Response of Tomato (Solanum lycopersicum L.) Genotypes to Heat Stress Using Morphological and Expression Study
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Growth Parameters
4.3. Total RNA Extraction
4.4. cDNA Synthesis
4.5. Potential Candidate Genes for Heat Shock Protein in Tomato
4.6. Quantitative Real-Time PCR
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Tanksley, S.D. Chromosomal evolution in the plant family Solanaceae. BMC Genom. 2010, 11, 182. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, A.K.; Thakur, S.; Suri, V. Technology transfer model on integrated nutrient management technology for sustainable crop production in high-value cash crops and vegetables in northwestern Himalayas. Commun. Soil Sci. Plant Anal. 2013, 44, 1684–1699. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). FAOSTAT Database. 2016. Available online: http://faostat.fao.org/site/291/default.aspx (accessed on 12 October 2021).
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Blum, A. Plant Breeding for Stress Environments; CRC Press: Boca Raton, FL, USA, 1988. [Google Scholar]
- Reddy, K.R.; Kakani, V. Screening Capsicum species of different origins for high temperature tolerance by in vitro pollen germination and pollen tube length. Sci. Hortic. 2007, 112, 130–135. [Google Scholar] [CrossRef]
- Karapanos, I.; Akoumianakis, K.; Olympios, C.; Passam, H.C. Tomato pollen respiration in relation to in vitro germination and pollen tube growth under favourable and stress-inducing temperatures. Sex. Plant Reprod. 2010, 23, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Lohar, D.; Peat, W. Floral characteristics of heat-tolerant and heat-sensitive tomato (Lycopersicon esculentum Mill.) cultivars at high temperature. Sci. Hortic. 1998, 73, 53–60. [Google Scholar] [CrossRef]
- Hedhly, A.; Hormaza, J.; Herrero, M. Flower emasculation accelerates ovule degeneration and reduces fruit set in sweet cherry. Sci. Hortic. 2009, 119, 455–457. [Google Scholar] [CrossRef]
- Peet, M.; Sato, S.; Gardner, R. Comparing heat stress effects on male-fertile and male-sterile tomatoes. Plant Cell Environ. 1998, 21, 225–231. [Google Scholar] [CrossRef]
- Qu, G.-Q.; Liu, X.; Zhang, Y.-L.; Yao, D.; Ma, Q.-M.; Yang, M.-Y.; Zhu, W.-H.; Yu, S.; Luo, Y.-B. Evidence for programmed cell death and activation of specific caspase-like enzymes in the tomato fruit heat stress response. Planta 2009, 229, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Zinn, K.E.; Tunc-Ozdemir, M.; Harper, J.F. Temperature stress and plant sexual reproduction: Uncovering the weakest links. J. Exp. Bot. 2010, 61, 1959–1968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, A.; Hayat, K.; Khan, A.; Iqbal, S. Heat tolerance studies in tomato (Lycopersicon esculentum Mill.). Int. J. Agric. Biol. 2007, 9, 649–652. [Google Scholar]
- Sung, S.S.; Brassington, A.-M.E.; Krakowiak, P.A.; Carey, J.C.; Jorde, L.B.; Bamshad, M. Mutations in TNNT3 cause multiple congenital contractures: A second locus for distal arthrogryposis type 2B. Am. J. Hum. Genet. 2003, 73, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Singh, M.K.; Singh, V.; Singh, R.; Raghuvanshi, T.; Singh, C. Debilitation in tomato (Solanum lycopersicum L.) as result of heat stress. J. Pharmacogn. Phytochem. 2017, 6, 1917–1922. [Google Scholar]
- Camejo, D.; Rodríguez, P.; Morales, M.A.; Dell’Amico, J.M.; Torrecillas, A.; Alarcón, J.J. High temperature effects on photosynthetic activity of two tomato cultivars with different heat susceptibility. J. Plant Physiol. 2005, 162, 281–289. [Google Scholar] [CrossRef]
- Driedonks, N. From Flower to Fruit in the Heat—Reproductive Thermotolerance in Tomato and Its Wild Relatives. Ph.D. Thesis, Radboud University Nijmegen, Nijmegen, The Netherlands, 2018. [Google Scholar]
- Raja, M.M.; Vijayalakshmi, G.; Naik, M.L.; Basha, P.O.; Sergeant, K.; Hausman, J.F.; Khan, P.S.S.V. Pollen development and function under heat stress: From effects to responses. Acta Physiol. Plant. 2019, 41, 1–20. [Google Scholar] [CrossRef]
- Alsamir, M.; Mahmood, T.; Trethowan, R.; Ahmad, N. An overview of heat stress in tomato (Solanum lycopersicum L.). Saudi J. Biol. Sci. 2021, 28, 1654. [Google Scholar] [CrossRef]
- Grover, A.; Mittal, D.; Negi, M.; Lavania, D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013, 205, 38–47. [Google Scholar] [CrossRef]
- Lin, Y.-X.; Jiang, H.-Y.; Chu, Z.-X.; Tang, X.-L.; Zhu, S.-W.; Cheng, B.-J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Czarnecka-Verner, E.; Yuan, C.-X.; Fox, P.C.; Gurley, W.B. Isolation and characterization of six heat shock transcription factor cDNA clones from soybean. Plant Mol. Biol. 1995, 29, 37–51. [Google Scholar] [CrossRef]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Döring, P.; Treuter, E.; Kistner, C.; Lyck, R.; Chen, A.; Nover, L. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 2000, 12, 265–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; Von Koskull-Döring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Shim, D.; Hwang, J.-U.; Lee, J.; Lee, S.; Choi, Y.; An, G.; Martinoia, E.; Lee, Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 2009, 21, 4031–4043. [Google Scholar] [CrossRef] [Green Version]
- Takayama, K.; Tsutsumi, S.; Katayama, S.; Okayama, T.; Horie-Inoue, K.; Ikeda, K.; Urano, T.; Kawazu, C.; Hasegawa, A.; Ikeo, K. Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells. Oncogene 2011, 30, 619–630. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Thalor, S.K.; Takahashi, Y.; Berberich, T.; Kusano, T. An inhibitory effect of the sequence-conserved upstream open-reading frame on the translation of the main open-reading frame of HsfB1 transcripts in Arabidopsis. Plant Cell Environ. 2012, 35, 2014–2030. [Google Scholar] [CrossRef]
- Song, X.; Liu, G.; Duan, W.; Liu, T.; Huang, Z.; Ren, J.; Li, Y.; Hou, X. Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genom. 2014, 289, 541–551. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, Y.; Gao, Y.; Zhou, Y.; Zhang, E.; Hu, Y.; Yuan, Y.; Liang, G.; Xu, C. Adaptive evolution and divergent expression of heat stress transcription factors in grasses. BMC Evol. Biol. 2014, 14, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ro, S.; Chea, L.; Ngoun, S.; Stewart, Z.P.; Roeurn, S.; Theam, P.; Lim, S.; Sor, R.; Kosal, M.; Roeun, M. Response of tomato genotypes under different high temperatures in field and greenhouse conditions. Plants 2021, 10, 449. [Google Scholar] [CrossRef]
- Ayenan, M.A.T.; Danquah, A.; Hanson, P.; Asante, I.K.; Danquah, E.Y. Identification of new sources of heat tolerance in cultivated and wild tomatoes. Euphytica 2021, 217, 1–16. [Google Scholar] [CrossRef]
- Ayenan, M.A.T.; Danquah, A.; Hanson, P.; Asante, I.K.; Danquah, E.Y. Tomato (Solanum lycopersicum L.) Genotypes Respond Differently to Long-Term Dry and Humid Heat Stress. Horticulturae 2022, 8, 118. [Google Scholar] [CrossRef]
- Scharf, K.-D.; Rose, S.; Zott, W.; Schöffl, F.; Nover, L.; Schöff, F. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990, 9, 4495–4501. [Google Scholar] [CrossRef]
- Morimoto, R. Cells in stress: Transcriptional activation of heat shock genes. Science 1993, 259, 1409–1411. [Google Scholar] [CrossRef]
- Neta-Sharir, I.; Isaacson, T.; Lurie, S.; Weiss, D. Dual role for tomato heat shock protein 21: Protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell 2005, 17, 1829–1838. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta BBA Gene Struct. Expr. 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- Sun, W.; Bernard, C.; Van De Cotte, B.; Van Montagu, M.; Verbruggen, N. At-HSP17. 6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 2001, 27, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.J.; Vierling, E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 2000, 122, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Zhu, W.; Zhang, H.; Liu, N.; Tian, S. Heat shock factors in tomatoes: Genome-wide identification, phylogenetic analysis and expression profiling under development and heat stress. PeerJ 2016, 4, e1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, C.H.; Nam, H.G.; Park, Y.S. Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 2003, 36, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Arce, D.; Spetale, F.; Krsticevic, F.; Cacchiarelli, P.; De Las Rivas, J.; Ponce, S.; Pratta, G.; Tapia, E. Regulatory motifs found in the small heat shock protein (sHSP) gene family in tomato. BMC Genom. 2018, 19, 860. [Google Scholar] [CrossRef] [Green Version]
- Marko, D.; El-Shershaby, A.; Carriero, F.; Summerer, S.; Petrozza, A.; Iannacone, R.; Schleiff, E.; Fragkostefanakis, S. Identification and characterization of a thermotolerant TILLING allele of heat shock binding protein 1 in tomato. Genes 2019, 10, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.; Meeley, R.; Scanlon, M.J. Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 2002, 14, 3119–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.; Rogowsky, P.; Nover, L.; Scanlon, M.J. The maize heat shock factor-binding protein paralogs EMP2 and HSBP2 interact non-redundantly with specific heat shock factors. Planta 2006, 224, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Rana, R.M.; Dong, S.; Tang, H.; Ahmad, F.; Zhang, H. Functional analysis of OsHSBP1 and OsHSBP2 revealed their involvement in the heat shock response in rice (Oryza sativa L.). J. Exp. Bot. 2012, 63, 6003–6016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knapp, S.; Consortium, T.G. The tomato genome sequence provides insights into fleshy fruit evolution. Res. Lett. 2012, 485, 635–641. [Google Scholar]
- Giorno, F.; Wolters-Arts, M.; Grillo, S.; Scharf, K.-D.; Vriezen, W.H.; Mariani, C. Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot. 2010, 61, 453–462. [Google Scholar] [CrossRef] [Green Version]
- Fragkostefanakis, S.; Mesihovic, A.; Simm, S.; Paupière, M.J.; Hu, Y.; Paul, P.; Mishra, S.K.; Tschiersch, B.; Theres, K.; Bovy, A. HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues. Plant Physiol. 2016, 170, 2461–2477. [Google Scholar] [CrossRef]
- Frank, G.; Pressman, E.; Ophir, R.; Althan, L.; Shaked, R.; Freedman, M.; Shen, S.; Firon, N. Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J. Exp. Bot. 2009, 60, 3891–3908. [Google Scholar] [CrossRef]
- Hsu, S.-F.; Lai, H.-C.; Jinn, T.-L. Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis. Plant Physiol. 2010, 153, 773–784. [Google Scholar] [CrossRef] [Green Version]
- Al-Gaadi, K.A.; Tola, E.; Madugundu, R.; Fulleros, R.B. Sentinel-2 images for effective mapping of soil salinity in agricultural fields. Curr. Sci. 2021, 121, 384. [Google Scholar]
- Pathak, T.B.; Stoddard, C.S. Climate change effects on the processing tomato growing season in California using growing degree day model. Modeling Earth Syst. Environ. 2018, 4, 765–775. [Google Scholar] [CrossRef]
- Scholberg, J.; McNeal, B.L.; Boote, K.J.; Jones, J.W.; Locascio, S.J.; Olson, S.M. Nitrogen stress effects on growth and nitrogen accumulation by field-grown tomato. Agron. J. 2000, 92, 159–167. [Google Scholar] [CrossRef]
- Sturn, A.; Quackenbush, J.; Trajanoski, Z. Genesis: Cluster analysis of microarray data. Bioinformatics 2002, 18, 207–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
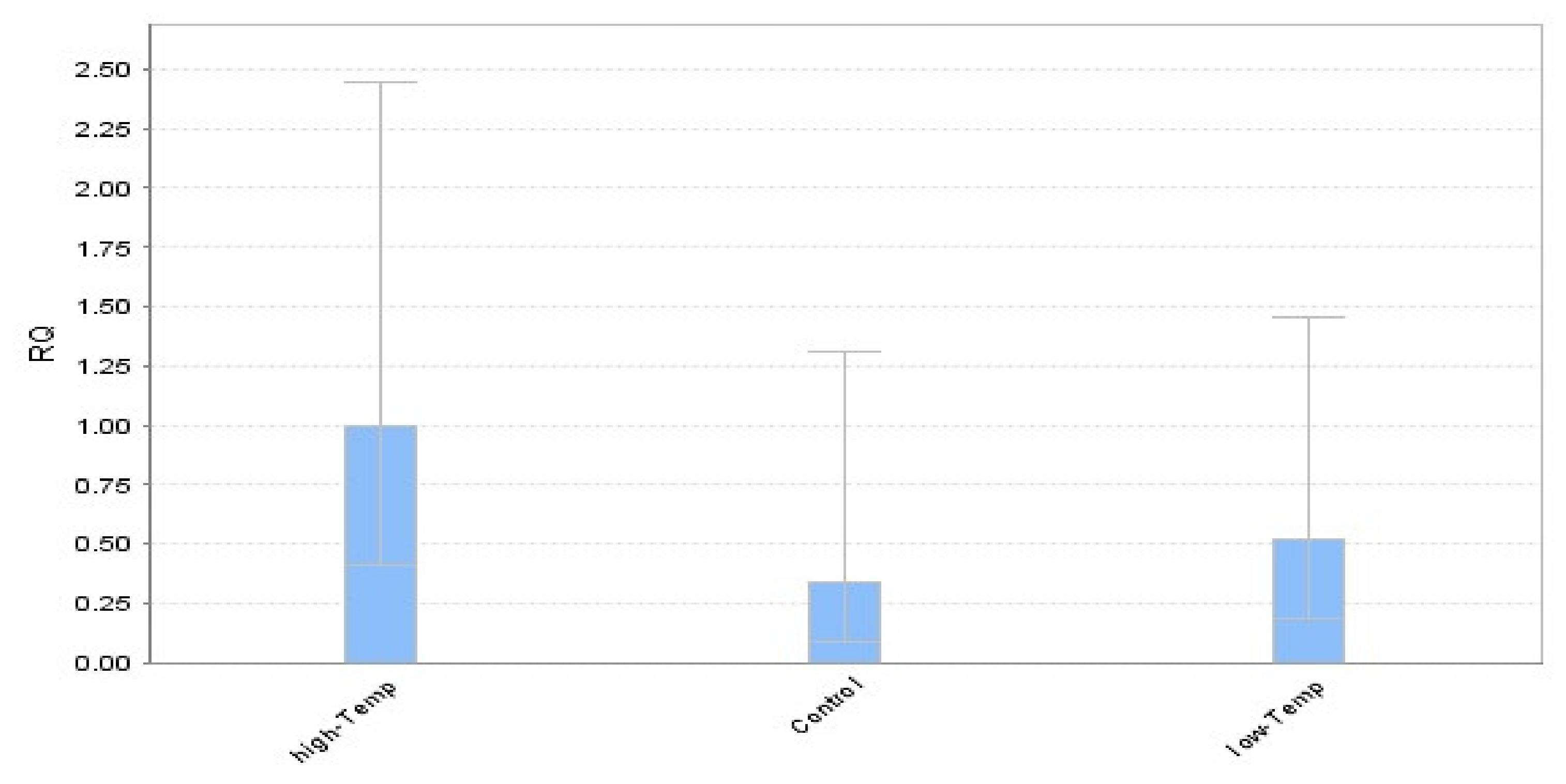


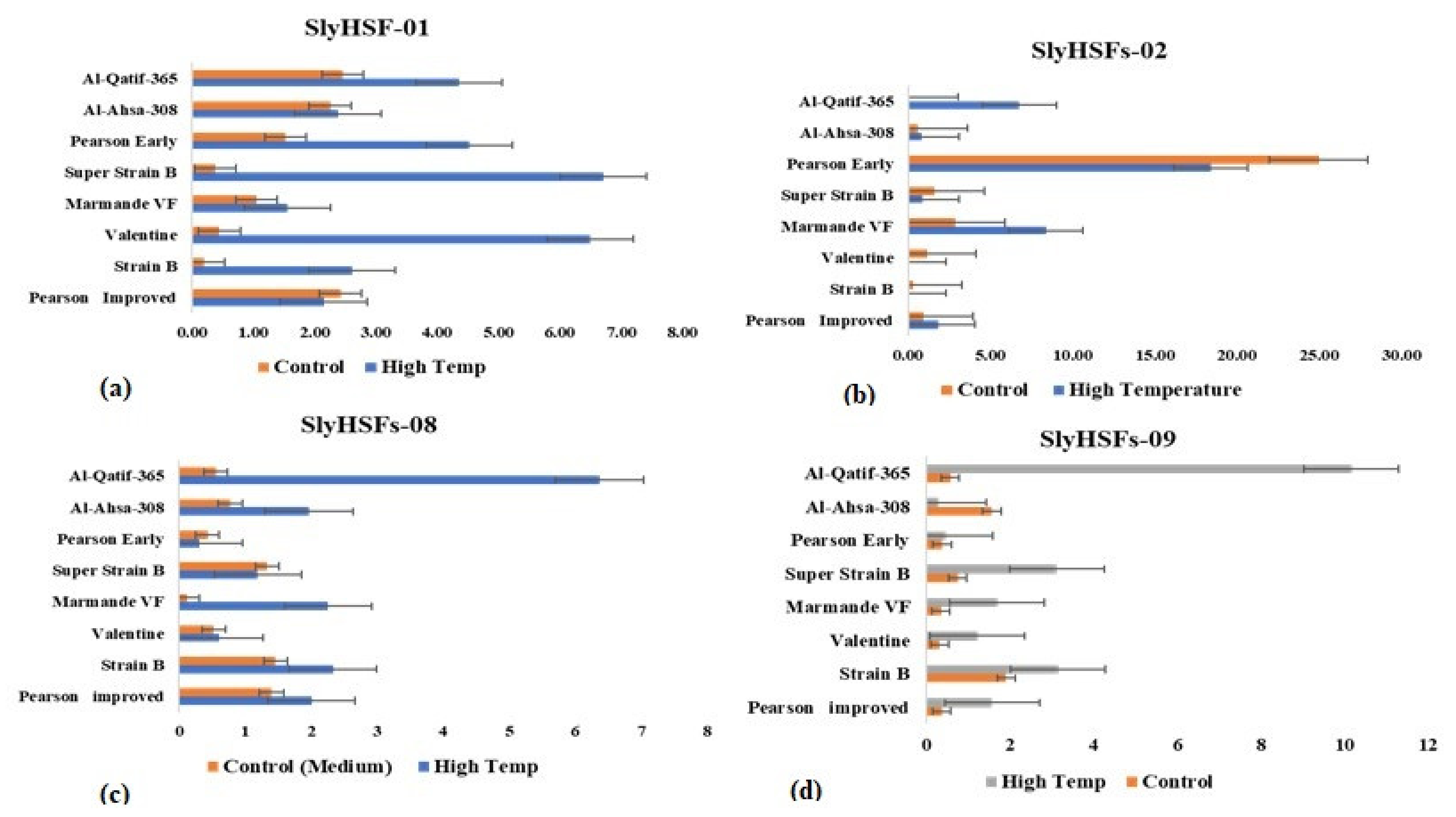
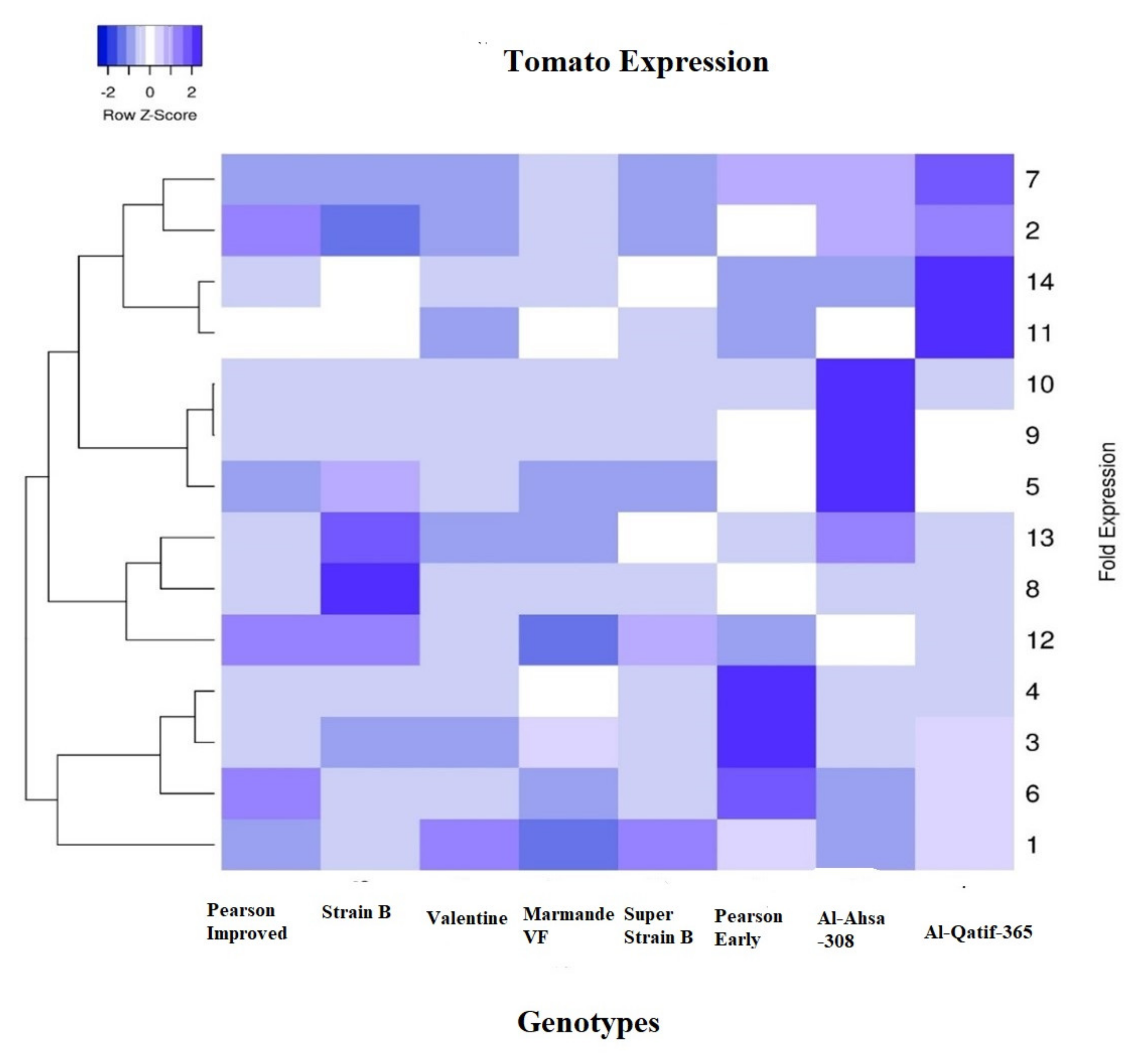
| Traits | Mean | SD | Minimum | Maximum |
|---|---|---|---|---|
| Plant height (cm) | 66.86 | 6.7 | 52.90 | 83 |
| DWL (g) | 64.70 | 8.82 | 48 | 85.49 |
| DWS (g) | 45.17 | 6.22 | 34.26 | 60.53 |
| FWL(g) | 527.28 | 73.81 | 396.10 | 699 |
| FW/p (g) | 838.06 | 117.26 | 633.73 | 1114 |
| FWS (g) | 310.80 | 43.43 | 234.90 | 415 |
| NoB | 6.96 | 0.86 | 5.6 | 8.90 |
| LA/p (cm2) | 5614.4 | 778.85 | 4248.3 | 7445.8 |
| ID | Name | Chr # | Start | End | Gene (bp) | # cDNA | # Amino Acid |
|---|---|---|---|---|---|---|---|
| SlyHSF-01 | Solyc11g064990.1 | SL2.40ch11 | 47,389,718 | 47,391,840 | 2123 | 756 | 251 |
| SlyHSF-02 | Solyc08g005170.2 | SL2.40ch08 | 111,412 | 116,839 | 5428 | 1949 | 527 |
| SlyHSF-03 | Solyc03g026020.2 | SL2.40ch03 | 7,810,489 | 7,812,280 | 1792 | 1594 | 338 |
| SlyHSF-04 | Solyc03g097120.2 | SL2.40ch03 | 52,901,766 | 52,904,929 | 3164 | 1874 | 491 |
| SlyHSF-05 | Solyc02g090820.2 | SL2.40ch02 | 46,880,125 | 46,883,382 | 3258 | 1611 | 301 |
| SlyHSF-06 | Solyc09g065660.2 | SL2.40ch09 | 59,473,864 | 59,475,995 | 2132 | 1405 | 372 |
| SlyHSF-07 | Solyc04g078770.2 | SL2.40ch04 | 61,036,586 | 61,037,903 | 1318 | 1230 | 360 |
| SlyHSF-08 | Solyc06g072750.2 | SL2.40ch06 | 41,255,352 | 41,258,348 | 2997 | 1613 | 482 |
| SlyHSF-09 | Solyc12g098520.1 | SL2.40ch12 | 48,549,454 | 48,552,229 | 2776 | 1437 | 478 |
| SlyHSF-10 | Solyc08g080540.2 | SL2.40ch08 | 60,985,869 | 60,987,278 | 1410 | 1329 | 325 |
| SlyHSF-11 | Solyc04g016000.2 | SL2.40ch04 | 6,594,909 | 6,598,451 | 3543 | 1320 | 237 |
| Soil Texture | pH | EC dS m−1 | Anions (mEq L−1) | Cations (mEq L−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clay (%) | Silt (%) | Sand (%) | Soil Type | Ca | Mg | K | Na | HCO3 | Cl | SO4 | ||
| 8.45 | 7.83 | 83.72 | Sandy Loam | 7.8 | 1.98 | 10.50 | 4.50 | 1.32 | 6.97 | 2.30 | 2.65 | 18.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldubai, A.A.; Alsadon, A.A.; Migdadi, H.H.; Alghamdi, S.S.; Al-Faifi, S.A.; Afzal, M. Response of Tomato (Solanum lycopersicum L.) Genotypes to Heat Stress Using Morphological and Expression Study. Plants 2022, 11, 615. https://doi.org/10.3390/plants11050615
Aldubai AA, Alsadon AA, Migdadi HH, Alghamdi SS, Al-Faifi SA, Afzal M. Response of Tomato (Solanum lycopersicum L.) Genotypes to Heat Stress Using Morphological and Expression Study. Plants. 2022; 11(5):615. https://doi.org/10.3390/plants11050615
Chicago/Turabian StyleAldubai, Abdulhakim A., Abdullah A. Alsadon, Hussein H. Migdadi, Salem S. Alghamdi, Sulieman A. Al-Faifi, and Muhammad Afzal. 2022. "Response of Tomato (Solanum lycopersicum L.) Genotypes to Heat Stress Using Morphological and Expression Study" Plants 11, no. 5: 615. https://doi.org/10.3390/plants11050615
APA StyleAldubai, A. A., Alsadon, A. A., Migdadi, H. H., Alghamdi, S. S., Al-Faifi, S. A., & Afzal, M. (2022). Response of Tomato (Solanum lycopersicum L.) Genotypes to Heat Stress Using Morphological and Expression Study. Plants, 11(5), 615. https://doi.org/10.3390/plants11050615






