catalyzes the Final Step in Abscisic Acid Biosynthesis in Leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef] [Green Version]
- Miransari, M.; Smith, D.L. Plant Hormones and Seed Germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Souter, M.; Lindsey, K. Polarity and Signalling in Plant Embryogenesis. J. Exp. Bot. 2000, 51, 971–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, J.; Giraudat, J. Abscisic Acid Signal Transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 199–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, R.S.P.; Møller, I.M. Pattern of Occurrence and Occupancy of Carbonylation Sites in Proteins. Proteomics 2011, 11, 4166–4173. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Balmer, Y. Redox Regulation: A Broadening Horizon. Annu. Rev. Plant Biol. 2005, 56, 187–220. [Google Scholar] [CrossRef] [PubMed]
- Oien, D.B.; Carrasco, G.A.; Moskovitz, J. Decreased Phosphorylation and Increased Methionine Oxidation of α-Synuclein in the Methionine Sulfoxide Reductase A Knockout Mouse. J. Amino Acids 2011, 2011, 721094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardin, S.C.; Larue, C.T.; Oh, M.-H.; Jain, V.; Huber, S.C. Coupling Oxidative Signals to Protein Phosphorylation via Methionine Oxidation in Arabidopsis. Biochem. J. 2009, 422, 305–312. [Google Scholar] [CrossRef] [Green Version]
- Rao, R.; Xu, D.; Thelen, J.J.; Miernyk, J.A. Circles within Circles: Crosstalk between Protein Ser/Thr/Tyr-Phosphorylation and Met Oxidation. BMC Bioinform. 2013, 14, S14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aussel, L.; Ezraty, B. Methionine Redox Homeostasis in Protein Quality Control. Front. Mol. Biosci. 2021, 8, 665492. [Google Scholar] [CrossRef]
- Chibani, K.; Ali-Rachedi, S.; Job, C.; Job, D.; Jullien, M.; Grappin, P. Proteomic Analysis of Seed Dormancy in Arabidopsis. Plant Physiol. 2006, 142, 1493–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, P.; Poland, J.; Schnölzer, M.; Rabilloud, T. A New Silver Staining Apparatus and Procedure for Matrix-Assisted Laser Desorption/Ionization-Time of Flight Analysis of Proteins after Two-Dimensional Electrophoresis. Proteomics 2001, 1, 835–840. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durek, P.; Schmidt, R.; Heazlewood, J.L.; Jones, A.; MacLean, D.; Nagel, A.; Kersten, B.; Schulze, W.X. PhosPhAt: The Arabidopsis thaliana Phosphorylation Site Database. An Update. Nucleic Acids Res. 2010, 38, D828–D834. [Google Scholar] [CrossRef] [PubMed]
- Källberg, M.; Margaryan, G.; Wang, S.; Ma, J.; Xu, J. RaptorX Server: A Resource for Template-Based Protein Structure Modeling. Methods Mol. Biol. 2014, 1137, 17–27. [Google Scholar] [CrossRef] [PubMed]

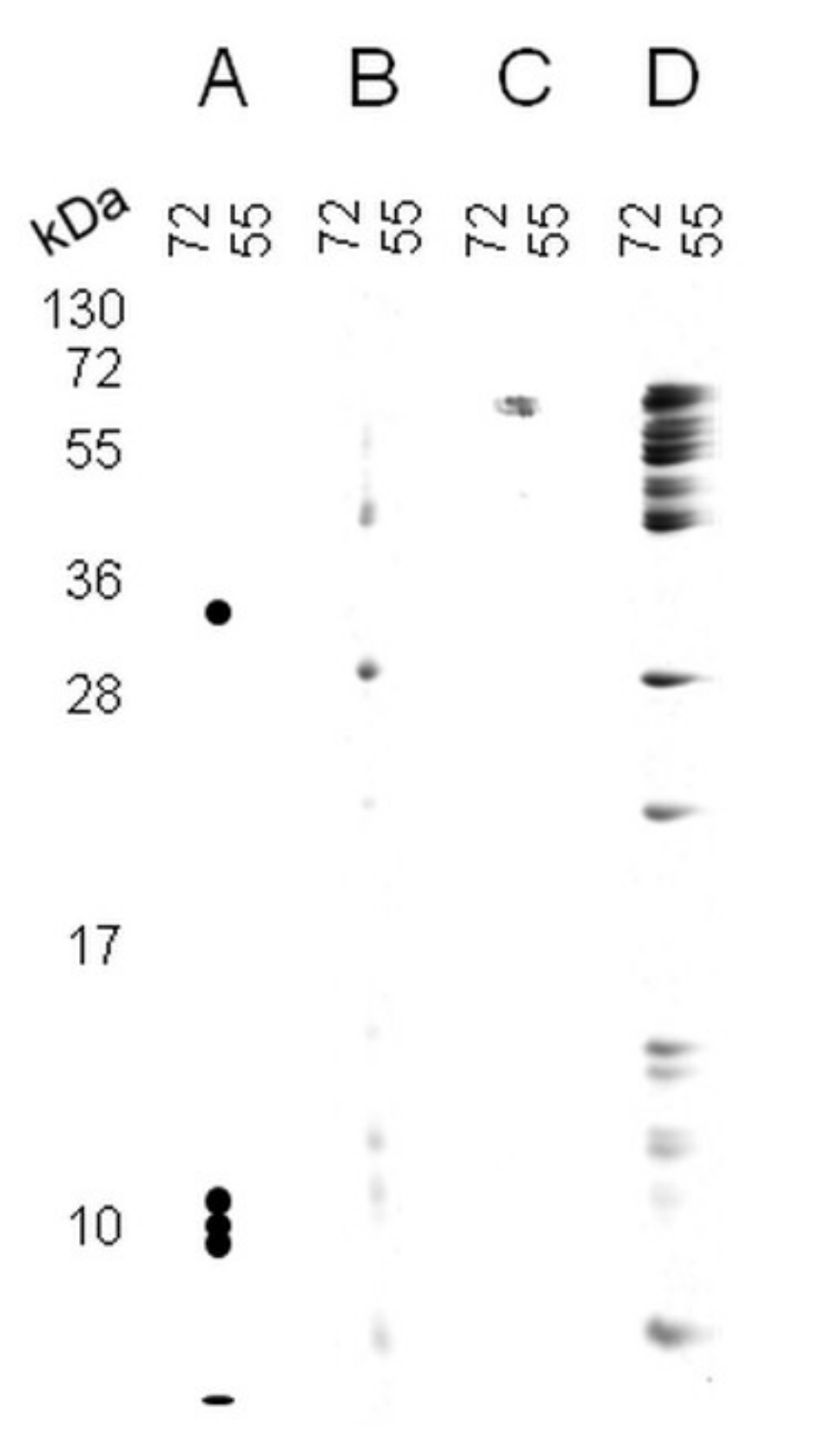


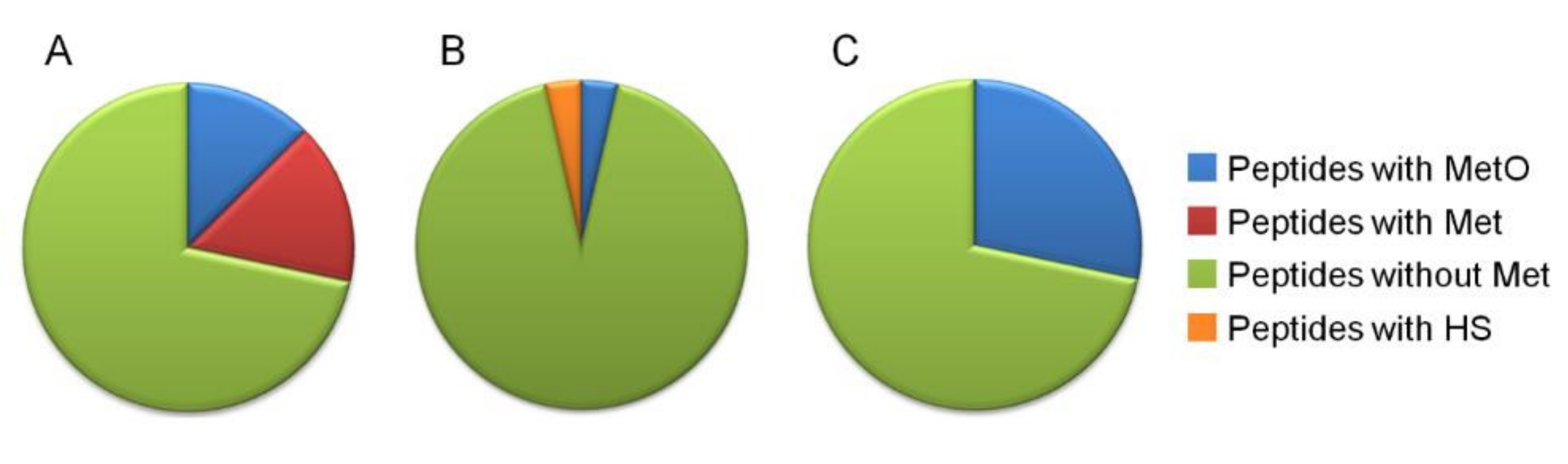
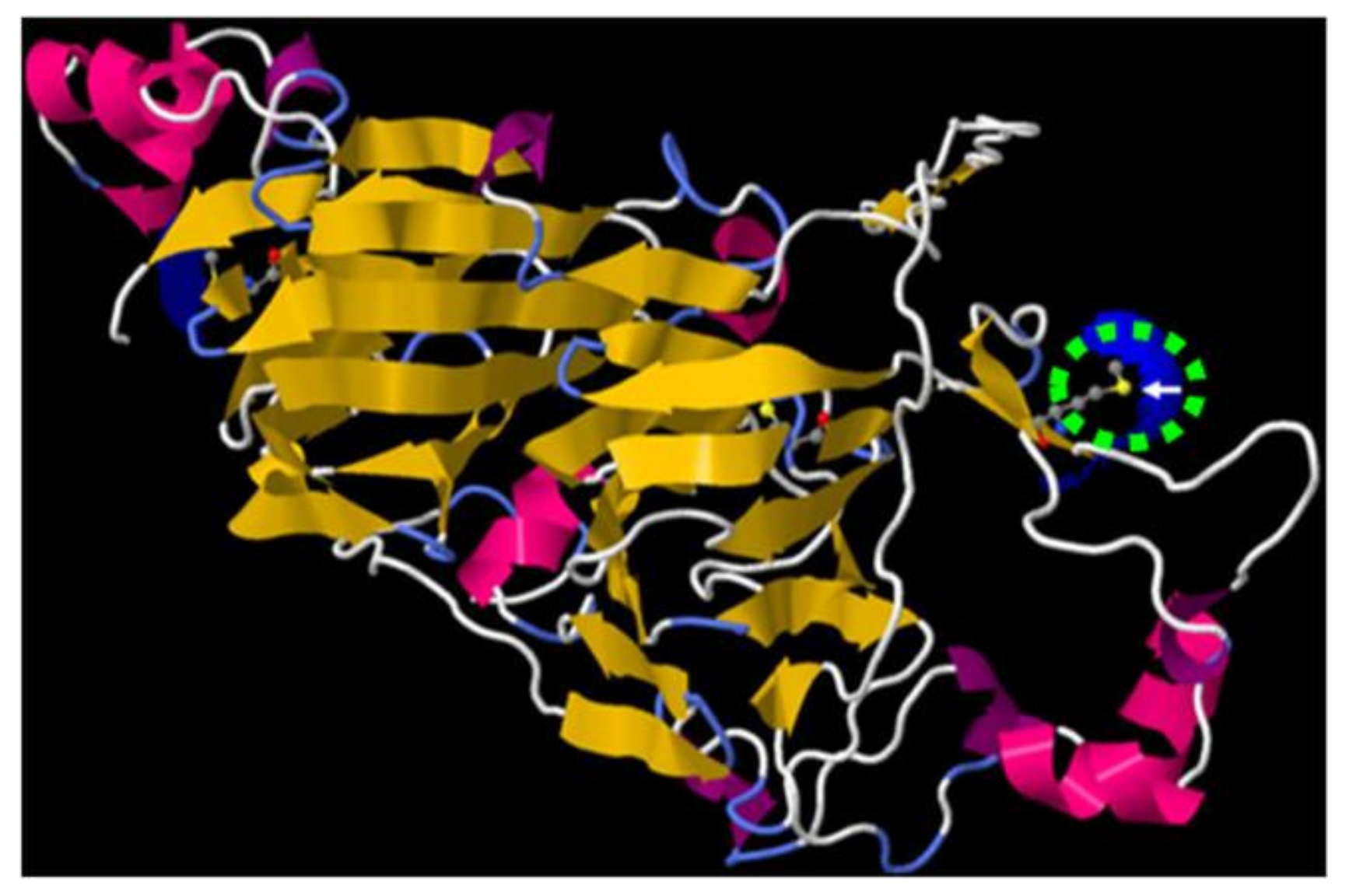
| Table | Protein Name | Peptide Sequence | Met Position |
|---|---|---|---|
| AT2G42560.1 | Late embryogenesis abundant domain | GSNMPVSDEGEGETK DQEMHQGGEEEKQPGFVSGAR THEHGTTDPDYMR | M65 M487 M628 |
| AT1G07920.1 | Elongation factor 1-alpha 3 | VETGMIKPGMVVTFAPTGLTTEVK PGMVVTFAPTGLTTEVK MTPTKPMVVETFSEYPPLGR | M259, 264 M264 M398 |
| AT4G24620.1 | Glucose-6-phosphate isomerase 1, chloroplastic | VGFTDEFVAEMEPR YLQQLVMESLGK | M107 M391 |
| AT2G36640.1 | Late embryogenesis abundant protein | LTMPSDIVEETR | M382 |
| AT1G21750.1 | Protein disulfide isomerase-like 1–1 | LSGSEFDSFMAIAEK FPKLSGSEFDSFMAIAEK | M180 M180 |
| AT3G17240.1 | Dihydrolipoyl dehydrogenase 2, mitochondrial | VSSVEVDLPAMLAQK | M124 |
| AT3G22500.1 | Late embryogenesis abundant protein in group 5 | GGPAAVMQSAATTNIR | M58 |
| AT3G48990.1 | 4-Coumarate-CoA ligase-like 10 | SSNPLPEEGPHKPGSVGKP VGQEMAILNEK | M345 |
| AT3G53040.1 | Late embryogenesis abundant protein-like | TTTTEPERPGLIGSVMK | M59 |
| AT3G54400.1 | Aspartyl protease family protein | ASGTSLPAQGLMGLGR | M209 |
| AT4G12290.1 | Amine oxidase | VGLSGILMVK | M491 |
| AT5G20960.1 | Indole-3-acetaldehyde oxidase | VPAVYAVNMR | M1046 |
| AT5G44120.3 | 12S seed storage protein CRA1 | DMHQKVEHIR | M138 |
| AT5G52300.1 | Low-temperature-induced 65 kDa protein | MESQLTRPYGHEQAEEPIR | M1 |
| AT2G31670.1 | Uncharacterized protein | DLSEMEAVDAQK | M223 |
| AT1G54100.1 | Aldehyde dehydrogenase family 7 member B4 | VGSMVQQTVNAR | M251 |
| AT1G77510.1 | Protein disulfide isomerase-like 1–2 | LSGDEFDSFMALAEK | M179 |
| AT2G36530.1 | Bifunctional enolase 2/transcriptional activator | VVIGMDVAASEFYSEDK | M249 |
| AT1G47710.1 | Cysteine protease inhibitor/ serine-type endopeptidase inhibitor | ESISLQNQVSMNLAK | M15 |
| TAIR acc. | Protein Name | Met Position | Ser, Thr, and Tyr Position * |
|---|---|---|---|
| AT1G07920.1 | Elongation factor 1-alpha 3 | M264 | S315 |
| AT4G12290.1 | Amine oxidase | M491 | Y243 |
| AT5G20960.1 | Indole-3-acetaldehyde oxidase | M1046 | Y1179 |
| AT5G44120.3 | 12S seed storage protein CRA1 | M138 | T115 ** |
| AT1G54100.1 | Aldehyde dehydrogenase family 7 member B4 | M251 | S250 |
| AT1G47710.1 | Cysteine protease inhibitor/serine-type endopeptidase inhibitor | M15 | Y239 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalemba, E.M.; Valot, B.; Job, D.; Bailly, C.; Meimoun, P. Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsis thaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis. Plants 2022, 11, 569. https://doi.org/10.3390/plants11040569
Kalemba EM, Valot B, Job D, Bailly C, Meimoun P. Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsis thaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis. Plants. 2022; 11(4):569. https://doi.org/10.3390/plants11040569
Chicago/Turabian StyleKalemba, Ewa Marzena, Benoît Valot, Dominique Job, Christophe Bailly, and Patrice Meimoun. 2022. "Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsis thaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis" Plants 11, no. 4: 569. https://doi.org/10.3390/plants11040569
APA StyleKalemba, E. M., Valot, B., Job, D., Bailly, C., & Meimoun, P. (2022). Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsis thaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis. Plants, 11(4), 569. https://doi.org/10.3390/plants11040569









