Tracing the Evolution of the Angiosperm Genome from the Cytogenetic Point of View
Abstract
1. Introduction
2. Genome Size Analyses
2.1. The Patterns and Directions of Genome Size Evolution
2.2. The Repetitive Sequences—The Main Players in Genome Size Evolution
3. Why Is the Chromosome Number So Variable in Angiosperms?
3.1. Changes in the Chromosome Number against the Phylogenetic Background
3.2. Genome Evolution from the Cytogenetic Point of View
4. Polyploidy in Angiosperms
4.1. Introduction to Polyploidy: ‘Definitions’ and ‘Numbers’
4.2. Cytogenetic Approaches on Duty in Polyploid Research
4.2.1. ‘Hunting’ for Polyploids—Cytogenetic Methods in Polyploid Identification
4.2.2. Polyploidisation Events and What Happens Next?
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harris, H. The cells of the body: A history of somatic cell genetics. J. Hist. Biol. 1998, 31, 295–296. [Google Scholar]
- Paweletz, N. Walther Flemming: Pioneer of mitosis research. Nat. Rev. Mol. Cell Biol. 2001, 2, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Sharma, A. 17—Nucleic acid and its components. In Chromosome Techniques, 3rd ed.; Sharma, A.K., Sharma, A., Eds.; Butterworth-Heinemann: Oxford, UK, 1980; pp. 509–542. [Google Scholar]
- Vosa, C.G. A Modified Aceto-Orcein Method for Pollen Mother Cells. Caryologia 1961, 14, 107–110. [Google Scholar] [CrossRef]
- Jellen, E.N. C-Banding of Plant Chromosomes. In Plant Cytogenetics: Methods and Protocols; Kianian, S.F., Kianian, P.M.A., Eds.; Springer New York: New York, NY, USA, 2016; pp. 1–5. [Google Scholar]
- Schwarzacher, T.; Heslop-Harrison, J.; Heslop-Harrison, P. Practical in Situ Hybridization; BIOS: Oxford, UK, 2000. [Google Scholar]
- Borowska-Zuchowska, N.; Robaszkiewicz, E.; Wolny, E.; Betekhtin, A.; Hasterok, R. Ribosomal DNA loci derived from Brachypodium stacei are switched off for major parts of the life cycle of Brachypodium hybridum. J. Exp. Bot. 2019, 70, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Contreras-Moreira, B.; Levy, J.J.; Djamei, A.; Czedik-Eysenberg, A.; Tartaglio, V.S.; Session, A.; Martin, J.; Cartwright, A.; Katz, A.; et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat. Commun. 2020, 11, 3670. [Google Scholar] [CrossRef]
- Hloušková, P.; Mandáková, T.; Pouch, M.; Trávníček, P.; Lysak, M.A. The large genome size variation in the Hesperis clade was shaped by the prevalent proliferation of DNA repeats and rarer genome downsizing. Ann. Bot. 2019, 124, 103–120. [Google Scholar] [CrossRef]
- Mandáková, T.; Joly, S.; Krzywinski, M.; Mummenhoff, K.; Lysak, M.A. Fast Diploidization in Close Mesopolyploid Relatives of Arabidopsis. Plant Cell 2010, 22, 2277–2290. [Google Scholar] [CrossRef]
- Mandáková, T.; Lysak, M.A. Chromosomal Phylogeny and Karyotype Evolution in x = 7 Crucifer Species (Brassicaceae). Plant Cell 2008, 20, 2559–2570. [Google Scholar] [CrossRef]
- Faizullah, L.; Morton, J.A.; Hersch-Green, E.I.; Walczyk, A.M.; Leitch, A.R.; Leitch, I.J. Exploring environmental selection on genome size in angiosperms. Trends Plant Sci. 2021, 26, 1039–1049. [Google Scholar] [CrossRef]
- Greilhuber, J.; Dolezel, J.; Lysák, M.A.; Bennett, M.D. The Origin, Evolution and Proposed Stabilization of the Terms ’Genome Size’ and ’C-Value’ to Describe Nuclear DNA Contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef]
- Dolezel, J.; Bartos, J.; Voglmayr, H.; Greilhuber, J. Nuclear DNA content and genome size of trout and human. Cytometry A 2003, 51, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.D.; Smith, J.B. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. B. 1991, 334, 309–345. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Jang, T.-S.; Parker, J.S.; Weiss-Schneeweiss, H. Euchromatic Supernumerary Chromosomal Segments—Remnants of Ongoing Karyotype Restructuring in the Prospero autumnale Complex? Genes 2018, 9, 468. [Google Scholar] [CrossRef]
- Senderowicz, M.; Nowak, T.; Rojek-Jelonek, M.; Bisaga, M.; Papp, L.; Weiss-Schneeweiss, H.; Kolano, B. Descending dysploidy and bidirectional changes in genome size accompanied Crepis (Asteraceae) Evolution. Genes 2021, 12, 1436. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N.; Rees, H. The Influence of B-chromosomes upon the nuclearphenotype in rye. Chromosoma 1968, 24, 158–176. [Google Scholar] [CrossRef]
- Maluszynska, J. B Chromosomes of Crepis capillaris (L.) Waller. In Vivo and In Vitro; Wydawnictwo UŚ: Katowice, Poland, 1990. [Google Scholar]
- Rosado, T.B.; Clarindo, W.R.; Carvalho, C.R. An integrated cytogenetic, flow and image cytometry procedure used to measure the DNA content of Zea mays A and B chromosomes. Plant Sci. 2009, 176, 154–158. [Google Scholar] [CrossRef]
- Kersey, P.J. Plant genome sequences: Past, present, future. Curr. Opin. Plant Biol. 2019, 48, 1–8. [Google Scholar] [CrossRef]
- Pellicer, J.; Leitch, I.J. The Plant DNA C-values database (release 7.1): An updated online repository of plant genome size data for comparative studies. New Phytol. 2020, 226, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, A.; Michael, T.P.; Rivadavia, F.; Sousa, A.; Wang, W.; Temsch, E.M.; Greilhuber, J.; Müller, K.F.; Heubl, G. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Ann. Bot. 2014, 114, 1651–1663. [Google Scholar] [CrossRef]
- Pellicer, J.; Fay, M.F.; Leitch, I.J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010, 164, 10–15. [Google Scholar] [CrossRef]
- Leitch, I.J.; Chase, M.W.; Bennett, M.D. Phylogenetic Analysis of DNA C-values provides Evidence for a Small Ancestral Genome Size in Flowering Plants. Ann. Bot. 1998, 82, 85–94. [Google Scholar] [CrossRef]
- Leitch, I.J.; Leitch, A.R. Genome size diversity and evolution in land plants. In Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Vienna, Austria, 2013; pp. 307–322. [Google Scholar]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.J.; Renny-Byfield, S.; Pellicer, J.; Macas, J.; Novak, P.; Neumann, P.; Lysak, M.; Day, P.D.; Berger, M.; Fay, M.F.; et al. Analysis of the giant genomes of Fritillaria (L iliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 2015, 208, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, J.; Kelly, L.J.; Leitch, I.J.; Zomlefer, W.B.; Fay, M.F. A universe of dwarfs and giants: Genome size and chromosome evolution in the monocot family Melanthiaceae. New Phytol. 2014, 201, 1484–1497. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M. New Record Holders for Maximum Genome Size in Eudicots and Monocots. J. Bot. 2010, 2010, 527357. [Google Scholar] [CrossRef]
- Puttick, M.N.; Clark, J.; Donoghue, P. Size is not everything: Rates of genome size evolution, not C -value, correlate with speciation in angiosperms. Proc. R. Soc. B Boil. Sci. 2015, 282, 20152289. [Google Scholar] [CrossRef]
- Leitch, I.J.; Johnston, E.; Pellicer, J.; Hildago, O.; Bennett, M.D. Plant DNA C-Values Database. Available online: https://cvalues.science.kew.org/ (accessed on 18 February 2022).
- Lysak, M.A.; Koch, M.A.; Beaulieu, J.M.; Meister, A.; Leitch, I.J. The Dynamic Ups and Downs of Genome Size Evolution in Brassicaceae. Mol. Biol. Evol. 2008, 26, 85–98. [Google Scholar] [CrossRef]
- Sears, C.J.; Whitton, J. A reexamination of the North American Crepis agamic complex and comparison with the findings of Babcock and Stebbins’ classic biosystematic monograph. Am. J. Bot. 2016, 103, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Vitales, D.; Álvarez, I.; Garcia, S.; Hidalgo, O.; Feliner, G.N.; Pellicer, J.; Vallès, J.; Garnatje, T. Genome size variation at constant chromosome number is not correlated with repetitive DNA dynamism in Anacyclus (Asteraceae). Ann. Bot. 2020, 125, 611–623. [Google Scholar] [CrossRef]
- Vitales, D.; Fernández, P.; Garnatje, T.; Garcia, S. Progress in the study of genome size evolution in Asteraceae: Analysis of the last update. Database 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Ambrožová, K.; Mandáková, T.; Bureš, P.; Neumann, P.; Leitch, I.J.; Koblížková, A.; Macas, J.; Lysak, M.A. Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Ann. Bot. 2011, 107, 255–268. [Google Scholar] [CrossRef]
- Ng, C.H.; Lee, S.L.; Tnah, L.H.; Ng, K.K.S.; Lee, C.T.; Madon, M. Genome size variation and evolution in Dipterocarpaceae. Plant Ecol. Divers. 2016, 9, 437–446. [Google Scholar] [CrossRef]
- Leitch, I.J.; Bennett, M.D. Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 2004, 82, 651–663. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, Z.; Li, Y.; Hu, H.; Wang, Z.; Du, X.; Zhang, S.; Zhu, M.; Dong, L.; Ren, G.; et al. Which factors contribute most to genome size variation within angiosperms? Ecol. Evol. 2021, 11, 2660–2668. [Google Scholar] [CrossRef] [PubMed]
- Elliott, T.A.; Gregory, T.R. What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140331. [Google Scholar] [CrossRef]
- Novák, P.; Guignard, M.S.; Neumann, P.; Kelly, L.J.; Mlinarec, J.; Koblížková, A.; Dodsworth, S.; Kovařík, A.; Pellicer, J.; Wang, W.; et al. Repeat-sequence turnover shifts fundamentally in species with large genomes. Nat. Plants 2020, 6, 1325–1329. [Google Scholar] [CrossRef]
- Grover, C.E.; Wendel, J.F. Recent Insights into Mechanisms of Genome Size Change in Plants. J. Bot. 2010, 2010, 382732. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Macas, J.; Novák, P.; Pellicer, J.; Čížková, J.; Koblížková, A.; Neumann, P.; Fuková, I.; Doležel, J.; Kelly, L.J.; Leitch, I.J. In Depth Characterization of Repetitive DNA in 23 Plant Genomes Reveals Sources of Genome Size Variation in the Legume Tribe Fabeae. PLoS ONE 2015, 10, e0143424. [Google Scholar] [CrossRef]
- Gaiero, P.; Vaio, M.; Peters, S.A.; Schranz, M.E.; De Jong, H.; Speranza, P. Comparative analysis of repetitive sequences among species from the potato and the tomato clades. Ann. Bot. 2019, 123, 521–532. [Google Scholar] [CrossRef]
- Mascagni, F.; Giordani, T.; Ceccarelli, M.; Cavallini, A.; Natali, L. Genome-wide analysis of LTR-retrotransposon diversity and its impact on the evolution of the genus Helianthus (L.). BMC Genom. 2017, 18, 634. [Google Scholar] [CrossRef] [PubMed]
- Sader, M.; Vaio, M.; Cauz-Santos, L.A.; Dornelas, M.C.; Vieira, M.L.C.; Melo, N.; Pedrosa-Harand, A. Large vs small genomes in Passiflora: The influence of the mobilome and the satellitome. Planta 2021, 253, 86. [Google Scholar] [CrossRef] [PubMed]
- McCann, J.; Macas, J.; Novak, P.; Stuessy, T.F.; Villaseñor, J.L.; Weiss-Schneeweiss, H. Differential Genome Size and Repetitive DNA Evolution in Diploid Species of Melampodium sect. Melampodium (Asteraceae). Front. Plant Sci. 2020, 11, 362. [Google Scholar] [CrossRef] [PubMed]
- Sader, M.A.; Amorim, B.S.; Costa, L.; Souza, G.; Pedrosa-Harand, A. The role of chromosome changes in the diversification of Passiflora L. (Passifloraceae). Syst. Biodivers. 2019, 17, 7–21. [Google Scholar] [CrossRef]
- Divashuk, M.G.; Karlov, G.I.; Kroupin, P.Y. Copy Number Variation of Transposable Elements in Thinopyrum intermedium and Its Diploid Relative Species. Plants 2020, 9, 15. [Google Scholar] [CrossRef]
- Oliver, K.R.; McComb, J.A.; Greene, W. Transposable Elements: Powerful Contributors to Angiosperm Evolution and Diversity. Genome Biol. Evol. 2013, 5, 1886–1901. [Google Scholar] [CrossRef]
- Nystedt, B.; Street, N.; Wetterbom, A.; Zuccolo, A.; Lin, Y.-C.; Scofield, D.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, J.; Fernández, P.; Fay, M.F.; Michálková, E.; Leitch, I.J. Genome Size Doubling Arises From the Differential Repetitive DNA Dynamics in the Genus Heloniopsis (Melanthiaceae). Front. Genet. 2021, 12, 726211. [Google Scholar] [CrossRef]
- Ågren, J.A.; Greiner, S.; Johnson, M.T.J.; Wright, S. No evidence that sex and transposable elements drive genome size variation in evening primroses. Evolution 2015, 69, 1053–1062. [Google Scholar] [CrossRef]
- Hasterok, R.; Draper, J.; Jenkins, G. Laying the Cytotaxonomic Foundations of a New Model Grass, Brachypodium distachyon (L.) Beauv. Chromosom. Res. 2004, 12, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Orzechowska, M.; Majka, M.; Weiss-Schneeweiss, H.; Kovarik, A.; Borowska-Zuchowska, N.; Kolano, B. Organization and evolution of two repetitive sequences, 18–24J and 12–13P, in the genome of Chenopodium (Amaranthaceae). Genome 2018, 61, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.; Glick, L.; Abadi, S.; Einhorn, M.; Kopelman, N.M.; Salman-Minkov, A.; Mayzel, J.; Chay, O.; Mayrose, I. The Chromosome Counts Database (CCDB)—A community resource of plant chromosome numbers. New Phytol. 2015, 206, 19–26. [Google Scholar] [CrossRef]
- Leach, C.R.; Donald, T.M.; Franks, T.K.; Spiniello, S.S.; Hanrahan, C.F.; Timmis, J.N. Organisation and origin of a B chromosome centromeric sequence from Brachycome dichromosomatica. Chromosoma 1995, 103, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Uhl, C.H. Chromosomes of mexican Sedum ii. Section Pachysedum. Rhodora 1978, 80, 491–512. [Google Scholar]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011, 27, 207–216. [Google Scholar] [CrossRef]
- Siljak-Yakovlev, S. La dysploïdie et l’évolution du caryotype. Bocconea 1996, 5, 211–220. [Google Scholar]
- Mayrose, I.; Lysak, M.A. The Evolution of Chromosome Numbers: Mechanistic Models and Experimental Approaches. Genome Biol. Evol. 2020, 13, 13. [Google Scholar] [CrossRef]
- Soltis, P.S.; Liu, X.; Marchant, D.B.; Visger, C.J.; Soltis, D.E. Polyploidy and novelty: Gottlieb’s legacy. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130351. [Google Scholar] [CrossRef]
- Kolano, B.; Saracka, K.; Broda-Cnota, A.; Maluszynska, J. Localization of ribosomal DNA and CMA3/DAPI heterochromatin in cultivated and wild Amaranthus species. Sci. Hortic. 2013, 164, 249–255. [Google Scholar] [CrossRef]
- Pal, M.; Pandey, R.M.; Khoshoo, T.N. Evolution and improvement of cultivated amaranths: IX. Cytogenetic relationship between the two basic chromosome numbers. J. Hered. 1982, 73, 353–356. [Google Scholar] [CrossRef]
- Islam-Faridi, N.; Sakhnokho, H.F.; Nelson, C.D. New chromosome number and cyto-molecular characterization of the African Baobab (Adansonia digitata L.)—“The Tree of Life”. Sci. Rep. 2020, 10, 13174. [Google Scholar] [CrossRef] [PubMed]
- Grant, V. Plant Speciation; Columbia University Press: New York, NY, USA, 1981. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold: London, UK, 1971. [Google Scholar]
- Carta, A.; Bedini, G.; Peruzzi, L. A deep dive into the ancestral chromosome number and genome size of flowering plants. New Phytol. 2020, 228, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Chiavegatto, R.B.; Carta, A.; Pereira, D.G.S.; Benites, F.R.G.; Techio, V.H.; Peruzzi, L. Reconstructing ancestral chromosome numbers and inflorescence features in Eleusininae (Poaceae: Chloridoideae: Cynodonteae). Bot. J. Linn. Soc. 2020, 193, 402–418. [Google Scholar] [CrossRef]
- Moeglein, M.K.; Chatelet, D.S.; Donoghue, M.J.; Edwards, E.J. Evolutionary dynamics of genome size in a radiation of woody plants. Am. J. Bot. 2020, 107, 1527–1541. [Google Scholar] [CrossRef]
- Mota, L.; Torices, R.; Loureiro, J. The Evolution of Haploid Chromosome Numbers in the Sunflower Family. Genome Biol. Evol. 2016, 8, 3516–3528. [Google Scholar] [CrossRef]
- Cusimano, N.; Sousa, A.; Renner, S.S. Maximum likelihood inference implies a high, not a low, ancestral haploid chromosome number in Araceae, with a critique of the bias introduced by ‘x’. Ann. Bot. 2012, 109, 681–692. [Google Scholar] [CrossRef]
- Barrett, C.F.; McKain, M.R.; Sinn, B.T.; Ge, X.-J.; Zhang, Y.; Antonelli, A.; Bacon, C.D. Ancient Polyploidy and Genome Evolution in Palms. Genome Biol. Evol. 2019, 11, 1501–1511. [Google Scholar] [CrossRef]
- Bowers, J.; Chapman, B.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Albert, V.A.; Barbazuk, W.B.; dePamphilis, C.W.; Der, J.P.; Leebens-Mack, J.; Ma, H.; Palmer, J.D.; Rounsley, S.; Sankoff, D.; Schuster, S.C.; et al. The Amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef]
- Huang, C.-H.; Zhang, C.; Liu, M.; Hu, Y.; Gao, T.; Qi, J.; Ma, H. Multiple Polyploidization Events across Asteraceae with Two Nested Events in the Early History Revealed by Nuclear Phylogenomics. Mol. Biol. Evol. 2016, 33, 2820–2835. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Robinson, S.J.; Nixon, J.; Xiao, R.; Huebert, T.; Condie, J.; Kessler, D.; Clarke, W.E.; Edger, P.P.; Links, M.; et al. Polyploid Evolution of the Brassicaceae during the Cenozoic Era. Plant Cell 2014, 26, 2777–2791. [Google Scholar] [CrossRef]
- Cannon, S.B.; McKain, M.R.; Harkess, A.; Nelson, M.N.; Dash, S.; Deyholos, M.K.; Peng, Y.; Joyce, B.; Stewart, C.N., Jr.; Rolf, M.; et al. Multiple Polyploidy Events in the Early Radiation of Nodulating and Nonnodulating Legumes. Mol. Biol. Evol. 2014, 32, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Mandáková, T.; Guo, X.; Özüdoğru, B.; Mummenhoff, K.; Lysak, M.A. Hybridization-facilitated genome merger and repeated chromosome fusion after 8 million years. Plant J. 2018, 96, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Berr, A.; Pecinka, A.; Schmidt, R.; McBreen, K.; Schubert, I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 2006, 103, 5224–5229. [Google Scholar] [CrossRef]
- Mandáková, T.; Pouch, M.; Brock, J.R.; Al-Shehbaz, I.A.; Lysak, M.A. Origin and evolution of diploid and allopolyploid Camelina genomes were accompanied by chromosome shattering. Plant Cell 2019, 31, 2596–2612. [Google Scholar] [CrossRef]
- Escudero, M.; Martín-Bravo, S.; Mayrose, I.; Fernández-Mazuecos, M.; Fiz-Palacios, O.; Hipp, A.L.; Pimentel, M.; Jiménez-Mejías, P.; Valcarcel, V.; Vargas, P.; et al. Karyotypic Changes through Dysploidy Persist Longer over Evolutionary Time than Polyploid Changes. PLoS ONE 2014, 9, e85266. [Google Scholar] [CrossRef]
- Braz, G.T.; He, L.; Zhao, H.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Torres, G.A.; Jiang, J. Comparative Oligo-FISH Mapping: An Efficient and Powerful Methodology to Reveal Karyotypic and Chromosomal Evolution. Genetics 2018, 208, 513–523. [Google Scholar] [CrossRef]
- Kolano, B.; McCann, J.A.M.I.E.; Oskędra, M.; Chrapek, M.; Rojek, M.; Nobis, A.; Weiss-Schneeweiss, H. Parental origin and genome evolution of several Eurasian hexaploid species of Chenopodium (Chenopodiaceae). Phytotaxa 2019, 392, 163–185. [Google Scholar] [CrossRef]
- Kolano, B.; Siwinska, D.; McCann, J.; Weiss- Schneeweiss, H. The evolution of genome size and rDNA in diploid species of Chenopodium s.l. (Amaranthaceae). Bot. J. Linn. Soc. 2015, 179, 218–235. [Google Scholar] [CrossRef]
- Wu, F.; Tanksley, S.D. Chromosomal evolution in the plant family Solanaceae. BMC Genom. 2010, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Hollingshead, L.; Babcock, E. Chromosomes and Phylogeny in Crepis; University of California Press: Berkeley, CA, USA, 1930; Volume 6, pp. 1–53. [Google Scholar]
- Lusinska, J.; Betekhtin, A.; Lopez-Alvarez, D.; Catalan, P.; Jenkins, G.; Wolny, E.; Hasterok, R. Comparatively Barcoded Chromosomes of Brachypodium Perennials Tell the Story of Their Karyotype Structure and Evolution. Int. J. Mol. Sci. 2019, 20, 5557. [Google Scholar] [CrossRef] [PubMed]
- Weiss-Schneeweiss, H.; Stuessy, T.F.; Villaseñor, J.L. Chromosome Numbers, Karyotypes, and Evolution in Melampodium (Asteraceae). Int. J. Plant Sci. 2009, 170, 1168–1182. [Google Scholar] [CrossRef]
- Winterfeld, G.; Ley, A.; Hoffmann, M.H.; Paule, J.; Röser, M. Dysploidy and polyploidy trigger strong variation of chromosome numbers in the prayer-plant family (Marantaceae). Plant Syst. Evol. 2020, 306, 36. [Google Scholar] [CrossRef]
- Liu, X.; Sun, S.; Wu, Y.; Zhou, Y.; Gu, S.; Yu, H.; Yi, C.; Gu, M.; Jiang, J.; Liu, B.; et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J. 2020, 101, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Koch, M.A. Phylogeny, Genome, and Karyotype Evolution of Crucifers (Brassicaceae). In Genetics and Genomics of the Brassicaceae; Schmidt, R., Bancroft, I., Eds.; Springer: New York, NY, USA, 2011; pp. 1–31. [Google Scholar]
- Lusinska, J.; Majka, J.; Betekhtin, A.; Susek, K.; Wolny, E.; Hasterok, R. Chromosome identification and reconstruction of evolutionary rearrangements in Brachypodium distachyon, B. stacei and B. hybridum. Ann. Bot. 2018, 122, 445–459. [Google Scholar] [CrossRef]
- Pont, C.; Wagner, S.; Kremer, A.; Orlando, L.; Plomion, C.; Salse, J. Paleogenomics: Reconstruction of plant evolutionary trajectories from modern and ancient DNA. Genome Biol. 2019, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Cerbah, M.; Souza-Chies, T.; Jubier, M.F.; Lejeune, B.; Siljak-Yakovlev, S. Molecular phylogeny of the genus Hypochaeris using internal transcribed spacers of nuclear rDNA: Inference for chromosomal evolution. Mol. Biol. Evol. 1998, 15, 345–354. [Google Scholar] [CrossRef][Green Version]
- Siljak-Yakovlev, S.; Godelle, B.; Zoldoš, V.; Vallès, J.; Garnatje, T.; Hidalgo, O. Evolutionary implications of heterochromatin and rDNA in chromosome number and genome size changes during dysploidy: A case study in Reichardia genus. PLoS ONE 2017, 12, e0182318. [Google Scholar] [CrossRef]
- Jiang, J. Fluorescence in situ hybridization in plants: Recent developments and future applications. Chromosom. Res. 2019, 27, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-Specific Painting in Cucumis Species Using Bulked Oligonucleotides. Genetics 2015, 200, 771–779. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, T.; Wu, Y.; Zhang, W.; Zhang, P.; Xi, M.; Jiang, J. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J. 2020, 101, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Doležalová, A.; Sládeková, L.; Šimoníková, D.; Holušová, K.; Karafiátová, M.; Varshney, R.K.; Doležel, J.; Hřibová, E. Karyotype Differentiation in Cultivated Chickpea Revealed by Oligopainting Fluorescence in situ Hybridization. Front. Plant Sci. 2022, 12. [Google Scholar] [CrossRef]
- Zhao, Q.; Meng, Y.; Wang, P.; Qin, X.; Cheng, C.; Zhou, J.; Yu, X.; Li, J.; Lou, Q.; Jahn, M.; et al. Reconstruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J. 2021, 107, 1243–1259. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhao, H.; He, J.; Yang, Z.; Guan, B.; Chen, K.; Hong, Q.; Wang, J.; Liu, J.; Jiang, J. Extraordinarily conserved chromosomal synteny of Citrus species revealed by chromosome-specific painting. Plant J. 2020, 103, 2225–2235. [Google Scholar] [CrossRef]
- Maravilla, A.J.; Rosato, M.; Rosselló, J.A. Interstitial telomeric-like repeats (ITR) in seed plants as assessed by molecular cytogenetic techniques: A review. Plants 2021, 10, 2541. [Google Scholar] [CrossRef] [PubMed]
- Kolano, B.; McCann, J.; Orzechowska, M.; Siwinska, D.; Temsch, E.; Weiss-Schneeweiss, H. Molecular and cytogenetic evidence for an allotetraploid origin of Chenopodium quinoa and C. berlandieri (Amaranthaceae). Mol. Phylogenetics Evol. 2016, 100, 109–123. [Google Scholar] [CrossRef]
- Weiss-Schneeweiss, H.; Tremetsberger, K.; Schneeweiss, G.M.; Parker, J.S.; Stuessy, T.F. Karyotype Diversification and Evolution in Diploid and Polyploid South American Hypochaeris (Asteraceae) Inferred from rDNA Localization and Genetic Fingerprint Data. Ann. Bot. 2008, 101, 909–918. [Google Scholar] [CrossRef]
- Winterfeld, G.; Becher, H.; Voshell, S.; Hilu, K.; Röser, M. Karyotype evolution in Phalaris (Poaceae): The role of reductional dysploidy, polyploidy and chromosome alteration in a wide-spread and diverse genus. PLoS ONE 2018, 13, e0192869. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Kwasniewski, M.; Hasterok, R. Cytomolecular Analysis of Ribosomal DNA Evolution in a Natural Allotetraploid Brachypodium hybridum and Its Putative Ancestors—Dissecting Complex Repetitive Structure of Intergenic Spacers. Front. Plant Sci. 2016, 7, 7. [Google Scholar] [CrossRef]
- Breda, E.; Wolny, E.; Hasterok, R. Intraspecific polymorphism of ribosomal DNA loci number and morphology in Brachypodium pinnatum and Brachypodium sylvaticum. Cell. Mol. Biol. Lett. 2012, 17, 526–541. [Google Scholar] [CrossRef]
- Sousa, A.; Renner, S.S. Interstitial telomere-like repeats in the monocot family Araceae. Bot. J. Linn. Soc. 2015, 177, 15–26. [Google Scholar] [CrossRef]
- He, L.; Liu, J.; Torres, G.A.; Zhang, H.; Jiang, J.; Xie, C. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Res. 2013, 21, 5–13. [Google Scholar] [CrossRef]
- Volkov, R.; Medina, F.; Zentgraf, U.; Hemleben, V. Organization and molecular evolution of rDNA nucleolar dominance and nucleolus structure. In Progress in Botany; Esser, K., Luttge, U., Beyschlag, W., Murata, J., Eds.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2004; Volume 65. [Google Scholar]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Kovarik, A.; Matyasek, R.; Chase, M.W.; Knapp, S.; McCarthy, E.; Clarkson, J.J.; Leitch, A.R. Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. Plant J. 2006, 48, 907–919. [Google Scholar] [CrossRef]
- Song, Z.; Dai, S.; Bao, T.; Zuo, Y.; Xiang, Q.; Li, J.; Liu, G.; Yan, Z. Analysis of Structural Genomic Diversity in Aegilops umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata by Fluorescence In Situ Hybridization Karyotyping. Front. Plant Sci. 2020, 11, 710. [Google Scholar] [CrossRef]
- Waminal, N.E.; Pellerin, R.J.; Kang, S.-H.; Kim, H.H. Chromosomal Mapping of Tandem Repeats Revealed Massive Chromosomal Rearrangements and Insights into Senna tora Dysploidy. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Alix, K.; Gérard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S.P. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F. The wondrous cycles of polyploidy in plants. Am. J. Bot. 2015, 102, 1753–1756. [Google Scholar] [CrossRef] [PubMed]
- Masterson, J. Stomatal Size in Fossil Plants: Evidence for Polyploidy in Majority of Angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, G.L. Types of Polyploids: Their Classification and Significance. Adv. Genet. 1947, 1, 403–429. [Google Scholar] [CrossRef] [PubMed]
- Kihara, H.; Ono, T. Chromosomenzahlen und systematische Gruppierung der Rumex-Arten. Z. Zellforch. Microsk. Anat. 1926, 4, 475–481. [Google Scholar] [CrossRef]
- Mason, A.S.; Wendel, J.F. Homoeologous Exchanges, Segmental Allopolyploidy, and Polyploid Genome Evolution. Front. Genet. 2020, 11, 1014. [Google Scholar] [CrossRef]
- Spoelhof, J.P.; Soltis, P.S.; Soltis, D.E. Pure polyploidy: Closing the gaps in autopolyploid research. J. Syst. Evol. 2017, 55, 340–352. [Google Scholar] [CrossRef]
- Glombik, M.; Bačovský, V.; Hobza, R.; Kopecký, D. Competition of Parental Genomes in Plant Hybrids. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wendel, J.F.; Jackson, S.A.; Meyers, B.C.; Wing, R.A. Evolution of plant genome architecture. Genome Biol. 2016, 17, 37. [Google Scholar] [CrossRef]
- Del Pozo, J.C.; Ramirez-Parra, E. Whole genome duplications in plants: An overview from Arabidopsis. J. Exp. Bot. 2015, 66, 6991–7003. [Google Scholar] [CrossRef]
- Dodsworth, S.; Chase, M.W.; Leitch, A.R. Is post-polyploidization diploidization the key to the evolutionary success of angiosperms? Bot. J. Linn. Soc. 2016, 180, 1–5. [Google Scholar] [CrossRef]
- Mandáková, T.; Lysak, M.A. Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol. 2018, 42, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A. Allopolyploidy—A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005, 109, 250–258. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. The Role of Hybridization in Plant Speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef]
- Hias, N.; Leen, L.; Davey, M.W.; Vanderzande, S.; Van Huylenbroeck, J.; Keulemans, J. Effect of polyploidization on morphology in two apple (Malus × domestica) genotypes. Hortic. Sci. 2017, 44, 55–63. [Google Scholar]
- Pei, Y.; Yao, N.; He, L.; Deng, D.; Li, W.; Zhang, W. Comparative study of the morphological, physiological and molecular characteristics between diploid and tetraploid radish (Raphunas sativus L.). Sci. Hortic. 2019, 257, 108739. [Google Scholar] [CrossRef]
- Murray, B.G.; Hammett, K.R.W.; Standring, L.S. Genomic constancy during the development of Lathyrus odoratus cultivars. Heredity 1992, 68, 321–327. [Google Scholar] [CrossRef]
- Bennett, M.D.; Leitch, I.J. Nuclear DNA Amounts in Angiosperms: Progress, Problems and Prospects. Ann. Bot. 2005, 95, 45–90. [Google Scholar] [CrossRef]
- Dart, S.; Kron, P.; Mable, B.K. Characterizing polyploidy in Arabidopsis lyrata using chromosome counts and flow cytometry. Can. J. Bot. 2004, 82, 185–197. [Google Scholar] [CrossRef]
- Bory, S.; Catrice, O.; Brown, S.; Leitch, I.J.; Gigant, R.; Chiroleu, F.; Grisoni, M.; Duval, M.-F.; Besse, P. Natural polyploidy in Vanilla planifolia (Orchidaceae). Genome 2008, 51, 816–826. [Google Scholar] [CrossRef]
- Robertson, I.H. Chromosome numbers in Brachypodium Beauv. (Gramineae). Genetica 1981, 56, 55–60. [Google Scholar] [CrossRef]
- Hasterok, R.; Marasek, A.; Donnison, I.S.; Armstead, I.; Thomas, A.; King, I.P.; Wolny, E.; Idziak, D.; Draper, J.; Jenkins, G. Alignment of the Genomes of Brachypodium distachyon and Temperate Cereals and Grasses Using Bacterial Artificial Chromosome Landing with Fluorescence in Situ Hybridization. Genetics 2006, 173, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Catalán, P.; Müller, J.; Hasterok, R.; Jenkins, G.; Mur, L.A.J.; Langdon, T.; Betekhtin, A.; Siwinska, D.; Pimentel, M.; López-Alvarez, D. Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann. Bot. 2012, 109, 385–405. [Google Scholar] [CrossRef]
- Schwarzacher, T.; Leitch, A.R.; Bennett, M.D.; Heslop-Harrison, J.S. In situ localization of parental genomes in a wide hybrid. Ann. Bot. 1989, 64, 315–324. [Google Scholar] [CrossRef]
- Marková, M.; Vyskot, B. New Horizons of Genomic in situ Hybridization. Cytogenet. Genome Res. 2009, 126, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.B.; Lysak, M.A.; Schubert, I. Genomic in situ hybridization in plants with small genomes is feasible and elucidates the chromosomal parentage in interspecific Arabidopsis hybrids. Genome 2004, 47, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Hasterok, R.; Ksiazczyk, T.; Wolny, E.; Maluszynska, J. FISH and GISH analysis of Brassica genomes. Acta Biol. Crac. Ser. Bot. 2005, 47, 185–192. [Google Scholar]
- Maluszynska, J.; Hasterok, R. Identification of individual chromosomes and parental genomes in Brassica juncea using GISH and FISH. Cytogenet. Genome Res. 2005, 109, 310–314. [Google Scholar] [CrossRef]
- Barre, P.; Layssac, M.; D’Hont, A.; Louarn, J.; Charrier, A.; Hamon, S.; Noirot, M. Relationship between parental chromosomic contribution and nuclear DNA content in the coffee interspecific hybrid C. pseudozanguebariae × C. liberica var ‘dewevrei’. Theor. Appl. Genet. 1998, 96, 301–305. [Google Scholar] [CrossRef]
- Yu-Xiang, W.; Jin-Hong, C.; Qiu-Ling, H.; Shui-Jin, Z. Parental origin and genomic evolution of tetraploid Gossypium species by molecular marker and GISH analyses. Caryologia 2013, 66, 368–374. [Google Scholar] [CrossRef]
- Chase, M.W.; Knapp, S.; Cox, A.V.; Clarkson, J.J.; Butsko, Y.; Joseph, J.; Savolainen, V.; Parokonny, A.S. Molecular Systematics, GISH and the Origin of Hybrid Taxa in Nicotiana (Solanaceae). Ann. Bot. 2003, 92, 107–127. [Google Scholar] [CrossRef]
- Chester, M.; Riley, R.K.; Soltis, P.S.; Soltis, D.E. Patterns of chromosomal variation in natural populations of the neoallotetraploid Tragopogon mirus (Asteraceae). Heredity 2015, 114, 309–317. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Ainouche, M.; Leitch, I.J.; Lim, K.Y.; Le Comber, S.C.; Leitch, A.R. Flow cytometry and GISH reveal mixed ploidy populations and Spartina nonaploids with genomes of S. alterniflora and S. maritima origin. Ann. Bot. 2010, 105, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Vega, J.M.; Han, F.; Lamb, J.C.; Birchler, J.A. Advances in plant chromosome identification and cytogenetic techniques. Curr. Opin. Plant Biol. 2005, 8, 148–154. [Google Scholar] [CrossRef]
- Mukai, Y.; Nakahara, Y.; Yamamoto, M. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 1993, 36, 489–494. [Google Scholar] [CrossRef]
- Amosova, A.V.; Badaeva, E.D.; Muravenko, O.V.; Zelenin, A.V. An improved method of genomic in situ hybridization (GISH) for distinguishing closely related genomes of tetraploid and hexaploid wheat species. Russ. J. Dev. Biol. 2009, 40, 90–94. [Google Scholar] [CrossRef]
- Zhang, P.; Li, W.; Friebe, B.; Gill, B.S. Simultaneous painting of three genomes in hexaploid wheat by BAC-FISH. Genome 2004, 47, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Morton, J.A.; Pellicer, J.; Leitch, I.J.; Leitch, A.R. Genome downsizing after polyploidy: Mechanisms, rates and selection pressures. Plant J. 2021, 107, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Martin, S.L.; Bekele, W.A.; Latta, R.G.; Diederichsen, A.; Peng, Y.; Tinker, N.A. Genome size variation in the genus Avena. Genome 2016, 59, 209–220. [Google Scholar] [CrossRef]
- Mun, J.-H.; Kwon, S.-J.; Yang, T.-J.; Seol, Y.-J.; Jin, M.; Kim, J.-A.; Lim, M.-H.; Kim, J.S.; Baek, S.; Choi, B.-S.; et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009, 10, R111. [Google Scholar] [CrossRef]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef]
- Eilam, T.; Anikster, Y.; Millet, E.; Manisterski, J.; Feldman, M. Genome Size in Diploids, Allopolyploids, and Autopolyploids of Mediterranean Triticeae. J. Bot. 2010, 2010, 341380. [Google Scholar] [CrossRef][Green Version]
- Feldman, M.; Levy, A.A. Genome Evolution Due to Allopolyploidization in Wheat. Genetis 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Liu, B.; Vega, J.M.; Segal, G.; Abbo, S.; Rodova, M.; Feldman, M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 1998, 41, 272–277. [Google Scholar] [CrossRef]
- Leitch, I.J.; Hanson, L.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Clarkson, J.J.; Leitch, A.R. The Ups and Downs of Genome Size Evolution in Polyploid Species of Nicotiana (Solanaceae). Ann. Bot. 2008, 101, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Urbanska, K.M.; Hurka, H.; Landolt, E.; Neuffer, B.; Mummenhoff, K. Hybridization and evolution in Cardamine (Brassicaceae) at Urnerboden, Central Switzerland: Biosystematic and molecular evidence. Plant Syst. Evol 1997, 204, 233–256. [Google Scholar] [CrossRef]
- Abbott, R.J.; Lowe, A. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biol. J. Linn. Soc. 2004, 82, 467–474. [Google Scholar] [CrossRef]
- Ainouche, M.L.; Baumel, A.; Salmon, A. Spartina anglica C. E. Hubbard: A natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biol. J. Linn. Soc. 2004, 82, 475–484. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S.; Pires, J.C.; Kovarik, A.; Tate, J.A.; Mavrodiev, E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): Cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc. 2004, 82, 485–501. [Google Scholar] [CrossRef][Green Version]
- Ownbey, M. Natural Hybridization and Amphiploidy in the Genus Tragopogon. Am. J. Bot. 1950, 37, 487–499. [Google Scholar] [CrossRef]
- Lim, K.Y.; Soltis, D.E.; Soltis, P.S.; Tate, J.; Matyasek, R.; Srubarova, H.; Kovarik, A.; Pires, J.C.; Xiong, Z.; Leitch, A.R. Rapid Chromosome Evolution in Recently Formed Polyploids in Tragopogon (Asteraceae). PLoS ONE 2008, 3, e3353. [Google Scholar] [CrossRef] [PubMed]
- Chester, M.; Gallagher, J.P.; Symonds, V.V.; da Silva, A.V.C.; Mavrodiev, E.V.; Leitch, A.R.; Soltis, P.S.; Soltis, D.E. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 2012, 109, 1176–1181. [Google Scholar] [CrossRef] [PubMed]
- Gaeta, R.T.; Pires, J.C.; Iniguez-Luy, F.; Leon, E.; Osborn, T.C. Genomic Changes in Resynthesized Brassica napus and Their Effect on Gene Expression and Phenotype. Plant Cell 2007, 19, 3403–3417. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Matyasek, R.; Kovarik, A.; Leitch, A.R. Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 2004, 82, 599–606. [Google Scholar] [CrossRef]
- Skalická, K.; Lim, K.Y.; Matyasek, R.; Matzke, M.; Leitch, A.R.; Kovarik, A. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol. 2005, 166, 291–303. [Google Scholar] [CrossRef]
- Szadkowski, E.; Eber, F.; Huteau, V.; Lode, M.; Huneau, C.; Belcram, H.; Coriton, O.; Manzanares-Dauleux, M.J.; Delourme, R.; King, G.J.; et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 2010, 186, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.; Ainouche, M.L.; Wendel, J.F. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 2005, 14, 1163–1175. [Google Scholar] [CrossRef]
- Liu, B.; Brubaker, C.L.; Mergeai, G.; Cronn, R.C.; Wendel, J.F. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 2001, 44, 321–330. [Google Scholar] [CrossRef]
- Fang, L.; Guan, X.; Zhang, T. Asymmetric evolution and domestication in allotetraploid cotton (Gossypium hirsutum L.). Crop J. 2017, 5, 159–165. [Google Scholar] [CrossRef]
- Wang, S.; Chen, J.; Zhang, W.; Hu, Y.; Chang, L.; Fang, L.; Wang, Q.; Lv, F.; Wu, H.; Si, Z.; et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015, 16, 108. [Google Scholar] [CrossRef]
- Yang, Z.; Ge, X.; Yang, Z.; Qin, W.; Sun, G.; Wang, Z.; Li, Z.; Liu, J.; Wu, J.; Wang, Y.; et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat. Commun. 2019, 10, 2989. [Google Scholar] [CrossRef]
- Ardalani, S.; Mirzaghaderi, G.; Badakhshan, H. A Robertsonian translocation from Thinopyrum bessarabicum into bread wheat confers high iron and zinc contents. Plant Breed. 2016, 135, 286–290. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosomal rearrangements in wheat: Their types and distribution. Genome 2007, 50, 907–926. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Dedkova, O.S.; Pukhalskyi, V.A.; Zelenin, A.V. Chromosomal changes over the course of polyploid wheat evolution and domestication. In Advances in Wheat Genetics: From Genome to Field; Springer: Tokyo, Japan, 2015; pp. 83–89. [Google Scholar]
- Friebe, B.; Jiang, J.; Tuleen, N.; Gill, B.S. Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theor. Appl. Genet. 1995, 90, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, H.; Li, B.; Yu, Z.; Li, G.; Zhang, J.; Yang, Z. Molecular Cytogenetic Characterization of New Wheat—Dasypyrum breviaristatum Introgression Lines for Improving Grain Quality of Wheat. Front. Plant Sci. 2018, 9, 365. [Google Scholar] [CrossRef]
- Li, N.; Xu, C.; Zhang, A.; Lv, R.; Meng, X.; Lin, X.; Gong, L.; Wendel, J.F.; Liu, B. DNA methylation repatterning accompanying hybridization, whole genome doubling and homoeolog exchange in nascent segmental rice allotetraploids. New Phytol. 2019, 223, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Lu, P.; Tang, K.; Osborn, T.C. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 7719–7723. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.C. Polyploidy and Diploidy: New Perspectives on Chromosome Pairing and Its Evolutionary Implications. Am. J. Bot. 1982, 69, 1512–1523. [Google Scholar] [CrossRef]
- Svačina, R.; Sourdille, P.; Kopecký, D.; Bartoš, J. Chromosome Pairing in Polyploid Grasses. Front. Plant Sci. 2020, 11, 1056. [Google Scholar] [CrossRef]
- Bhullar, R.; Nagarajan, R.; Bennypaul, H.; Sidhu, G.K.; Sidhu, G.; Rustgi, S.; von Wettstein, D.; Gill, K.S. Silencing of a metaphase I-specific gene results in a phenotype similar to that of the Pairing homeologous 1 gene mutations. Proc. Natl. Acad. Sci. USA 2014, 111, 14187–14192. [Google Scholar] [CrossRef]
- Feldman, M. Cytogenetic Activity and Mode of Action of the Pairing Homoeologous (Ph1) Gene of Wheat. Crop Sci. 1993, 33, 894–897. [Google Scholar] [CrossRef]
- Griffiths, S.; Sharp, R.; Foote, T.N.; Bertin, I.; Wanous, M.; Reader, S.; Colas, I.; Moore, G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 2006, 439, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Bomblies, K.; Jones, G.H.; Franklin, C.S.; Zickler, D.; Kleckner, N. The challenge of evolving stable polyploidy: Could an increase in “crossover interference distance” play a central role? Chromosoma 2016, 125, 287–300. [Google Scholar] [CrossRef]
- Morgan, C.; White, M.A.; Franklin, F.C.H.; Zickler, D.; Kleckner, N.; Bomblies, K. Evolution of crossover interference enables stable autopolyploidy by ensuring pairwise partner connections in Arabidopsis arenosa. Curr. Biol. 2021, 31, 4713–4726.e4. [Google Scholar] [CrossRef]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005, 41, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Gaeta, R.T.; Edger, P.P.; Cao, Y.; Zhao, K.; Zhang, S.; Pires, J.C. Chromosome inheritance and meiotic stability in allopolyploid Brassica napus. G3 2021, 11, jkaa011. [Google Scholar] [CrossRef]
- Buggs, R.J.; Chamala, S.; Wu, W.; Tate, J.A.; Schnable, P.S.; Soltis, D.E.; Soltis, P.S.; Barbazuk, W.B. Rapid, Repeated, and Clustered Loss of Duplicate Genes in Allopolyploid Plant Populations of Independent Origin. Curr. Biol. 2012, 22, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Buggs, R.J.A.; Doust, A.N.; Tate, J.A.; Koh, J.; Soltis, K.; Feltus, F.A.; Paterson, A.H.; Soltis, P.S.; Soltis, D.E. Gene loss and silencing in Tragopogon miscellus (Asteraceae): Comparison of natural and synthetic allotetraploids. Heredity 2009, 103, 73–81. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Kovarik, A.; Robaszkiewicz, E.; Tuna, M.; Tuna, G.S.; Gordon, S.; Vogel, J.P.; Hasterok, R. The fate of 35S rRNA genes in the allotetraploid grass Brachypodium hybridum. Plant J. 2020, 103, 1810–1825. [Google Scholar] [CrossRef] [PubMed]
- Kolano, B.; Tomczak, H.; Molewska, R.; Jellen, E.; Maluszynska, J. Distribution of 5S and 35S rRNA gene sites in 34 Chenopodium species (Amaranthaceae). Bot. J. Linn. Soc. 2012, 170, 220–231. [Google Scholar] [CrossRef]
- Kovarik, A.; Pires, J.C.; Leitch, A.R.; Lim, K.Y.; Sherwood, A.M.; Matyasek, R.; Rocca, J.; Soltis, D.E.; Soltis, P.S. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics 2005, 169, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Malinska, H.; Tate, J.A.; Matyasek, R.; Leitch, A.R.; Soltis, D.E.; Soltis, P.S.; Kovarik, A. Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids. BMC Evol. Biol. 2010, 10, 291. [Google Scholar] [CrossRef] [PubMed]
- Sochorova, J.; Coriton, O.; Kuderova, A.; Lunerova, J.; Chevre, A.M.; Kovarik, A. Gene conversion events and variable degree of homogenization of rDNA loci in cultivars of Brassica napus. Ann. Bot. 2017, 119, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Symonová, R. Integrative rDNAomics-Importance of the Oldest Repetitive Fraction of the Eukaryote Genome. Genes 2019, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Komarova, N.Y.; Hemleben, V. Ribosomal DNA in plant hybrids: Inheritance, rearrangement, expression. Syst. Biodivers. 2007, 5, 261–276. [Google Scholar] [CrossRef]
- Feliner, G.N.; Rosselló, J.A. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol. Phylogenetics Evol. 2007, 44, 911–919. [Google Scholar] [CrossRef]
- Kotseruba, V.; Pistrick, K.; Blattner, F.R.; Kumke, K.; Weiss, O.; Rutten, T.; Fuchs, J.; Endo, T.; Nasuda, S.; Ghukasyan, A.; et al. The evolution of the hexaploid grass Zingeria kochii (Mez) Tzvel. (2n=12) was accompanied by complex hybridization and uniparental loss of ribosomal DNA. Mol. Phylogenetics Evol. 2010, 56, 146–155. [Google Scholar] [CrossRef]
- Lim, K.Y.; Matyasek, R.; Kovarik, A.; Leitch, A. Parental Origin and Genome Evolution in the Allopolyploid Iris versicolor. Ann. Bot. 2007, 100, 219–224. [Google Scholar] [CrossRef]
- Wendel, J.F.; Schnabel, A.; Seelanan, T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA 1995, 92, 280–284. [Google Scholar] [CrossRef]
- Volkov, R.A.; Kostyshin, S.S.; Panchuk, I.I. rDNA organization in species from subfamily Prunoideae. Mol. Biol. 1993, 27, 1356–1367. [Google Scholar]
- Dadejová, M.; Lim, K.Y.; Součkova, K.; Matyášek, R.; Grandbastien, M.-A.; Leitch, A.R.; Kovařík, A. Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids. New Phytol. 2007, 174, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Panchuk, I.I.; Borisjuk, N.V.; Hosiawa-Baranska, M.; Maluszynska, J.; Hemleben, V. Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biol. 2017, 17, 21. [Google Scholar] [CrossRef]
- Clarkson, J.J.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Knapp, S.; Leitch, A.R. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol. 2005, 168, 241–252. [Google Scholar] [CrossRef]
- Kovarik, A.; Dadejova, M.; Lim, Y.K.; Chase, M.W.; Clarkson, J.J.; Knapp, S.; Leitch, A.R. Evolution of rDNA in Nicotiana Allopolyploids: A Potential Link between rDNA Homogenization and Epigenetics. Ann. Bot. 2008, 101, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Kovarik, A.; Matýăsek, R.; Bezděek, M.; Lichtenstein, C.; Leitch, A. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma 2000, 109, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Mahelka, V.; Kopecký, D.; Baum, B.R. Contrasting Patterns of Evolution of 45S and 5S rDNA Families Uncover New Aspects in the Genome Constitution of the Agronomically Important Grass Thinopyrum intermedium (Triticeae). Mol. Biol. Evol. 2013, 30, 2065–2086. [Google Scholar] [CrossRef] [PubMed]
- Borisjuk, N.V.; Momot, V.P.; Gleba, Y. Novel class of rDNA repeat units in somatic hybrids between Nicotiana and Atropa. Theor. Appl. Genet. 1988, 76, 108–112. [Google Scholar] [CrossRef]
- Kovarik, A.; Matyasek, R.; Lim, K.Y.; Skalická, K.; Koukalová, B.; Knapp, S.; Chase, M.; Leitch, A.R. Concerted evolution of 18-5.8-26S rDNA repeats in Nicotiana allotetraploids. Biol. J. Linn. Soc. 2004, 82, 615–625. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Eickbush, D.G. Finely Orchestrated Movements: Evolution of the Ribosomal RNA Genes. Genetics 2007, 175, 477–485. [Google Scholar] [CrossRef]
- Nei, M.; Rooney, A.P. Concerted and Birth-and-Death Evolution of Multigene Families. Annu. Rev. Genet. 2005, 39, 121–152. [Google Scholar] [CrossRef]
- Cronn, R.; Zhao, X.; Paterson, A.H.; Wendell, J.F. Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J. Mol. Evol. 1996, 42, 685–705. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.; Appels, R. Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 1995, 140, 325–343. [Google Scholar] [CrossRef] [PubMed]
- Parisod, C.; Alix, K.; Just, J.; Petit, M.; Sarilar, V.; Mhiri, C.; Ainouche, M.; Chalhoub, B.; Grandbastien, M.-A. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010, 186, 37–45. [Google Scholar] [CrossRef]
- Wang, J.; Tian, L.; Lee, H.-S.; Wei, N.E.; Jiang, H.; Watson, B.; Madlung, A.; Osborn, T.C.; Doerge, R.W.; Comai, L.; et al. Genomewide Nonadditive Gene Regulation in Arabidopsis Allotetraploids. Genetics 2006, 172, 507–517. [Google Scholar] [CrossRef]
- Lloyd, A.; Blary, A.; Charif, D.; Charpentier, C.; Tran, J.; Balzergue, S.; Delannoy, E.; Rigaill, G.; Jenczewski, E. Homoeologous exchanges cause extensive dosage-dependent gene expression changes in an allopolyploid crop. New Phytol. 2018, 217, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhong, L.; Wu, X.; Fang, X.; Wang, J. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 2009, 229, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Sehrish, T.; Symonds, V.V.; Soltis, D.E.; Soltis, P.S.; Tate, J.A. Gene silencing via DNA methylation in naturally occurring Tragopogon miscellus (Asteraceae) allopolyploids. BMC Genom. 2014, 15, 701. [Google Scholar] [CrossRef][Green Version]
- Yoo, M.-J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 2013, 110, 171–180. [Google Scholar] [CrossRef]
- Chagué, V.; Just, J.; Mestiri, I.; Balzergue, S.; Tanguy, A.; Huneau, C.; Huteau, V.; Belcram, H.; Coriton, O.; Jahier, J.; et al. Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 2010, 187, 1181–1194. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Li, Y.; Zhang, Z.; Li, L.; Liu, B. Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA -sequencing. New Phytol. 2016, 209, 1264–1277. [Google Scholar] [CrossRef]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Yaakov, B.; Kashkush, K. Methylation, Transcription, and Rearrangements of Transposable Elements in Synthetic Allopolyploids. Int. J. Plant Genom. 2011, 2011, 569826. [Google Scholar] [CrossRef]
- Navashin, M. Chromosomal alterations caused by hybridization and their bearing upon certain general genetic problems. Cytologia 1934, 5, 169–203. [Google Scholar] [CrossRef]
- Ge, X.H.; Ding, L.; Li, Z.Y. Nucleolar dominance and different genome behaviors in hybrids and allopolyploids. Plant Cell Rep. 2013, 32, 1661–1673. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S. Nucleolar dominance: Uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol. Biol. 2000, 43, 163–177. [Google Scholar] [CrossRef]
- Idziak, D.; Hasterok, R. Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome 2008, 51, 387–391. [Google Scholar] [CrossRef]
- Hizume, M.; Sato, S.; Tanaka, A. A highly reproducible method of nucleolus organizing regions staining in plants. Stain Technol. 1980, 55, 87–90. [Google Scholar]
- Chen, Z.J.; Pikaard, C. Transcriptional analysis of nucleolar dominance in polyploid plants: Biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. USA 1997, 94, 3442–3447. [Google Scholar] [CrossRef]
- Hasterok, R.; Maluszynska, J. Nucleolar dominance does not occur in root tip cells of allotetraploid Brassica species. Genome 2000, 43, 574–579. [Google Scholar] [CrossRef]
- Pontes, O.; Lawrence, R.J.; da Silva, M.G.; Preuss, S.; Costa-Nunes, P.; Earley, K.; Neves, N.; Viegas, W.; Pikaard, C.S. Postembryonic Establishment of Megabase-Scale Gene Silencing in Nucleolar Dominance. PLoS ONE 2007, 2, e1157. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Robaszkiewicz, E.; Mykhailyk, S.; Wartini, J.; Pinski, A.; Kovarik, A.; Hasterok, R. To Be or Not to Be Expressed: The First Evidence of a Nucleolar Dominance Tissue-Specificity in Brachypodium hybridum. Front. Plant Sci. 2021, 12, 768347. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Hasterok, R. Epigenetics of the preferential silencing of Brachypodium stacei-originated 35S rDNA loci in the allotetraploid grass Brachypodium hybridum. Sci. Rep. 2017, 7, 5260. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Pikaard, C.S. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997, 11, 2124–2136. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.; Lawrence, R.J.; Pontes, O.; Reuther, R.; Enciso, A.J.; Silva, M.; Neves, N.; Gross, M.; Viegas, W.; Pikaard, C.S. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006, 20, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.Y.; Grabe, T.; Huigen, D.J.; Hemleben, V.; Volkov, R.A. Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol. Biol. 2004, 56, 439–463. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, R.J.; Earley, K.; Pontes, O.; Silva, M.; Chen, Z.J.; Neves, N.; Viegas, W.; Pikaard, C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 2004, 13, 599–609. [Google Scholar] [CrossRef]
- Neves, N.; Heslop-Harrison, J.S.; Viegas, W. rRNA gene activity and control of expression mediated by methylation and imprinting during embryo development in wheat x rye hybrids. Theor. Appl. Genet. 1995, 91, 529–533. [Google Scholar] [CrossRef]
- Pontvianne, F.; Blevins, T.; Chandrasekhara, C.; Feng, W.; Stroud, H.; Jacobsen, S.E.; Michaels, S.D.; Pikaard, C.S. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 2012, 26, 945–957. [Google Scholar] [CrossRef]
- Vieira, R.; Mellosampayo, T.; Viegas, W.S. 1R chromosome nucleolus organizer region activation by 5-azacytidine in wheat x rye hybrids. Genome 1990, 33, 707–712. [Google Scholar] [CrossRef]
- Guo, X.; Han, F. Asymmetric Epigenetic Modification and Elimination of rDNA Sequences by Polyploidization in Wheat. Plant Cell 2014, 26, 4311–4327. [Google Scholar] [CrossRef]
- Costa-Nunes, P.; Pontes, O.; Preuss, S.B.; Pikaard, C.S. Extra views on RNA-dependent DNA methylation and MBD6-dependent heterochromatin formation in nucleolar dominance. Nucleus 2010, 1, 254–259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Preuss, S.B.; Costa-Nunes, P.; Tucker, S.; Pontes, O.; Lawrence, R.J.; Mosher, R.; Kasschau, K.D.; Carrington, J.; Baulcombe, D.; Viegas, W.; et al. Multimegabase Silencing in Nucleolar Dominance Involves siRNA-Directed DNA Methylation and Specific Methylcytosine-Binding Proteins. Mol. Cell 2008, 32, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Herklotz, V.; Kovařík, A.; Lunerová, J.; Lippitsch, S.; Groth, M.; Ritz, C.M. The fate of ribosomal RNA genes in spontaneous polyploid dogrose hybrids [Rosa L. sect. Caninae (DC.) Ser.] exhibiting non-symmetrical meiosis. Plant J. 2018, 94, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Mohannath, G.; Pontvianne, F.; Pikaard, C.S. Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proc. Natl. Acad. Sci. USA 2016, 113, 13426–13431. [Google Scholar] [CrossRef] [PubMed]
- Nicoloff, H.; Anastassova-Kristeva, M.; Rieger, R.; Künzel, G. ‘Nucleolar dominance’ as observed in barley translocation lines with specifically reconstructed SAT chromosomes. Theor. Appl. Genet. 1979, 55, 247–251. [Google Scholar] [CrossRef]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef]
- Potapova, T.A.; Unruh, J.R.; Yu, Z.; Rancati, G.; Li, H.; Stampfer, M.R.; Gerton, J.L. Superresolution microscopy reveals linkages between ribosomal DNA on heterologous chromosomes. J. Cell Biol. 2019, 218, 2492–2513. [Google Scholar] [CrossRef]
- Sepsi, A.; Fábián, A.; Jäger, K.; Heslop-Harrison, J.S.; Schwarzacher, T. ImmunoFISH: Simultaneous Visualisation of Proteins and DNA Sequences Gives Insight into Meiotic Processes in Nuclei of Grasses. Front. Plant Sci. 2018, 9, 1193. [Google Scholar] [CrossRef] [PubMed]
- Dreissig, S.; Schiml, S.; Schindele, P.; Weiss, O.; Rutten, T.; Schubert, V.; Gladilin, E.; Mette, M.F.; Puchta, H.; Houben, A. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017, 91, 565–573. [Google Scholar] [CrossRef]

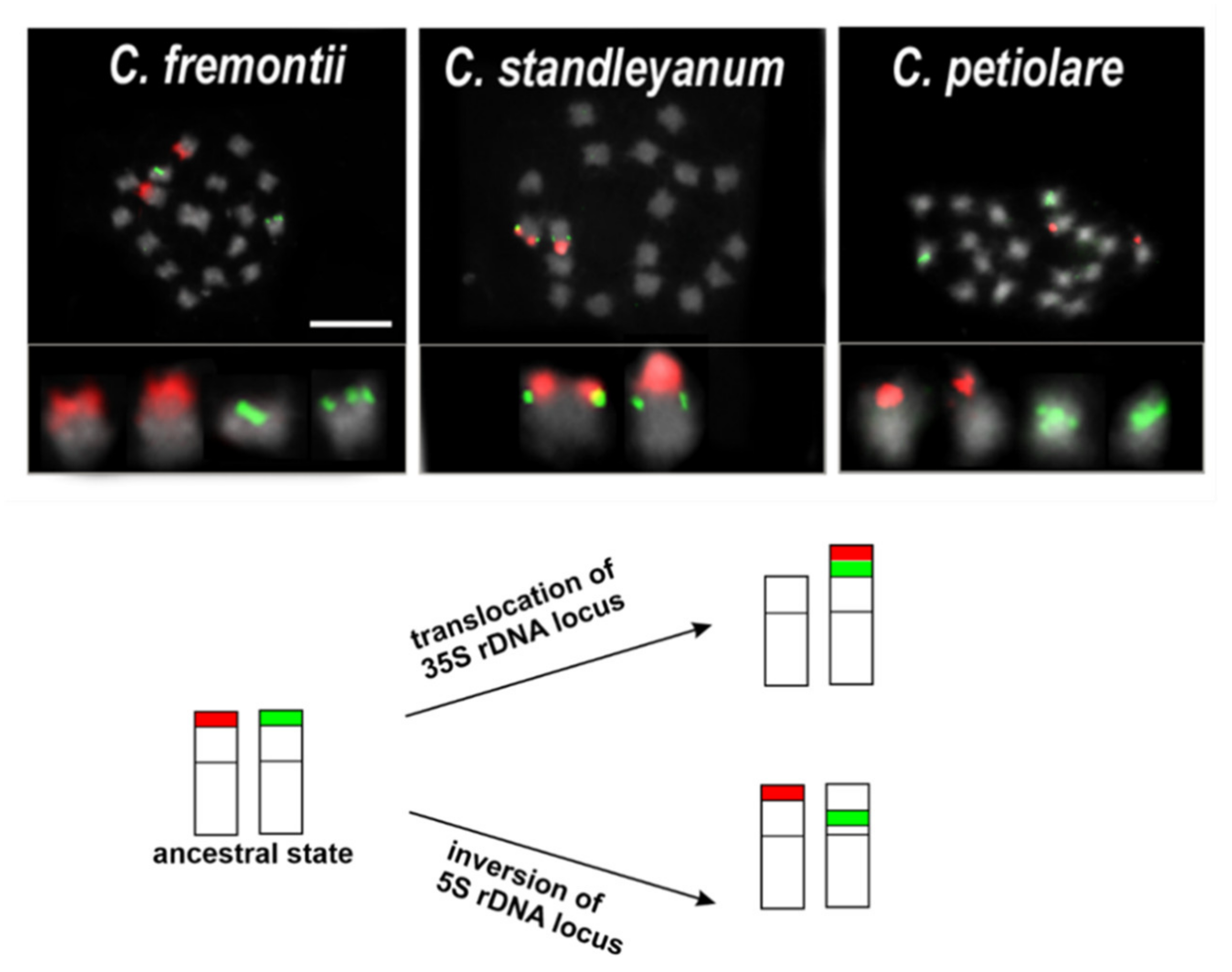

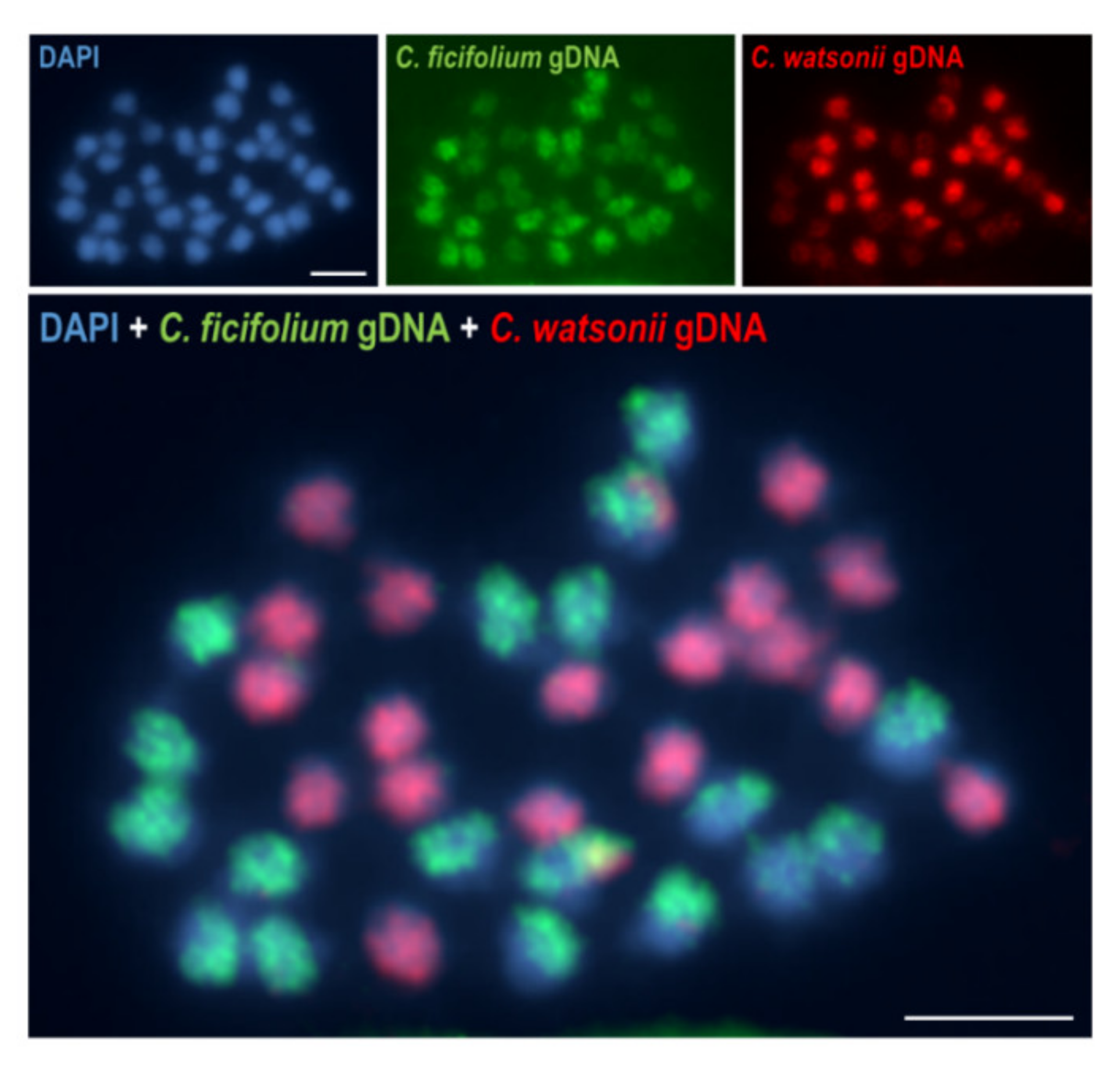
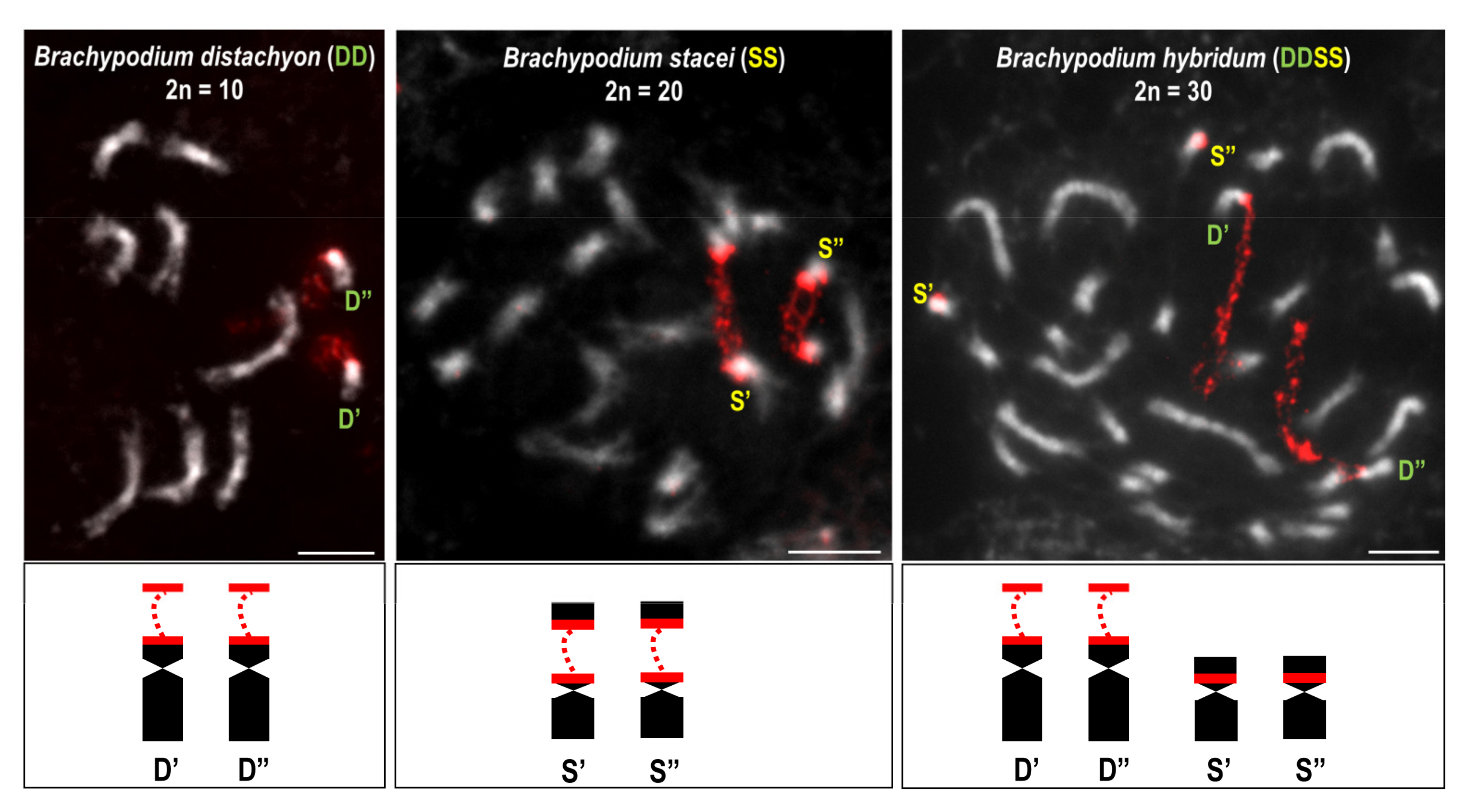
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borowska-Zuchowska, N.; Senderowicz, M.; Trunova, D.; Kolano, B. Tracing the Evolution of the Angiosperm Genome from the Cytogenetic Point of View. Plants 2022, 11, 784. https://doi.org/10.3390/plants11060784
Borowska-Zuchowska N, Senderowicz M, Trunova D, Kolano B. Tracing the Evolution of the Angiosperm Genome from the Cytogenetic Point of View. Plants. 2022; 11(6):784. https://doi.org/10.3390/plants11060784
Chicago/Turabian StyleBorowska-Zuchowska, Natalia, Magdalena Senderowicz, Dana Trunova, and Bozena Kolano. 2022. "Tracing the Evolution of the Angiosperm Genome from the Cytogenetic Point of View" Plants 11, no. 6: 784. https://doi.org/10.3390/plants11060784
APA StyleBorowska-Zuchowska, N., Senderowicz, M., Trunova, D., & Kolano, B. (2022). Tracing the Evolution of the Angiosperm Genome from the Cytogenetic Point of View. Plants, 11(6), 784. https://doi.org/10.3390/plants11060784






