Impact of Nanomaterials on the Regulation of Gene Expression and Metabolomics of Plants under Salt Stress
Abstract
:1. Introduction
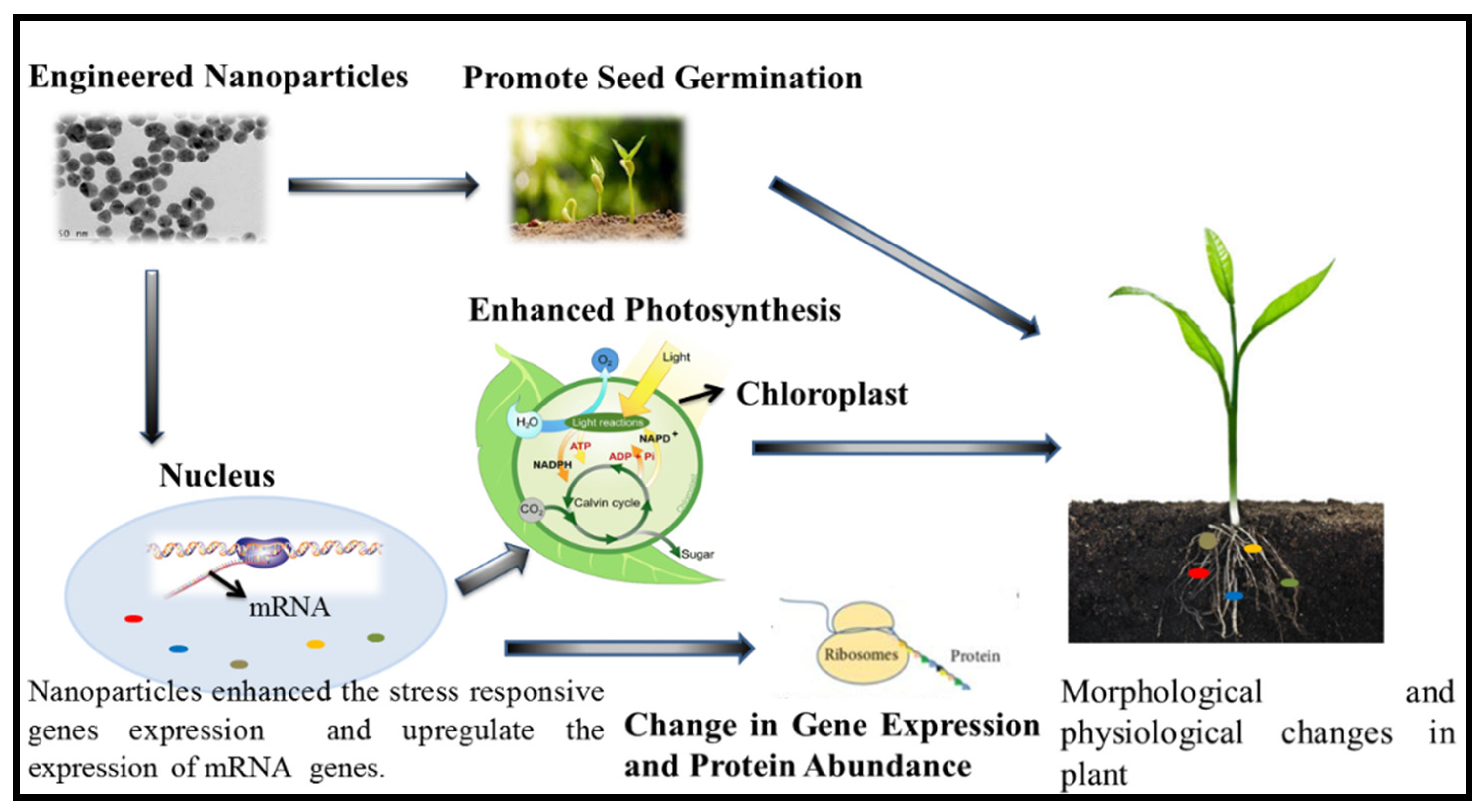
2. Engineered Nanoparticles and their Effect on Plant Salt Tolerance Genes: Enzymatic Expression
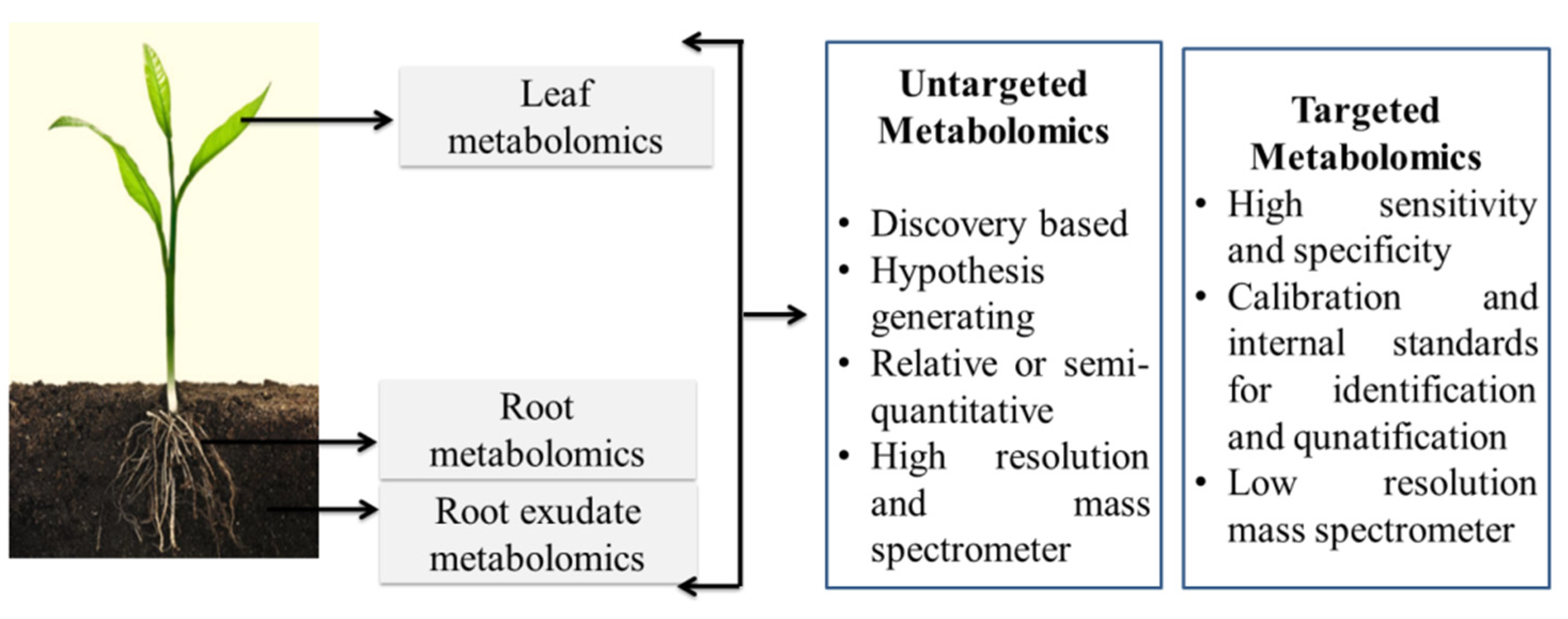
3. Plant Metabolomics and the Linkage of Molecular Functions to Nanomaterial Application
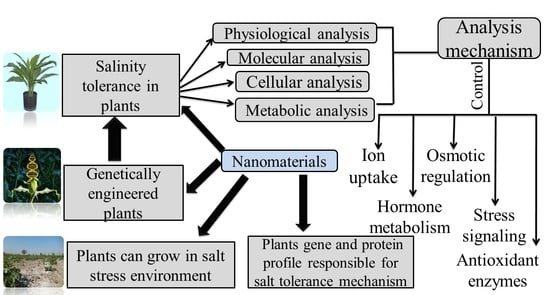
4. Plant Genetic Responses to Salinity Stress
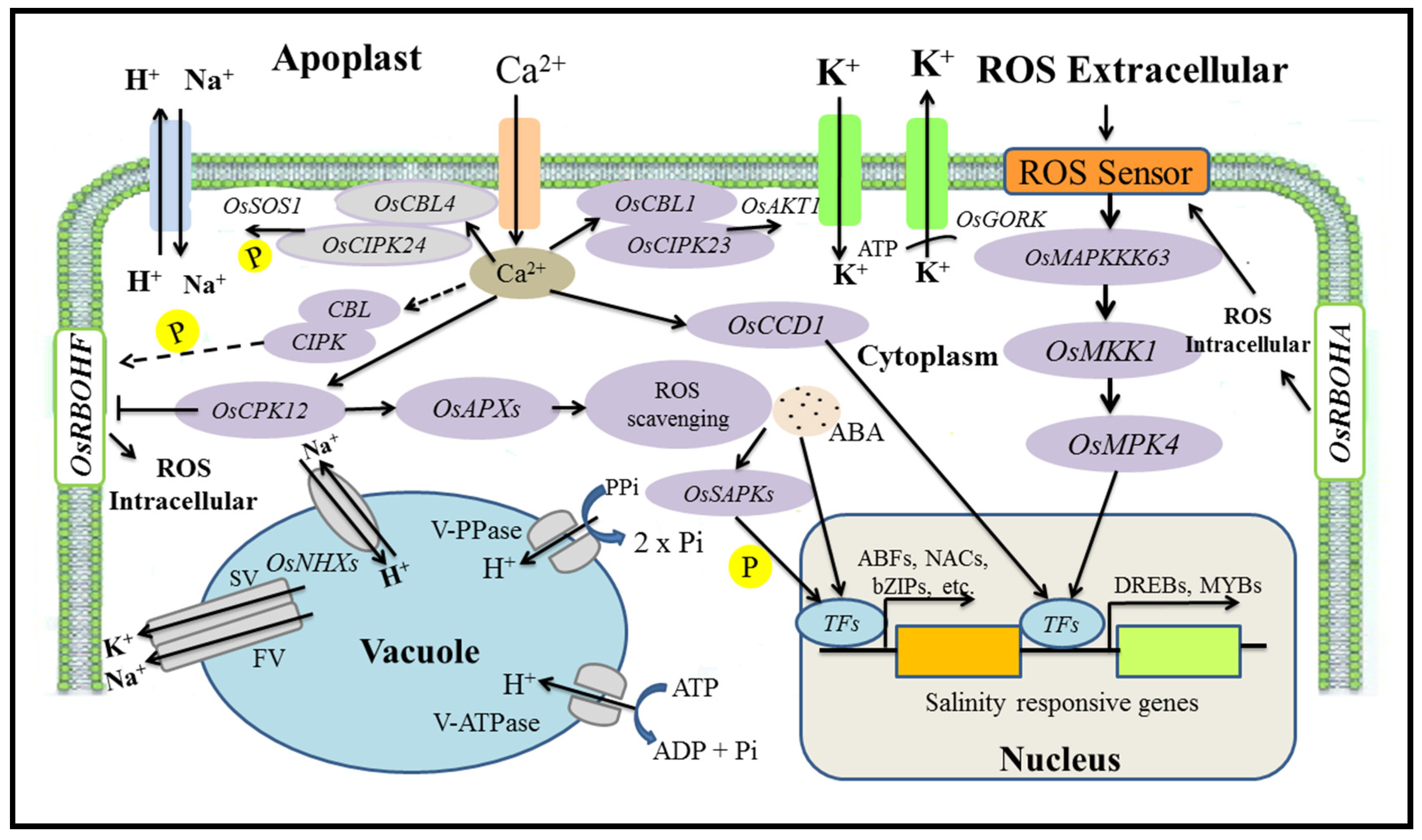
5. Mechanisms for the Regulation of Salt Tolerance Genes
5.1. Salinity Tolerance Mechanisms
5.2. Traditional Plant Breeding
6. Salt Responsive Genes Present in Halophytes
7. Promoters for Salt-Responsive Halophytic Genes
8. Transgenic Approach for Engineered Plants Having Enhanced Salt Tolerance
9. Development of Salt Tolerant Glycophytes using Halophytic Salt Tolerance Genes
10. MicroRNAs (miRNA), a New Target for Improving Plant Tolerance to Salt Stress
11. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abideen, Z.; Qasim, M.; Hussain, T.; Rasheed, A.; Gul, B.; Koyro, H.W.; Ansari, R.; Khan, M.A. Salinity improves growth, photosynthesis and bioenergy characteristics of Phragmites karka. Crop Pasture Sci. 2018, 69, 944–953. [Google Scholar] [CrossRef]
- Ehsen, S.; Abideen, Z.; Rizvi, R.F.; Gulzar, S.; Aziz, I.; Gul, B.; Khan, M.A.; Ansari, R. Ecophysiological adaptations and anti-nutritive status of sustainable cattle feed Haloxylon stocksii under saline conditions. Flora 2019, 257, 151425. [Google Scholar] [CrossRef]
- Shoukat, E.; Ahmed, M.Z.; Abideen, Z.; Azeem, M.; Ibrahim, M.; Gul, B.; Khan, M.A. Short and long term salinity induced differences in growth and tissue specific ion regulation of Phragmites karka. Flora 2020, 263, 151550. [Google Scholar] [CrossRef]
- Hussain, M.I.; Abideen, Z.; Qureshi, A.S. Soil degradation, resilience, restoration and sustainable use. In Sustainable Agriculture Reviews; Springer: Cham, Switzerland, 2021; Volume 52, pp. 335–365. [Google Scholar] [CrossRef]
- Abideen, Z.; Koyro, H.W.; Huchzermeyer, B.; Ahmed, M.Z.; Gul, B.; Khan, M.A. Moderate salinity stimulates growth and photosynthesis of Phragmites karka by water relations and tissue specific ion regulation. Environ. Exp. Bot. 2014, 105, 70–76. [Google Scholar] [CrossRef]
- Shoukat, E.; Abideen, Z.; Ahmed, M.Z.; Gulzar, S.; Nielsen, B.L. Changes in growth and photosynthesis linked with intensity and duration of salinity in Phragmites karka. Environ. Exp. Bot. 2019, 162, 504–514. [Google Scholar] [CrossRef]
- Munir, N.; Hasnain, M.; Roessner, U.; Abideen, Z. Strategies in improving plant salinity resistance and use of salinity resistant plants for economic sustainability. Crit. Rev. Environ. Sci. Technol. 2021, 1–47. [Google Scholar] [CrossRef]
- Abideen, Z.; Koyro, H.W.; Huchzermeyer, B.; Ahmed, M.; Zulfiqar, F.; Egan, T.; Khan, M.A. Phragmites karka plants adopt different strategies to regulate photosynthesis and ion flux in saline and water deficit conditions. Plant Biosyst.-An Int. J. Deal. All Asp. Plant Biol. 2021, 155, 524–534. [Google Scholar] [CrossRef]
- Lohani, N.; Jain, D.; Singh, M.B.; Bhalla, P.L. Engineering multiple abiotic stress tolerance in Canola, Brassica napus. Front. Plant Sci. 2020, 11, 3–11. [Google Scholar] [CrossRef]
- Rajaee, B.S.; Iranbakhsh, A.; Ebadi, M.; Majd, A.; Ardebili, Z.O. Red elemental selenium nanoparticles mediated substantial variations in growth, tissue differentiation, metabolism, gene transcription, epigenetic cytosine DNA methylation, and callogenesis in Bittermelon (Momordica charantia); an In vitro experiment. PLoS ONE 2020, 15, e0235556. [Google Scholar]
- Derbali, W.; Manaa, A.; Spengler, B.; Goussi, R.; Abideen, Z.; Ghezellou, P.; Abdelly, C.; Forreiter, C.; Koyro, H.W. Comparative proteomic approach to study the salinity effect on the growth of two contrasting quinoa genotypes. Plant Physiol. Biochem. 2021, 163, 215–229. [Google Scholar] [CrossRef]
- Tang, W. Heterologous expression of transcription factor ATWRKY57 alleviates salt stress-induced oxidative damage. Open Biotechnol. J. 2018, 12, 204–218. [Google Scholar] [CrossRef] [Green Version]
- Yokotani, N.; Higuchi, M.; Kondou, Y.; Ichikawa, T.; Iwabuchi, M.; Hirochika, H.; Matsui, M.; Oda, K. A novel chloroplast protein, CEST induces tolerance to multiple environmental stresses and reduces photooxidative damage in transgenic Arabidopsis. J. Exp. Bot. 2011, 62, 557–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyaya, C.P.; Venkatesh, J.; Gururani, M.A.; Asnin, L.; Sharma, K.; Ajappala, H.; Park, S.W. Transgenic potato overproducing l-ascorbic acid resisted an increase in methylglyoxal under salinity stress via maintaining higher reduced glutathione level and glyoxalase enzyme activity. Biotechnol. Lett. 2011, 33, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Hossain, M.A.; Fujita, M. Selenium-induced up-regulation of the antioxidant defense and methylglyoxal detoxification system reduces salinity-induced damage in Rapeseed seedlings. Biol. Trace Elem. Res. 2011, 143, 1704–1721. [Google Scholar] [CrossRef]
- Auffan, M.; Rose, J.; Bottero, J.Y.; Lowry, G.V.; Jolivet, J.P.; Wiesner, M.R. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. 2009, 4, 634–641. [Google Scholar] [CrossRef]
- Maurer-Jones, M.A.; Gunsolus, I.L.; Murphy, C.J.; Haynes, C.L. Toxicity of engineered nanoparticles in the environment. Anal. Chem. 2013, 85, 3036–3049. [Google Scholar] [CrossRef] [Green Version]
- Radad, K.; Al-Shraim, M.; Moldzio, R.; Rausch, W.D. Recent advances in benefits and hazards of engineered nanoparticles. Environ. Toxicol. Pharmacol. 2012, 34, 661–672. [Google Scholar] [CrossRef]
- Chavali, M.S.; Nikolova, M.P. Metal oxide nanoparticles and their applications in nanotechnology. SN Appl. Sci. 2019, 1, 3199. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Dominguez, M.; Boutonnet, M.; Solans, C. A novel approach to metal and metal oxide nanoparticle synthesis: The oil-in-water microemulsion reaction method. J. Nanopart. Res. 2009, 11, 1823–1829. [Google Scholar] [CrossRef]
- Rana, A.; Yadav, K.; Jagadevan, S. A comprehensive review on green synthesis of nature-inspired metal nanoparticles: Mechanism, application and toxicity. J. Clean. Prod. 2020, 272, 122880. [Google Scholar] [CrossRef]
- Dumont, E.; Johnson, A.C.; Keller, V.D.; Williams, R.J. Nano silver and nano zinc-oxide in surface waters–exposure estimation for europe at high spatial and temporal resolution. Environ. Pollut. 2015, 196, 341–349. [Google Scholar] [CrossRef] [Green Version]
- Levard, C.; Hotze, E.M.; Lowry, G.V.; Brown, G.E., Jr. Environmental transformations of silver nanoparticles: Impact on stability and toxicity. Environ. Sci. Technol. 2012, 46, 6900–6914. [Google Scholar] [CrossRef]
- Barrios, A.C.; Rico, C.M.; Trujillo-Reyes, J.; Medina-Velo, I.A.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Effects of uncoated and citric acid coated cerium oxide nanoparticles, bulk cerium oxide, cerium acetate, and citric acid on Tomato plants. Sci. Total Environ. 2016, 563, 956–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardea-Torresdey, J.L.; Rico, C.M.; White, J.C. Trophic transfer, transformation, and impact of engineered nanomaterials in terrestrial environments. Environ. Sci. Technol. 2014, 48, 2526–2540. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, K.N.; Peters, R.J.; Klumpp, E.; Bohme, S.; Van der Ploeg, M.; Ritsema, C.; Geissen, V. Silver nanoparticles in soil: Aqueous extraction combined with single-particle ICP-MS for detection and characterization. Environ. Nanotechnol. Monit. Manag. 2017, 7, 24–33. [Google Scholar] [CrossRef]
- Tripathi, D.K.; Singh, S.; Singh, S.; Srivastava, P.K.; Singh, V.P.; Singh, S.; Prasad, S.M.; Singh, P.K.; Dubey, N.K.; Pandey, A.C. Nitric oxide alleviates silver nanoparticles (Ag-NPs)-induced phytotoxicity in Pisum sativum seedlings. Plant Physiol. Biochem. 2017, 110, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lombi, E.; Zhao, F.J.; Kopittke, P.M. Nanotechnology: A new opportunity in plant sciences. Trends Plant Sci. 2016, 21, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Shekhawat, G.S. Phytotoxicity and oxidative stress perspective of two selected nanoparticles in Brassica juncea. 3 Biotech 2016, 6, 244. [Google Scholar] [CrossRef] [Green Version]
- Taran, N.; Batsmanova, L.; Kovalenko, M.; Okanenko, A. Impact of metal nanoform colloidal solution on the adaptive potential of plants. Nanoscale Res. Lett. 2016, 1, 11–89. [Google Scholar] [CrossRef] [Green Version]
- Zohra, E.; Ikram, M.; Omar, A.A.; Hussain, M.; Satti, S.H.; Raja, N.I.; Ehsan, M. Potential applications of biogenic selenium nanoparticles in alleviating biotic and abiotic stresses in plants: A comprehensive insight on the mechanistic approach and future perspectives. Green Process. Synth. 2021, 10, 456–475. [Google Scholar] [CrossRef]
- Munir, N.; Hanif, M.; Dias, D.A.; Abideen, A. The role of halophytic nanoparticles towards the remediation of degraded and saline agricultural lands. Environ. Sci. Pollut. Res. 2021, 28, 60383–60405. [Google Scholar] [CrossRef]
- Sharma, P.; Bhatt, D.; Zaidi, M.; Saradhi, P.P.; Khanna, P.; Arora, S. Silver nanoparticle-mediated enhancement in growth and antioxidant status of Brassica juncea. Appl. Biochem. Biotechnol. 2012, 167, 2225–2233. [Google Scholar] [CrossRef]
- Gunjan, B.; Zaidi, M. Impact of gold nanoparticles on physiological and biochemical characteristics of Brassica juncea. J. Plant Biochem. Physiol. 2014, 2, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Mazumdar, H.; Ahmed, G. Phytotoxicity effect of silver nanoparticles on Oryza sativa. Int. J. ChemTech Res. 2011, 3, 1494–1500. [Google Scholar]
- Mirzajani, F.; Askari, H.; Hamzelou, S.; Schober, Y.; Römpp, A.; Ghassempour, A.; Spengler, B. Proteomics study of silver nanoparticles toxicity on Oryza sativa L. Ecotoxicol. Environ. Saf. 2014, 108, 335–339. [Google Scholar] [CrossRef]
- Khodakovskaya, M.; Dervishi, E.; Mahmood, M.; Xu, Y.; Li, Z.; Watanabe, F.; Biris, A.S. Carbon nanotubes are able to penetrate plant seed coat and dramatically affect seed germination and plant growth. ACS Nano 2009, 3, 3221–3227. [Google Scholar] [CrossRef]
- Tawfik, M.; Mohamed, M.H.; Sadak, M.S.; Thalooth, A.T. Iron oxide nanoparticles effect on growth, physiological traits and nutritional contents of Moringa oleifera grown in saline environment. Bull. Natl. Res. Cent. 2021, 45, 177. [Google Scholar] [CrossRef]
- Rodríguez-Celma, J.; Lattanzio, G.; Grusak, M.A.; Abadía, A.; Abadía, J.; López-Millán, A.F. Root responses of Medicago truncatula plants grown in two different iron deficiency conditions: Changes in root protein profile and riboflavin biosynthesis. J. Proteome Res. 2011, 10, 2590–2601. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.J.; Lindermayr, C.; Toerne, C.; Fink-Straube, C.; Durner, J.; Bauer, P. Iron and fer-like iron deficiency-induced transcription factor-dependent regulation of proteins and genes in Arabidopsis thaliana roots. Proteomics 2015, 15, 3030–3047. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, Y.K. Effect of nanoscale Fe3O4, TiO2 and carbon particles on cucumber seed germination. J. Environ. Sci. Health 2011, 46, 1732–1735. [Google Scholar] [CrossRef]
- Zhu, H.; Han, J.; Xiao, J.Q.; Jin, Y. Uptake, translocation, and accumulation of manufactured iron oxide nanoparticles by Pumpkin plants. J. Environ. Monit. 2008, 10, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, L.; Oukarroum, A.; Taher, L.B.; Smiri, L.S.; Abdelmelek, H.; Dewez, D. Effects of superparamagnetic iron oxide nanoparticles on photosynthesis and growth of the aquatic plant Lemna gibba. Arch. Environ. Contam. Toxicol. 2015, 68, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Shankramma, K.; Yallappa, S.; Shivanna, M.B.; Manjanna, J. Fe2O3 magnetic nanoparticles to enhance S. lycopersicum (tomato) plant growth and their biomineralization. Appl. Nanosci. 2016, 6, 983–990. [Google Scholar] [CrossRef] [Green Version]
- Rui, M.; Ma, C.; Hao, Y.; Guo, J.; Rui, Y.; Tang, X.; Zhao, Q.; Fan, X.; Zhang, Z.; Hou, T.; et al. Iron oxide nanoparticles as a potential iron fertilizer for peanut (Arachis hypogaea). Fron. Plant Sci. 2016, 7, 815. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Funct. Gen. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Hounsome, N.; Hounsome, B.; Tomos, D.; Edwards, J.G. Plant metabolites and nutritional quality of vegetables. J. Food Sci. 2008, 73, R48–R65. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112–121. [Google Scholar] [CrossRef] [Green Version]
- Attia-Ismail, S.A. Plant secondary metabolites of halophytes and salt tolerant plants. In Halophytic and Salt-Tolerant Feedstuffs; CRC Press: Boca Raton, FL, USA, 2015; Volume 1, pp. 127–142. [Google Scholar] [CrossRef]
- Guo, J.; Li, Y.; Han, G.; Song, J.; Wang, B. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 2018, 45, 350–361. [Google Scholar] [CrossRef]
- Yao, W.; Zhao, K.; Cheng, Z.; Li, X.; Zhou, B.; Jiang, T. Transcriptome analysis of Poplar under salt stress and over-expression of transcription factor NAC57 gene confers salt tolerance in transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 11–21. [Google Scholar] [CrossRef]
- Rao, A.Q.; Ud Din, S.; Akhtar, S.; Sarwar, M.B.; Ahmed, M.; Rashid, B.; Khan, M.A.U.; Qaisar, U.; Shahid, A.A.; Nasir, I.A. Genomics of salinity tolerance in plants. In Plant Genomic; InTechOpen: London, UK, 2016; pp. 273–299. [Google Scholar]
- Johnson, R.R.; Wagner, R.L.; Verhey, S.D.; Walker-Simmons, M.K. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002, 130, 837–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zhang, S.; Yuan, X.; Chen, C.; Wang, X.F.; Hao, Y.J. Genome-wide analysis and identification of stress-responsive genes of the NAM–ATAF1, 2–CUC2 transcription factor family in Apple. Plant Physiol. Biochem. 2013, 71, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, G.; Jia, J.; Liu, X.; Kong, X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2012, 63, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Negra, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, L.; Wang, H.; Zhang, L.; Wang, F.; Yu, D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 2013, 74, 730–745. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.; Mieulet, D.; Hubberten, H.M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; San Segundo, B.; Guiderdoni, E.; Schippers, J.H. Salt-responsive ERF1 regulates reactive oxygen species–dependent signaling during the initial response to salt stress in Rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef] [Green Version]
- Steinfeld, B.; Scott, J.; Vilander, G.; Marx, L.; Quirk, M.; Lindberg, J.; Koerner, K. The role of lean process improvement in implementation of evidence-based practices in behavioral health care. J. Behav. Health Serv. Res. 2015, 42, 504–518. [Google Scholar] [CrossRef]
- Kakali, M.; Roy, C.; Bhaskar, G.; Sudhiranjan, G.; Sengupta, D. An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of Indica Rice. BMC Plant Biol. 2006, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, K.; Sairam, R.K.; Bhattacharya, R. Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiol. Biochem. 2012, 51, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.H.; Zhang, X.; Chung, M.S.; Lee, D.J.; Woo, Y.M.; Cheong, H.S.; Kim, C.S. A putative novel transcription factor, AtSKIP, is involved in abscisic acid signalling and confers salt and osmotic tolerance in Arabidopsis. New Phytol. 2010, 185, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17.0 and OSHSP23.7 enhances drought and salt tolerance in Rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Song, N.H.; Ahn, Y.J. DcHsp17.7, a small heat shock protein in Carrot, is tissue-specifically expressed under salt stress and confers tolerance to salinity. New Biotechnol. 2011, 28, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Liu, X.; Deng, H.; Shen, S. Over-expression of JcDREB, a putative AP2/EREBP domain-containing transcription factor gene in woody biodiesel plant Jatropha curcas, enhances salt and freezing tolerance in transgenic Arabidopsis thaliana. Plant Sci. 2011, 181, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Sottosanto, J.B.; Saranga, Y.; Blumwald, E. Impact of AtNHX1, a vacuolar Na+/H+ antiporter, upon gene expression during short-and long-term salt stress in Arabidopsis thaliana. BMC Plant Biol. 2007, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Ding, X.; Chang, T.; Wang, Z.; Liu, R.; Zeng, X.; Cai, Y.; Zhu, Y. Overexpression of a vesicle trafficking gene, OsRab7, enhances salt tolerance in Rice. Sci. World J. 2014, 2014, 483526. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Wang, W.; Sun, J.; Ding, M.; Zhao, R.; Deng, S.; Wang, F.; Hu, Y.; Wang, Y.; Lu, Y. Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic Tobacco plants. J. Exp. Bot. 2013, 64, 4225–4238. [Google Scholar] [CrossRef] [Green Version]
- Kurotani, K.I.; Hayashi, K.; Hatanaka, S.; Toda, Y.; Ogawa, D.; Ichikawa, H.; Ishimaru, Y.; Tashita, R.; Suzuki, T.; Ueda, M. Elevated levels of CYP94 family gene expression alleviate the Jasmonate response and enhance salt tolerance in Rice. Plant Cell Physiol. 2015, 56, 779–789. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Zhang, M.; Zhang, J.; Duan, L.; Li, Z. SOS1 gene overexpression increased salt tolerance in transgenic Tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 2012, 169, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Bhauso, T.D.; Thankappan, R.; Kumar, A.; Mishra, G.P.; Dobaria, J.R.; Rajam, M.V. Over-expression of bacterial MTLD gene confers enhanced tolerance to salt-stress and water-deficit stress in transgenic Peanut (Arachis hypogaea) through accumulation of mannitol. Aust. J. Crop Sci. 2014, 8, 413–424. [Google Scholar]
- Zhou, W.; Li, Y.; Zhao, B.C.; Ge, R.C.; Shen, Y.Z.; Wang, G.; Huang, Z.J. Overexpression of TaSTRG gene improves salt and drought tolerance in Rice. J. Plant Physiol. 2009, 166, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Lawson, S.S.; Michler, C.H. Overexpression of AtSTO1 leads to improved salt tolerance in Populus tremula × P. alba. Transgenic Res. 2014, 23, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Mangrauthia, S.K.; Agarwal, S.; Sailaja, B.; Madhav, M.S.; Voleti, S. MicroRNAs and their role in salt stress response in plants. In Salt Stress in Plants, 3rd ed.; Springer: Cham, Switzerland, 2013; pp. 15–46. [Google Scholar]
- Lu, S.; Sun, Y.H.; Chiang, V.L. Stress-responsive microRNAs in Populus. Plant J. 2008, 55, 131–151. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golldack, D.; Lüking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef]
- Popova, O.V.; Yang, O.; Dietz, K.J.; Golldack, D. Differential transcript regulation in Arabidopsis thaliana and the halotolerant lobularia maritima indicates genes with potential function in plant salt adaptation. Gene 2008, 423, 142–148. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Silva, P.; Gerós, H. Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange. Plant Signal. Behav. 2009, 4, 718–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flowers, T.; Koyama, M.; Flowers, S.; Sudhakar, C.; Singh, K.; Yeo, A. QTL: Their place in engineering tolerance of Rice to salinity. J. Exp. Bot. 2000, 51, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Alam, R.; Sazzad-ur-Rahman, M.; Seraj, Z.I.; Thomson, M.J.; Ismail, A.M.; Tumimbang-Raiz, E.; Gregorio, G.B. Investigation of seedling-stage salinity tolerance QTLS using backcross lines derived from Oryza sativa l. Pokkali. Plant Breed. 2011, 130, 430–437. [Google Scholar] [CrossRef]
- Frary, A.; Göl, D.; Keleş, D.; Ökmen, B.; Pınar, H.; Şığva, H.Ö.; Yemenicioğlu, A.; Doğanlar, S. Salt tolerance in Solanum pennellii: Antioxidant response and related QTL. BMC Plant Biol. 2010, 10, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Chen, Z.Z.; Zhou, X.F.; Yin, H.B.; Li, X.; Xin, X.F.; Hong, X.H.; Zhu, J.K.; Gong, Z. Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Ventura, Y.; Eshel, A.; Pasternak, D.; Sagi, M. The development of halophyte-based agriculture: Past and present. Ann. Bot. 2015, 115, 529–540. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Himabindu, Y.; Chakradhar, T.; Reddy, M.C.; Kanygin, A.; Redding, K.E.; Chandrasekhar, T. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 2016, 124, 39–63. [Google Scholar] [CrossRef] [Green Version]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant salt stress: Adaptive responses, tolerance mechanism and bioengineering for salt tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Udawat, P.; Mishra, A.; Jha, B. Heterologous expression of an uncharacterized universal stress protein gene (SbUSP) from the extreme halophyte, Salicornia brachiata, which confers salt and osmotic tolerance to E. coli. Gene 2014, 536, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Dinakar, C. Balancing salinity stress responses in halophytes and non-halophytes: A comparison between Thellungiella and Arabidopsis thaliana. Funct. Plant Biol. 2013, 40, 819–831. [Google Scholar] [CrossRef]
- Wu, H.J.; Zhang, Z.; Wang, J.Y.; Oh, D.H.; Dassanayake, M.; Liu, B.; Huang, Q.; Sun, H.X.; Xia, R.; Wu, Y. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. USA 2012, 109, 12219–12224. [Google Scholar] [CrossRef] [Green Version]
- Taji, T.; Seki, M.; Satou, M.; Sakurai, T.; Kobayashi, M.; Ishiyama, K.; Narusaka, Y.; Narusaka, M.; Zhu, J.K.; Shinozaki, K. Comparative genomics in salt tolerance between Arabidopsis and arabidopsis-related halophyte salt stress using Arabidopsis microarray. Plant Physiol. 2004, 135, 1697–1709. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.E.; Li, Y.; Labbe, A.; Guevara, D.; Nuin, P.; Whitty, B.; Diaz, C.; Golding, G.B.; Gray, G.R.; Weretilnyk, E.A. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006, 140, 1437–1450. [Google Scholar] [CrossRef] [Green Version]
- Rigó, G.; Valkai, I.; Faragó, D.; Kiss, E.; Van Houdt, S.; Van de Steene, N.; Hannah, M.A.; Szabados, L. Gene mining in halophytes: Functional identification of stress tolerance genes in Lepidium crassifolium. Plant, Cell Environ. 2016, 39, 167–181. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.; Patel, M.K.; Mishra, A.; Jha, B. In planta transformed Cumin (Cuminum cyminum l.) plants, overexpressing the SbNHX1 gene showed enhanced salt endurance. PLoS ONE 2016, 11, e0159349. [Google Scholar]
- Jha, B.; Singh, N.P.; Mishra, A. Proteome profiling of seed storage proteins reveals the nutritional potential of Salicornia brachiata Roxb., an extreme halophyte. J. Agric. Food Chem. 2012, 60, 4320–4326. [Google Scholar] [CrossRef]
- Garg, R.; Verma, M.; Agrawal, S.; Shankar, R.; Majee, M.; Jain, M. Deep transcriptome sequencing of wild halophyte Rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res. 2014, 21, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.H.; Su, Q.; An, L.J.; Wu, S. Characterization and expression of a vacuolar Na+/H+ antiporter gene from the monocot halophyte Aeluropus littoralis. Plant Physiol. Biochem. 2008, 46, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.X.; Zhu, Z.Q.; Chang, F.Q.; Li, Y.X. Transformation of Tomato with the BADH gene from Atriplex improves salt tolerance. Plant Cell Rep. 2002, 21, 141–146. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Minor, B.L.; Elliott, V.; Fernandez, M.; O’Neal, L.; McLeod, L.; Delacqua, G.; Delacqua, F.; Kirby, J. The REDCap consortium: Building an international community of software platform partners. J. Biomed. Inform. 2019, 95, 15–32. [Google Scholar] [CrossRef]
- Singh, N.; Mishra, A.; Jha, B. Over-expression of the peroxisomal ascorbate peroxidase (SbpAPX) gene cloned from halophyte Salicornia brachiata confers salt and drought stress tolerance in transgenic tobacco. Mar. Biotechnol. 2014, 16, 321–332. [Google Scholar] [CrossRef]
- Singh, N.; Mishra, A.; Jha, B. Ectopic over-expression of peroxisomal ascorbate peroxidase (SbpAPX) gene confers salt stress tolerance in transgenic Peanut (Arachis hypogaea). Gene 2014, 547, 119–125. [Google Scholar] [CrossRef]
- Li, W.; Wang, D.; Jin, T.; Chang, Q.; Yin, D.; Xu, S.; Liu, B.; Liu, L. The vacuolar Na+/H+ antiporter gene SSNHX1 from the halophyte Salsola soda confers salt tolerance in transgenic Alfalfa (Medicago sativa L.). Plant Mol. Biol. Rep. 2011, 29, 278–290. [Google Scholar] [CrossRef]
- Wu, W.; Su, Q.; Xia, X.; Wang, Y.; Luan, Y.; An, L. The Suaeda liaotungensis KITAG betaine aldehyde dehydrogenase gene improves salt tolerance of transgenic maize mediated with minimum linear length of DNA fragment. Euphytica 2008, 159, 17–25. [Google Scholar] [CrossRef]
- Qiu, W.; Liu, M.; Qiao, G.; Jiang, J.; Xie, L.; Zhuo, R. An isopentyl transferase gene driven by the stress-inducible RD29A promoter improves salinity stress tolerance in transgenic Tobacco. Plant Mol. Biol. Rep. 2012, 30, 519–528. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, H. Salt and paraquat stress tolerance results from co-expression of the Suaeda salsa glutathione s-transferase and catalase in transgenic Rice. Plant Cell Tissue Organ Cult. 2006, 86, 349–358. [Google Scholar] [CrossRef]
- Guo, S.; Yin, H.; Zhang, X.; Zhao, F.; Li, P.; Chen, S.; Zhao, Y.; Zhang, H. Molecular cloning and characterization of a vacuolar H+-pyrophos-phatase gene, SsVP, from the halophyte Suaeda salsa and its overexpression increases salt and drought tolerance of Arabidopsis. Plant Mol. Biol. 2006, 60, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.H.; Leidi, E.; Zhang, Q.; Hwang, S.M.; Li, Y.; Quintero, F.J.; Jiang, X.; D’Urzo, M.P.; Lee, S.Y.; Zhao, Y. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009, 151, 210–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.L.; Chen, A.P.; Zhong, N.Q.; Liu, N.; Wu, X.M.; Wang, F.; Yang, C.L.; Romero, M.F.; Xia, G.X. The Thellungiella salsuginea tonoplast aquaporin TsTIP1; 2 functions in protection against multiple abiotic stresses. Plant Cell Physiol. 2014, 55, 148–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, F.; Xu, J.; Shi, J.; Li, H.; Li, B. Molecular cloning and characterization of a novel glyoxalase I gene TaGly I in Wheat (Triticum aestivum L.). Mol. Biol. Rep. 2010, 37, 729–737. [Google Scholar] [CrossRef]
- Baisakh, N.; RamanaRao, M.V.; Rajasekaran, K.; Subudhi, P.; Janda, J.; Galbraith, D.; Vanier, C.; Pereira, A. Enhanced salt stress tolerance of Rice plants expressing a vacuolar H+-ATPase subunit c1 (SaVHAc1) gene from the halophyte grass Spartina alterniflora löisel. Plant Biotechnol. J. 2012, 10, 453–464. [Google Scholar] [CrossRef]
- Wu, S.; Su, Q.; An, L. Isolation of choline monooxygenase (CMO) gene from Salicornia europaea and enhanced salt tolerance of transgenic Tobacco with CMO genes. Ind. J. Biochem. Biophys. 2010, 47, 298–305. [Google Scholar]
- Yin, X.Y.; Yang, A.F.; Zhang, K.W.; Zhang, J.R. Production and analysis of transgenic maize with improved salt tolerance by the introduction of AtNHX1 gene. Acta Bot. Sin.-Engl. Ed. 2004, 46, 854–861. [Google Scholar]
- Ohta, M.; Hayashi, Y.; Nakashima, A.; Tsunetomi, N.; Hamada, A.; Tanaka, A.; Nakamura, T.; Hayakawa, T. Salt tolerance of Rice is conffered by introduction of a Na+/H+ antiporter gene from atriplex gmelini. Plant Cell Physiol. 2002, 279, 282. [Google Scholar] [CrossRef] [Green Version]
- Taji, T.; Komatsu, K.; Katori, T.; Kawasaki, Y.; Sakata, Y.; Tanaka, S.; Kobayashi, M.; Toyoda, A.; Seki, M.; Shinozaki, K. Comparative genomic analysis of 1047 completely sequenced cDNA’s from an arabidopsis-related model halophyte, Thellungiella halophila. BMC Plant Biol. 2010, 10, 261. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, V.; Patel, M.K.; Chaturvedi, A.K.; Mishra, A.; Jha, B. Functional characterization of the Tau class glutathione-s-transferases gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic stress. PLoS ONE 2016, 11, e0148494. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Zhao, Y.; Luo, D.; Zhang, H. Isolating the promoter of a stress-induced gene encoding betaine aldehyde dehydrogenase from the halophyte Atriplex centralasiatica Iljin. Biochim. Biophys. Acta-Gene Struct. Express 2002, 1577, 452–456. [Google Scholar] [CrossRef]
- Li, Q.L.; Xie, J.H.; Ma, X.Q.; Li, D. Molecular cloning of phosphoethanolamine n-methyltransferase (PEAMT) gene and its promoter from the halophyte Suaeda liaotungensis and their response to salt stress. Acta Physiol. Plant 2016, 38, 39–45. [Google Scholar] [CrossRef]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Schaeffer, H.J.; Forsthoefel, N.R.; Cushman, J.C. Identification of enhancer and silencer regions involved in salt-responsive expression of crassulacean acid metabolism (CAM) genes in the facultative halophyte Mesembryanthemum crystallinum. Plant Mol. Biol. 1995, 28, 205–218. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, W.; Xu, Y.P.; Cao, J.Y.; Braam, J.; Ca, X.Z. Genome-wide identification and functional analyses of Calmodulin genes in Solanaceous species. BMC Plant Biol. 2013, 13, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saad, R.B.; Romdhan, W.B.; Zouari, N.; Azaza, J.; Mieulet, D.; Verdeil, J.L.; Guiderdoni, E.; Hassairi, A. Promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs developmental-regulated, stress-inducible, and organ-specific gene expression in transgenic Tobacco. Transgenic Res. 2011, 20, 1003–1018. [Google Scholar] [CrossRef]
- Ben-Saad, R.; Meynard, D.; Ben-Romdhane, W.; Mieulet, D.; Verdeil, J.L.; Al-Doss, A.; Guiderdoni, E.; Hassairi, A. The promoter of the ALSAP gene from the halophyte grass Aeluropus littoralis directs a stress-inducible expression pattern in transgenic Rice plants. Plant Cell Rep. 2015, 34, 1791–1806. [Google Scholar] [CrossRef]
- Sun, Q.; Gao, F.; Zhao, L.; Li, K.; Zhang, J. Identification of a new 130 bp cis-acting element in the TSVP1 promoter involved in the salt stress response from Thellungiella halophila. BMC Plant Biol. 2010, 10, 90. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Yu, Y.; Xia, X.; Yin, W. Identification and functional characterisation of the promoter of the calcium sensor gene CBL1 from the xerophyte Ammopiptanthus mongolicus. BMC Plant Biol. 2010, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, V.; Chaturvedi, A.K.; Mishra, A.; Jha, B. The transcriptional regulatory mechanism of the peroxisomal ascorbate peroxidase (pAPX) gene cloned from an extreme halophyte, Salicornia brachiata. Plant Cell Physiol. 2014, 55, 201–217. [Google Scholar] [CrossRef] [Green Version]
- James, C. Global Status of Commercialized Biotech/GM Crops, 2nd ed; International Service for the Acquisition of Agri-Biotech Applications (ISAAA): Ithaca, NY, USA, 2007; pp. 22–45. [Google Scholar]
- Ashraf, M.; Athar, H.; Harris, P.; Kwon, T. Some prospective strategies for improving crop salt tolerance. Adv. Agron. 2008, 97, 45–110. [Google Scholar] [CrossRef]
- Goel, D.; Singh, A.K.; Yadav, V.; Babbar, S.B.; Murata, N.; Bansal, K.C. Transformation of Tomato with a bacterial CODA gene enhances tolerance to salt and water stresses. J. Plant Physiol. 2011, 168, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Hirji, R.; Zhang, L.; He, C.; Selvaraj, G.; Wu, R. Evaluation of the stress-inducible production of choline oxidase in transgenic Rice as a strategy for producing the stress-protectant glycine betaine. J. Exp. Bot. 2006, 57, 1129–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortina, C.; Culiáñez-Macià, F.A. Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 2005, 169, 75–82. [Google Scholar] [CrossRef]
- Almeida, A.M.; Villalobos, E.; Araújo, S.S.; Leyman, B.; Van Dijck, P.; Alfaro-Cardoso, L.; Fevereiro, P.S.; Torné, J.M.; Santos, D.M. Transformation of tobacco with an Arabidopsis thaliana gene involved in trehalose biosynthesis increases tolerance to several abiotic stresses. Euphytica 2005, 146, 165–176. [Google Scholar] [CrossRef]
- Abebe, T.; Guenzi, A.C.; Martin, B.; Cushman, J.C. Tolerance of mannitol-accumulating transgenic Wheat to water stress and salinity. Plant Physiol. 2003, 131, 1748–1755. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Tao, R.; Miura, K.; Dandekar, A.M.; Sugiura, A. Transformation of Japanese Persimmon (Diospyros kaki thunb.) with Apple cDNA encoding NADP-dependent sorbitol-6-phosphate dehydrogenase. Plant Sci. 2001, 160, 837–845. [Google Scholar] [CrossRef]
- Hmida-Sayari, A.; Gargouri-Bouzid, R.; Bidani, A.; Jaoua, L.; Savouré, A.; Jaoua, S. Overexpression of δ1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci. 2005, 169, 746–752. [Google Scholar] [CrossRef]
- Zhang, H.X.; Hodson, J.N.; Williams, J.P.; Blumwald, E. Engineering salt-tolerant Brassica plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. USA 2001, 98, 12832–12836. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zuo, K.; Wu, W.; Song, J.; Sun, X.; Lin, J.; Li, X.; Tang, K. Expression of a novel antiporter gene from Brassica napus resulted in enhanced salt tolerance in transgenic Tobacco plants. Biol. Plant 2004, 48, 509–515. [Google Scholar] [CrossRef]
- Singla-Pareek, S.; Reddy, M.; Sopory, S. Genetic engineering of the glyoxalase pathway in Tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. USA 2003, 100, 14672–14677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.Q.; Chen, M.; Xu, Z.S.; Zhao, C.P.; Li, L.; Xu, H.J.; Tang, Y.M.; Zhao, X.; Ma, Y.Z. The Soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 2011, 75, 537–553. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Park, J.B.; Cho, Y.J.; Jung, C.; Seo, H.S.; Park, S.K.; Nahm, B.H.; Song, J.T. Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol. Cells 2010, 30, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Montero-Barrientos, M.; Hermosa, R.; Cardoza, R.E.; Gutierrez, S.; Nicolas, C.; Monte, E. Transgenic expression of the Trichoderma harzianum HSP70 gene increases Arabidopsis resistance to heat and other abiotic stresses. J. Plant Physiol. 2010, 167, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xi, D.; Li, S.; Gao, Z.; Zhao, S.; Shi, J.; Wu, C.; Guo, X. A Cotton group c map kinase gene, GhMPK2, positively regulates salt and drought tolerance in Tobacco. Plant Mol. Biol. 2011, 77, 17–31. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. Ros homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Britto, D.T. Sodium transport in plants: A critical review. New Phytol. 2011, 189, 54–81. [Google Scholar] [CrossRef]
- Volkov, V. Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front. Plant Sci. 2015, 6, 873–884. [Google Scholar] [CrossRef]
- Mishra, A.; Tanna, B. Halophytes: Potential resources for salt stress tolerance genes and promoters. Front. Plant Sci. 2017, 8, 829. [Google Scholar] [CrossRef]
- Lan, T.; Duan, Y.; Wang, B.; Zhou, Y.; Wu, W. Molecular cloning and functional characterization of a Na+/H+ antiporter gene from halophyte Spartina anglica. Turk. J. Agric. For. 2011, 35, 535–543. [Google Scholar]
- Patel, M.K.; Joshi, M.; Mishra, A.; Jha, B. Ectopic expression of SbNHX1 gene in transgenic Castor (Ricinus communis l.) enhances salt stress by modulating physiological process. Plant Cell Tissue Organ Cult. 2015, 122, 477–490. [Google Scholar] [CrossRef]
- Udawat, P.; Jha, R.K.; Sinha, D.; Mishra, A.; Jha, B. Overexpression of a cytosolic abiotic stress responsive universal stress protein (SbUSP) mitigates salt and osmotic stress in transgenic Tobacco plants. Front. Plant Sci. 2016, 7, 518–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Lin, W.; Cai, W.; Arora, R. Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta 2007, 226, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Sahoo, A.; Devendran, R.; Jain, M. Over-expression of a Rice Tau class glutathione s-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS ONE 2014, 9, e92900. [Google Scholar]
- Oh, D.H.; Dassanayake, M.; Haas, J.S.; Kropornika, A.; Wright, C.; d’Urzo, M.P.; Hong, H.; Ali, S.; Hernandez, A.; Lambert, G.M. Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol. 2010, 154, 1040–1052. [Google Scholar] [CrossRef] [Green Version]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S. Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhu, P.; Kang, H.; Liu, L.; Cao, Q.; Sun, J. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genom. 2020, 21, 164. [Google Scholar] [CrossRef] [Green Version]
- Jodder, J.; Das, R.; Sarkar, D.; Bhattacharjee, P.; Kundu, P. Distinct transcriptional and processing regulations control miR167a level in tomato during stress. RNA Biol. 2018, 15, 130–143. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wang, Q.; Zhang, B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 2013, 530, 26–32. [Google Scholar] [CrossRef]
- Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Profiling the abiotic stress responsive microRNA landscape of Arabidopsis thaliana. Plants 2019, 8, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.; Shi, L.; Wang, Y.; Chen, L.; Cai, Z.; Wang, Y. Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean nodules by high-throughput sequencing. Int. J. Mol. Sci. 2013, 14, 2717–2738. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Luo, H. Role of microRNA319 in creeping bentgrass salinity and drought stress response. Plant Signal Behav. 2014, 9, e28700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, C.; Long, R.; Zhang, T.; Kang, J.; Wang, Z.; Wang, P. Genome-wide identification of microRNAs in response to salt/alkali stress in Medicago truncatula through high-throughput sequencing. Int. J. Mol. Sci. 2018, 19, 4076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.S.; Gudimella, R.; Wong, G.R.; Tammi, M.T.; Khalid, N.; Harikrishna, J.A. Transcripts and microRNAs responding to salt stress in Musa acuminata colla (AAA Group) cv. berangan roots. PLoS ONE 2015, 10, e0127526. [Google Scholar] [CrossRef]
- Tripathi, A.; Chacon, O.; Singla-Pareek, S.L.; Sopory, S.K.; Sanan-Mishra, N. Mapping the microRNA expression profiles in glyoxalase over-expressing salinity tolerant rice. Curr. Genom. 2018, 19, 21–35. [Google Scholar] [CrossRef]
- Xie, F.; Stewart, C.N., Jr.; Taki, F.A.; He, Q.; Liu, H.; Zhang, B. High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnol. J. 2014, 12, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Gharat, S.A.; Shaw, B.P. Novel and conserved miRNAs in the halophyte Suaeda maritima identified by deep sequencing and computational predictions using the ESTs of two mangrove plants. BMC Plant Biol. 2015, 15, 301. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Zhang, L.; Yang, Z.; Wei, Y.; Dong, T. Identification and functional characterization of Plant miRNA under salt stress shed light on salinity resistance improvement through miRNA manipulation in crops. Front. Plant Sci. 2021, 12, 665439. [Google Scholar] [CrossRef]
- Alzahrani, S.M.; Alaraidh, I.A.; Khan, M.A.; Migdadi, H.M.; Alghamdi, S.S.; Alsahli, A.A. Identification and characterization of salt-responsive microRNAs in Vicia faba by high-throughput sequencing. Genes 2019, 10, 303. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zhao, C.; Li, M.; Sun, W.; Liu, Y.; Xia, H. Genome-wide identification of Thellungiella salsuginea microRNAs with putative roles in the salt stress response. BMC Plant Biol. 2013, 13, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Xu, L.; Wang, Y.; Yu, R.; Zhu, X.; Luo, X. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom. 2015, 16, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, L.; Fan, Y.; Wang, L. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 2015, 55, 178–185. [Google Scholar] [CrossRef]
- Yuan, S.; Li, Z.; Li, D.; Yuan, N.; Hu, Q.; Luo, H. Constitutive expression of rice microRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bentgrass. Plant Physiol. 2015, 169, 576–593. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Zhao, J.; Li, Z.; Hu, Q.; Yuan, N.; Zhou, M. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic. Res. 2019, 6, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Guo, J.; Cai, Y.; Gong, X.; Xiong, X.; Qi, W. Genome-wide identification and characterization of Eutrema salsugineum microRNAs for salt tolerance. Physiol. Plant 2016, 157, 453–468. [Google Scholar] [CrossRef]
- Chiang, C.P.; Yim, W.C.; Sun, Y.H.; Ohnishi, M.; Mimura, T.; Cushman, J.C. Identification of ice plant (Mesembryanthemum crystallinum L.) microRNAs using RNA-seq and their putative roles in high salinity responses in seedlings. Front. Plant Sci. 2016, 7, 1143. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Zeng, Y.; Yi, X.; Zhao, L.; Zhang, Y. Small RNA deep sequencing reveals the important role of microRNAs in the halophyte Halostachys caspica. Plant Biotechnol. J. 2015, 13, 395–408. [Google Scholar] [CrossRef]




| Fodder Crops | Secondary Metabolites |
|---|---|
| Atriplex nummularia | Saponin, Alkaloids, Tannins, Nitrate |
| Atriplex leucoclada | Saponin, Alkaloids, Tannins |
| Atriplex halimus | Saponin, Flavonoids, Alkaloids, Tannins, Nitrate |
| Diplache fusca | Flavonoids, Alkaloids |
| Halocnemum strobilecum | Saponin, Flavonoids, Alkaloids, Tannins, Nitrate |
| Haloxylon salicornicum | Saponin, Flavonoids, Alkaloids, Tannins |
| Kochia eriophora | Alkaloids, Tannins |
| Juncus acutus | Flavonoids, Alkaloids, Tannins, Nitrate |
| Juncus arabicus | Alkaloids, Tannins |
| Juncus subulatus | Alkaloids, Tannins, Flavonoids |
| Limonium pruinosum | Saponin, Alkaloids, Tannins |
| Nitraria retusa | Saponin, Tannins |
| Salsola glauca | Saponin, Flavonoids, Alkaloids |
| Suaeda fruticosa | Alkaloids, Tannins, Nitrate |
| Tamarix aphylla | Saponin, Tannins |
| Salsola tetrandra | Nitrate |
| Tamarix mannifera | Saponin, Tannins |
| Zygophyllum album | Saponin, Flavonoids, Alkaloids, Tannins, Nitrate |
| Sesbania sesban | Saponin, Alkaloids |
| Gene Name | Plants | Gene Functions | References |
|---|---|---|---|
| SOS1,SOS2, AtNHX1 | Brassica campestris Brassica juncea | Na+/K+ plasma membrane antiporter, calcium-binding protein, and protein kinase | [65] |
| AtSKIP | Arabidopsis thaliana | Transcription factor, splicing, and polyadenylation | [66] |
| OsHsp17.0 OsHsp23.7 | Oryza sativa L. | Transporting proteins and heat-shock proteins | [67] |
| DcHsp17.7 | Daucus carota | Cell viability and membrane stability under heat stress | [68] |
| JcDREB | Arabidopsis thaliana | Transcription factor | [69] |
| AtNHX1 | Arabidopsis thaliana | Calcium-binding protein, vacuolar Na+/K+ antiporter | [70] |
| OsRab7 | Oryza sativa L. | Vesicle trafficking gene enhanced growth and proline | [71] |
| PeXTH | Populus euphratica | Higher cell viability, water holding capacity, and membrane integrity | [72] |
| CYP94 | Oryza sativa | Enhanced CYP94C2b expression | [73] |
| SOS1 | Nicotina tabacum | Plasma membrane, Na+/K+ and vacuolar Na+/K+ antiporter | [74] |
| mtlD | Escherichia coli | Higher mannitol 1 phosphate dehydrogenase levels | [75] |
| TaSTRG | Triticum aestivum | Increase salinity and water deficit resistance | [76] |
| AtSTO1 | Arabidopsis thaliana | Higher root, pith size, and photosynthesis | [77] |
| Halophytes | Genes | Description | Recipient Plants | References |
|---|---|---|---|---|
| Aeluropus littoralis | AlNHX1 | Vacuolar Na+/H+ antiporter | Nicotiana tabacum | [106] |
| Atriplex hortensis | AhBADH | Glycine betaine synthesis | Solanum lycopersicum | [107] |
| Avicennia marina | AmMDHAR | ROS scavenging | Nicotiana tabacum | [108] |
| Salicornia brachiata | SbASR1 | Ascorbate regeneration and ROS scavenging | Nicotiana tabacum | [108] |
| Salicornia brachiata | SbpAPX | Peroxisomal ascorbate peroxidase | Nicotiana tabacum | [109] |
| Salicornia brachiata | SbpAPX | Peroxisomal ascorbate peroxidase | Arachis hypogea | [110] |
| Salsola soda | SsNHX1 | Vacuolar Na+/H+ antiporter | Alfalfa | [111] |
| Suaeda liaotungensis | SlBADH | Glycine betaine synthesis | Zea mays | [112] |
| Suaeda salsa | SsCAX1 | Vacuolar H+/Ca2+ transporter | Arabidopsis | [113] |
| Suaeda salsa | SsGST | Glutathione S-transferase | Oryza sativa | [114] |
| Suaeda salsa | SsVP | Vacuolar-H+-pyrophosphatase | Arabidopsis | [115] |
| Thellungiella halophila | ThSOS1 | Salt overly sensitive gene | Arabidopsis | [116] |
| Thellungiella salsuginea | TsTIP1 | Tonoplast AQP gene | Arabidopsis | [117] |
| Tamarix androssowii | TaMnSOD | Manganese superoxide dismutase | Populus | [118] |
| Spartina alterniflora | SaVHAc1 | Vacuolar H+-ATPase subunit Cl | Oryza sativa | [119] |
| Salicornia europaea | SeCMO | Enhanced glycine betaine synthesis | Nicotiana tabacum | [120] |
| Kalidium foliatum | V-ATPase | Vacuolar-H+-pyrophosphatase | Arabidopsis | [121] |
| Atriplex gmelini | AgNHX1 | Vacuolar Na+/H+ antiporter | Oryza sativa | [122] |
| Transgene | Gene Isolated | Promoters | Transgenic Crop |
|---|---|---|---|
| Ion exclusion Na+/H+ antiporter (SOS1) | Arabidopsis | Constitutive | Nicotiana tabacum |
| Na+/H+ antiporter (SOD2) | Salicornia brachiata | Stress inducible | Oryza sativa |
| Tissue tolerance Na+/H+ antiporter (NHX) | Arabidopsis | Constitutive | Fagopyrum esculentum |
| Tissue tolerance Trehalose-6-phosphate synthase (TPS) | Yeast | Constitutive | Medicago sativa |
| Tissue tolerance Trehalose-6-phosphate phosphatase (TPP) | Rice | Stress inducible | Solanum lycopersicum |
| Mannitol-1-phosphate dehydrogenase (mt1D) | E.coli | Shoot expression | Oryza sativa |
| Myoinositol O-methyltransferase | M. crystallinum | Constitutive | Triticum aestivum |
| Tissue tolerance Ascorbate (APX) | Arabidopsis | Constitutive | Nicotiana tabacum |
| Glutathione S-transferase (GST) | Tomato | Protein targeted to chloroplast/cytosol | Oryza sativa |
| Mitogen activated protein kinase (MAPK) | Chickpea | Constitutive | Nicotiana tabacum |
| Sucrose protein kinase | Rice | Inducible | Oryza sativa |
| Transcription factors DREB | Pennisetum glaucum | Constitutive & inducible | Nicotiana tabacum |
| Desired Gene | Donor Plant | Target Plant | References |
|---|---|---|---|
| codA | Arthrobacter globiformis | Solanum lycopersicum | [137] |
| Cox | Arthrobacter pascens | Oryza sativa | [138] |
| TPS1 | Yeast | Solanum lycopersicum | [139] |
| AtTPS1 | Arabidopsis | Nicotiana tabacum | [140] |
| mtID | Triticum aestivum | Escherichia coli | [141] |
| S6PDH | Malus domestica | Japanese Persimmon | [142] |
| P5CS | Vigna acontifolia | Nicotiana tabacum | [143] |
| nhaA | E.coli | Arabidopsis | [141] |
| AtNHX1 | Arabidopsis | Solanum lycopersicum | [144] |
| BnNHX1 | Brassica | Nicotiana tabacum | [145] |
| GlyII | Oryza sativa | Nicotiana tabacum | [146] |
| GmbZIP1 | Soybean | Arabidopsis, Nicotiana tabacum | [147] |
| BrERF4 | Brassica | Arabidopsis | [148] |
| T30hsp70 | Trichoderma harzianum | Arabidopsis | [149] |
| GhMPK2 | Cotton | Nicotiana tabacum | [150] |
| Plants | NaCl Concentration | miRNA Number | References |
|---|---|---|---|
| Arabidopsis thaliana | 150 mM | 118 | [166] |
| Glycine max | 125 mM | 238 | [167] |
| Leymus chinensis | 100 mM | 148 | [168] |
| Medicago truncatula | 20 mM | 876 | [169] |
| Musa nana | 300 mM | 181 | [170] |
| Oryza sativa | 200 mM | 498 | [171] |
| Panicum virgatum | 0.5 % | 273 | [172] |
| Suaeda maritima | 255 mM | 147 | [173] |
| Zea mays | 250 mM | 1077 | [174] |
| Vicia faba | 150 mM | 693 | [175] |
| Thellungiella salsugniea | 200 mM | 246 | [176] |
| Raphanus sativus | 200 mM | 204 | [177] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abideen, Z.; Hanif, M.; Munir, N.; Nielsen, B.L. Impact of Nanomaterials on the Regulation of Gene Expression and Metabolomics of Plants under Salt Stress. Plants 2022, 11, 691. https://doi.org/10.3390/plants11050691
Abideen Z, Hanif M, Munir N, Nielsen BL. Impact of Nanomaterials on the Regulation of Gene Expression and Metabolomics of Plants under Salt Stress. Plants. 2022; 11(5):691. https://doi.org/10.3390/plants11050691
Chicago/Turabian StyleAbideen, Zainul, Maria Hanif, Neelma Munir, and Brent L. Nielsen. 2022. "Impact of Nanomaterials on the Regulation of Gene Expression and Metabolomics of Plants under Salt Stress" Plants 11, no. 5: 691. https://doi.org/10.3390/plants11050691
APA StyleAbideen, Z., Hanif, M., Munir, N., & Nielsen, B. L. (2022). Impact of Nanomaterials on the Regulation of Gene Expression and Metabolomics of Plants under Salt Stress. Plants, 11(5), 691. https://doi.org/10.3390/plants11050691