Genome-Wide Identification and Expression Profile Reveal Potential Roles of Peanut ZIP Family Genes in Zinc/Iron-Deficiency Tolerance
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of the AhZIP Family in Peanut
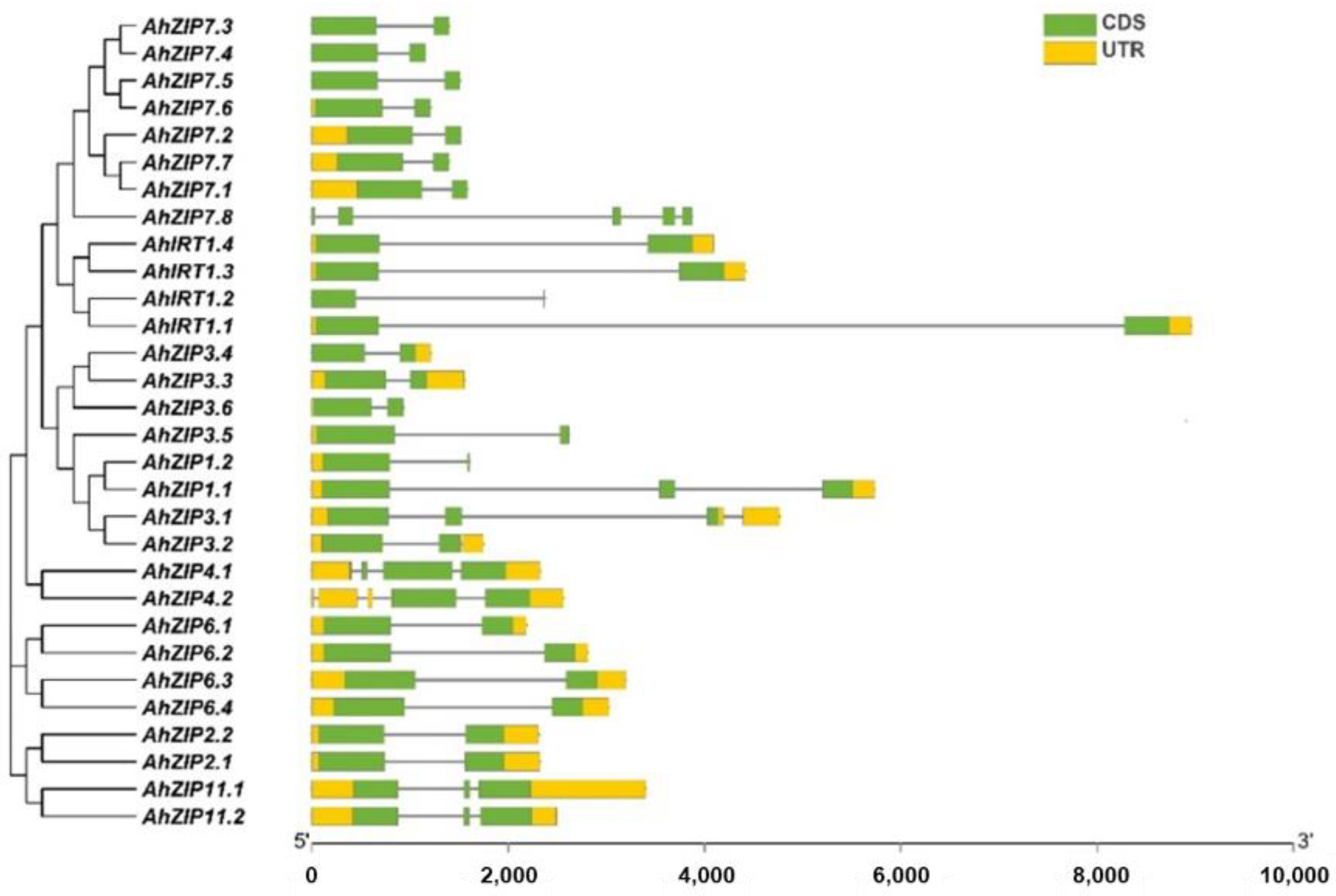
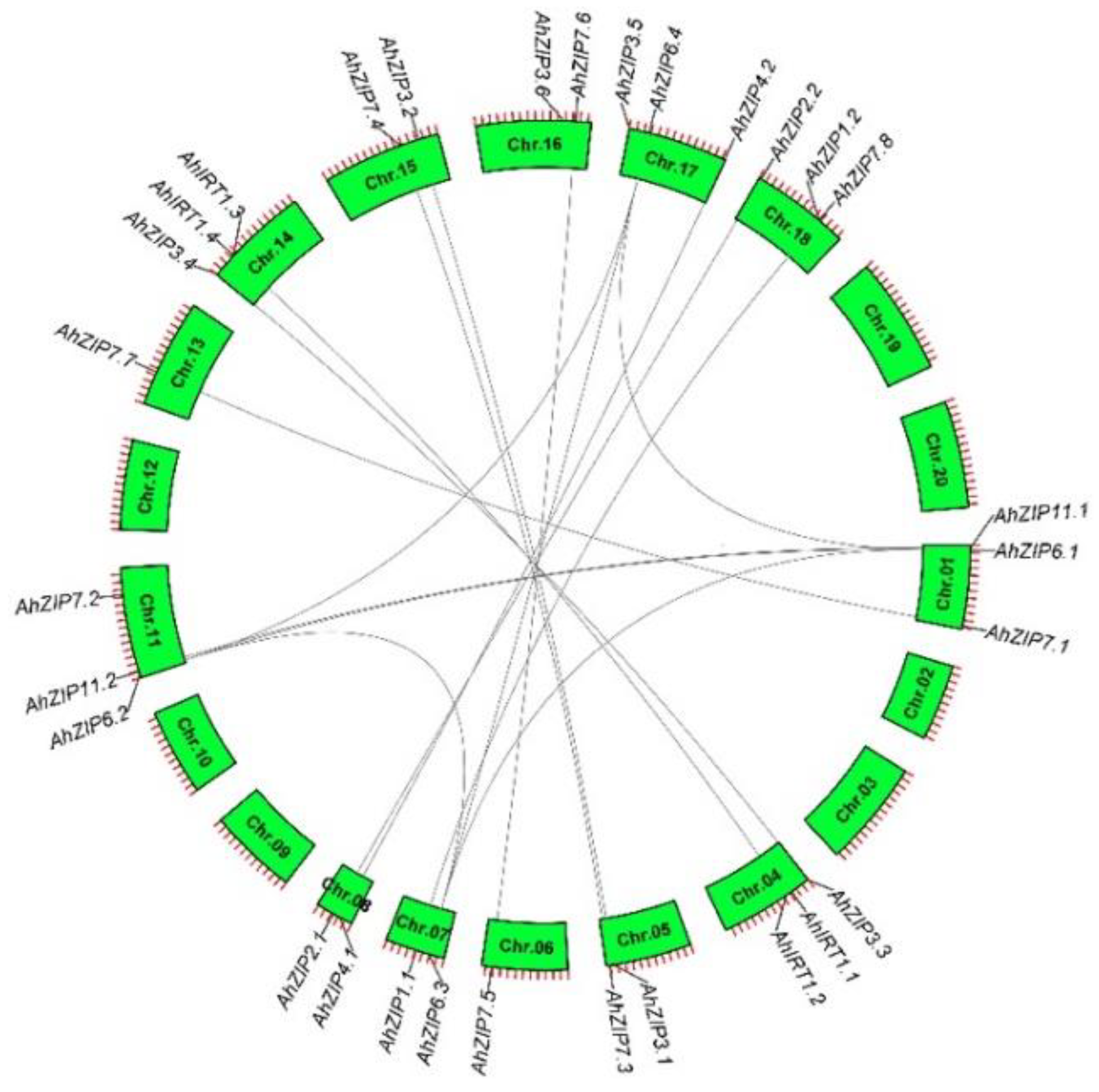
2.2. Gene Structure, Duplication and Ka/Ks of the AhZIP Family
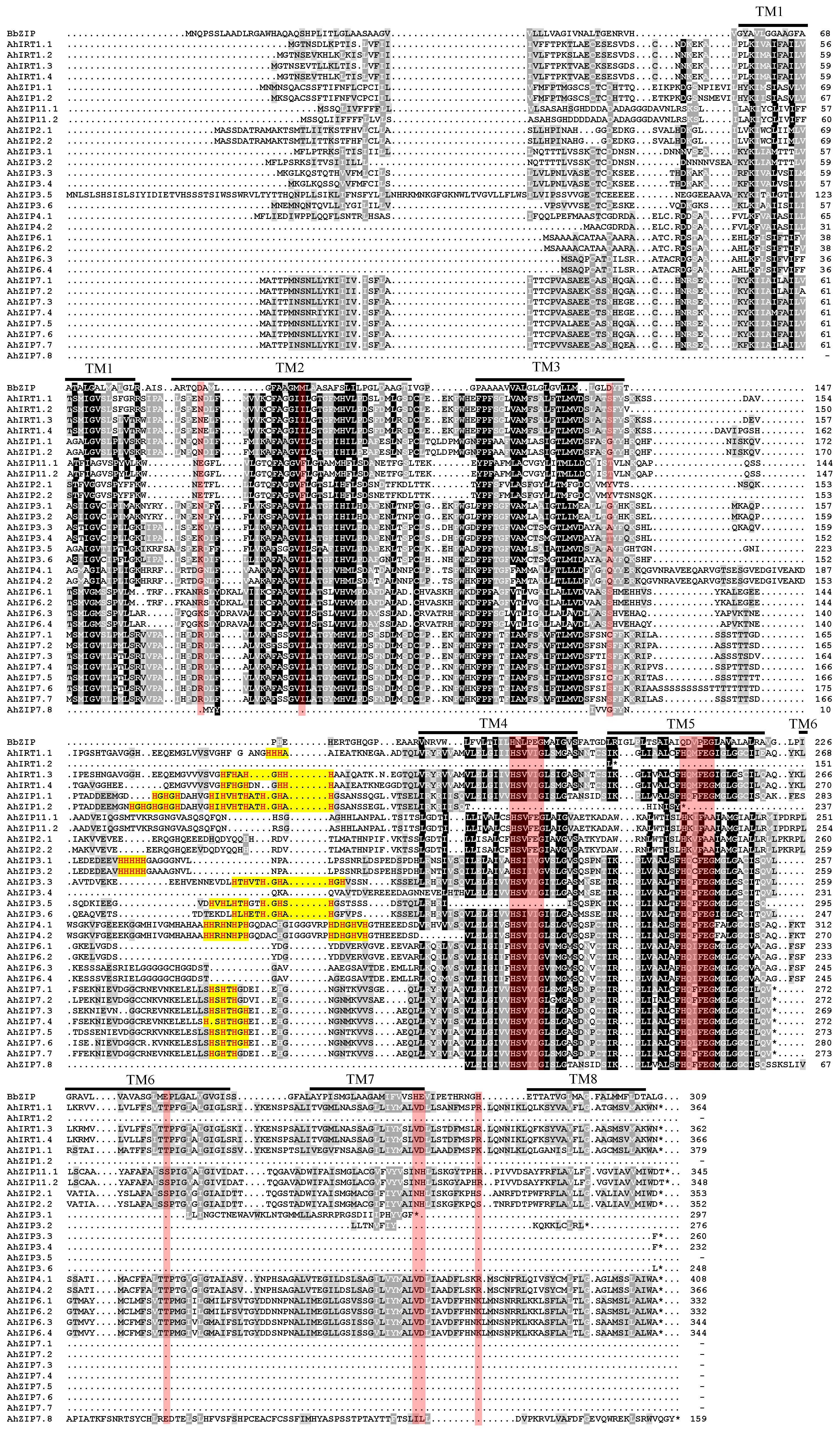
2.3. Conserved Motifs, Domain Architectures and Models of AhZIP Proteins
2.4. Expression Profiles of AhZIP Genes in Different Tissues of Peanut
2.5. Gene Expression of AhZIPs in Response to Fe- and Zn-Deficiency
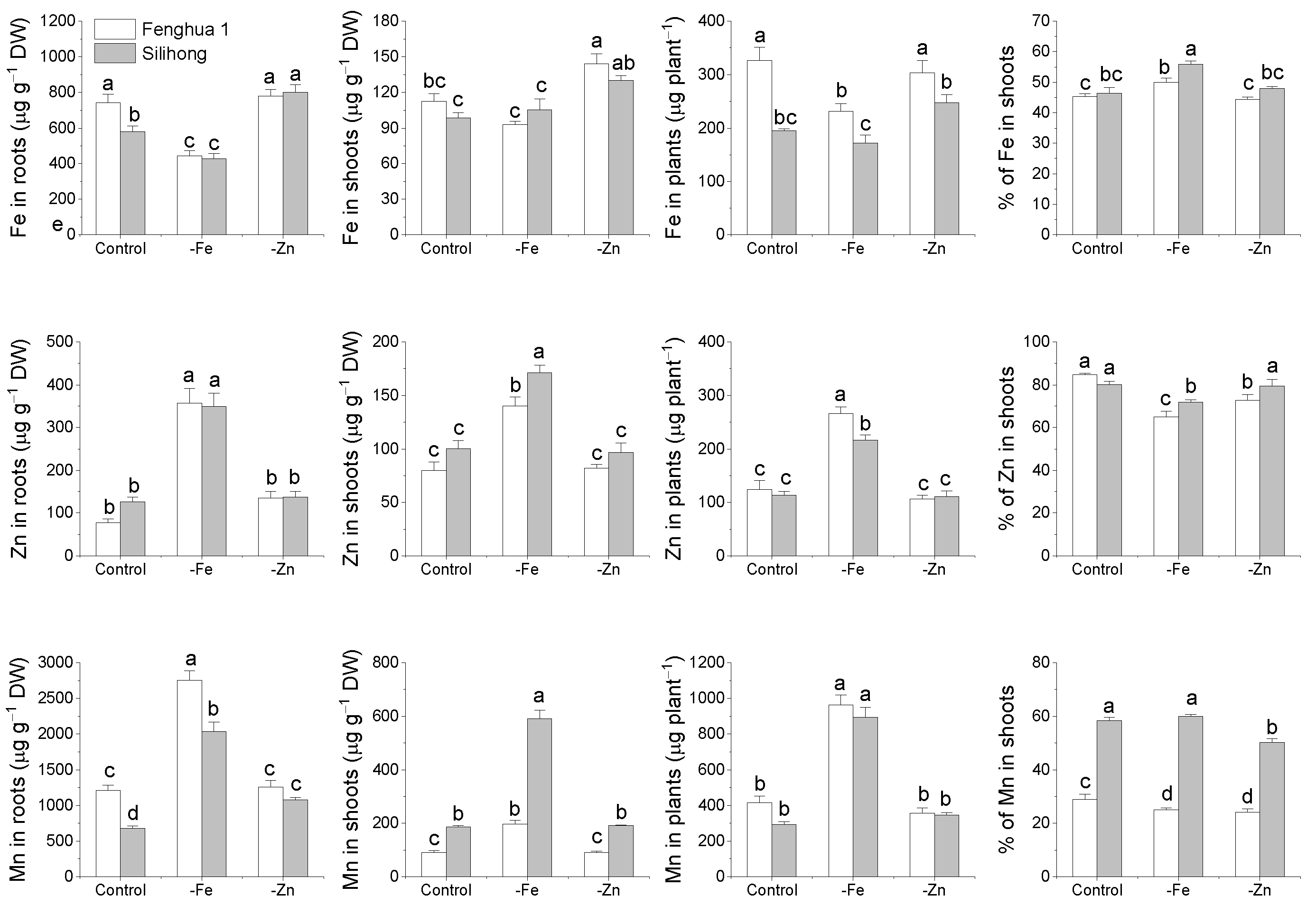
2.6. Metal Accumulation and Translocation in Response to Fe- or Zn-Deficiency
2.7. Relationship of Gene Expression of AhZIPs and Metal Accumulation
3. Discussion
4. Materials and Methods
4.1. Identification of ZIP Genes in Peanut
4.2. Phylogenetic and Structural Analysis of AhZIP Proteins
4.3. Structure, Duplication and Ka/Ks of AhZIP Genes
4.4. Gene Expression Analysis Based on RNA-Seq Data
4.5. Plant Growth, Metal Determination and RT-qPCR Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: Boston, MA, USA, 2012. [Google Scholar]
- Gómez-Galera, S.; Rojas, E.; Sudhakar, D.; Zhu, C.; Pelacho, A.M.; Capell, T.; Christou, P. Critical evaluation of strategies for mineral fortification of staple food crops. Transgenic Res. 2010, 19, 165–180. [Google Scholar] [CrossRef]
- Guerinot, M.L.; Yi, Y. Iron: Nutritious, noxious, and not readily available. Plant Physiol. 1994, 104, 815–820. [Google Scholar] [CrossRef] [Green Version]
- Alloway, B.J. Soil factors associated with zinc deficiency in crops and humans. Environ. Geochem. Health 2009, 31, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Jin, Y.; Li, Y.; Zhai, F.; Kok, F.J.; Jacobsen, E.; Yang, X. Iron and zinc deficiencies in China: What is a feasible and cost-effective strategy? Public Health Nutr. 2008, 11, 632–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korshunova, Y.O.; Eide, D.; Clark, W.G.; Guerinot, M.L.; Pakrasi, H.B. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 1999, 40, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.; Broderius, M.; Fett, J.; Guerinot, M.L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 5624–5628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, S.; Tsuzuki, C.; Kato, A.; Aisu, A.; Yoshida, J.; Mizuno, T. AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1433–1442. [Google Scholar] [CrossRef] [Green Version]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krausko, M.; Labajová, M.; Peterková, D.; Jásik, J. Specific expression of AtIRT1 in phloem companion cells suggests its role in iron translocation in aboveground plant organs. Plant Signal. Behav. 2021, 16, 1925020. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Barberon, M.; Zelazny, E.; Séguéla, M.; Briat, J.-F.; Curie, C. Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta 2009, 229, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Liang, H.-M.; Yang, S.-Y.; Boch, A.; Clemens, S.; Chen, C.-C.; Wu, J.-F.; Huang, J.-L.; Yeh, K.-C. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 2009, 182, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Milner, M.J.; Seamon, J.; Craft, E.; Kochian, L.V. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 2013, 64, 369–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grotz, N.; Fox, T.; Connolly, E.; Park, W.; Guerinot, M.L.; Eide, D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. USA 1998, 95, 7220–7224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Biophys. Acta (BBA)-Biomembr. 2000, 1465, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef]
- Nakanishi, H.; Ogawa, I.; Ishimaru, Y.; Mori, S.; Nishizawa, N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006, 52, 464–469. [Google Scholar] [CrossRef]
- Lee, S.; An, G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009, 32, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Feng, S.J.; Zhang, B.Q.; Wang, M.Q.; Cao, H.W.; Rono, J.K.; Chen, X.; Yang, Z.M. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 2019, 19, 283. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Suzuki, M.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 2005, 56, 3207–3214. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Masuda, H.; Suzuki, M.; Bashir, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J. Exp. Bot. 2007, 58, 2909–2915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Jeong, H.J.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 2010, 73, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 2010, 29, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Gindri, R.G.; Navarro, B.B.; da Cruz Dias, P.V.; Tarouco, C.P.; Nicoloso, F.T.; Brunetto, G.; Berghetti, L.P.; da Silva, L.O.S.; Fett, J.P.; Menguer, P.K.; et al. Physiological responses of rice (Oryza sativa L.) oszip7 loss-of-function plants exposed to varying Zn concentrations. Physiol. Mol. Biol. Plants 2020, 26, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Ricachenevsky, F.K.; Punshon, T.; Lee, S.; Oliveira, B.H.N.; Trenz, T.S.; Maraschin, F.D.S.; Hindt, M.N.; Danku, J.; Salt, D.E.; Fett, J.P.; et al. Elemental profiling of rice FOX lines leads to characterization of a new Zn plasma membrane trans-porter, OsZIP7. Front. Plant Sci. 2018, 9, 865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Zhu, Y.; Fan, T.; Peng, C.; Wang, J.; Sun, L.; Chen, C. OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun. 2019, 512, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.K.; Ganie, S.A.; Rana, M.K.; Sharma, T.R. Genome-wide analysis of zinc transporter genes of maize (Zea mays). Plant Mol. Biol. Rep. 2013, 32, 605–616. [Google Scholar] [CrossRef]
- Li, X.B.; Suo, H.C.; Liu, J.T.; Wang, L.; Li, C.C.; Liu, W. Genome-wide identification and expression analysis of the potato ZIP gene family under Zn-deficiency. Biol. Plant. 2020, 64, 845–855. [Google Scholar] [CrossRef]
- Migeon, A.; Blaudez, D.; Wilkins, O.; Montanini, B.; Campbell, M.M.; Richaud, P.; Thomine, S.; Chalot, M. Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cell. Mol. Life Sci. 2010, 67, 3763–3784. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, S.; Li, D.; Xu, X.; Li, C. Genome-wide analysis of the zrt, irt-like protein (zip) family and their responses to metal stress in Populus trichocarpa. Plant Mol. Biol. Rep. 2017, 35, 534–549. [Google Scholar] [CrossRef]
- Fu, X.Z.; Zhou, X.; Xing, F.; Ling, L.L.; Chun, C.P.; Cao, L.; Aarts, M.G.M.; Peng, L.Z. Genome-wide identification, cloning and functional analysis of the zinc/iron-regulated transporter-like protein (ZIP) gene family in trifoliate orange (Poncirus trifoliata L. Raf.). Front. Plant Sci. 2017, 8, 588. [Google Scholar] [CrossRef] [Green Version]
- Gaither, L.A.; Eide, D.J. Eukaryotic zinc transporters and their regulation. BioMetals 2001, 14, 251–270. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.; Kannan, P.; Prabukumar, G.; Govindaraj, M. Zinc deficiency in Indian soils with special focus to enrich zinc in peanut. Afr. J. Agric. Res. 2013, 8, 6681–6688. [Google Scholar]
- Lal, C.; Singh, A.L. Screening for high zinc density groundnut genotypes in India. In Proceedings of the Zinc Crops 2007 Conference for Improving Crop Production and Human Health, Istanbul, Turkey, 24–26 May 2007. [Google Scholar]
- Su, Y.; Zhang, Z.; Su, G.; Liu, J.; Liu, C.; Shi, G. Genotypic differences in spectral and photosynthetic response of peanut to iron deficiency. J. Plant Nutr. 2015, 38, 145–160. [Google Scholar] [CrossRef]
- Ding, H.; Duan, L.; Jing, L.; Yan, H.; Zhao, M.; Zhang, F.; Li, W.X. Cloning and functional analysis of the peanut iron trans-porter AhIRT1 during iron deficiency stress and intercropping with maize. J. Plant Physiol. 2010, 167, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Kobayashi, T.; Kakei, Y.; Senoura, T.; Nakazono, M.; Takahashi, H.; Nakanishi, H.; Shen, H.; Duan, P.; Guo, X.; et al. AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J. Exp. Bot. 2012, 63, 4437–4446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [Green Version]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, J.; Fellner, M.; Zhang, C.; Sui, D.; Hu, J. Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 2017, 3, e1700344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pattee, H.E.; Johns, E.B.; Singleton, J.A.; Sanders, T.H. Composition changes of peanut fruit parts during maturation1. Peanut Sci. 1974, 1, 57–62. [Google Scholar] [CrossRef]
- Bafaro, E.M.; Antala, S.; Nguyen, T.-V.; Dzul, S.P.; Doyon, B.; Stemmler, T.L.; Dempski, R.E. The large intracellular loop of hZIP4 is an intrinsically disordered zinc binding domain. Metallomics 2015, 7, 1319–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, W.; Liao, B.-Y.; Chang, A.Y.-F.; Zhang, J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010, 26, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Yamaji, N.; Mitani-Ueno, N.; Kashino, M.; Ma, J.F. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 2015, 84, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Qu, M.; Zhu, Y.; Peng, C.; Wang, J.; Gao, D.; Chen, C. Zinc transporter5 and zinc transporter9 function synergistically in zinc/cadmium uptake. Plant Physiol. 2020, 183, 1235–1249. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, P.G.; Sam, K.; Mathew, M.K. Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol. Biochem. 2015, 97, 165–174. [Google Scholar]
- Wu, J.; Zhao, F.J.; Ghandilyan, A.; Logoteta, B.; Guzman, M.O.; Schat, H.; Wang, X.; Aarts, M.G.M. Identification and func-tional analysis of two ZIP metal transporters of the hyperaccumulator Thlaspi caerulescens. Plant Soil. 2009, 325, 79. [Google Scholar] [CrossRef] [Green Version]
- Spielmann, J.; Ahmadi, H.; Scheepers, M.; Weber, M.; Nitsche, S.; Carnol, M.; Bosman, B.; Kroymann, J.; Motte, P.; Clemens, S.; et al. The two copies of the zinc and cadmium ZIP6 transporter of Arabidopsis halleri have distinct effects on cadmium tolerance. Plant Cell Environ. 2020, 43, 2143–2157. [Google Scholar] [CrossRef] [PubMed]
- Kozak, K.; Papierniak, A.; Barabasz, A.; Kendziorek, M.; Palusińska, M.; Williams, L.E.; Antosiewicz, D.M. NtZIP11, a new Zn transporter specifically upregulated in tobacco leaves by toxic Zn level. Environ. Exp. Bot. 2018, 157, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Cao, Q.; Xv, C.; Jiang, Q.; Wang, L.; Shi, G. Comparative transcriptome analysis reveals key genes responsible for the homeo-stasis of iron and other divalent metals in peanut roots under iron deficiency. Plant Soil. 2019, 445, 513–531. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular lo-calization. PLoS ONE. 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TB tools: An integrative toolkit developed for in-teractive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jin, J.P.; Guo, A.-Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Liu, J.; Lu, Z.; Wang, X.; Zhang, Z.; Shi, G. Effects of iron deficiency on subcellular distribution and chemical forms of cadmium in peanut roots in relation to its translocation. Environ. Exp. Bot. 2013, 97, 40–48. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Z.; Su, Y.; Liu, C.; Shi, G. Cultivar variation in morphological response of peanut roots to cadmium stress and its relation to cadmium accumulation. Ecotoxicol. Environ. Saf. 2013, 91, 147–155. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Gene ID | Gene Length (bp) | CDS (bp) | MW a (kDa) | Aa b | Instability | Aliphatic Index | GRAVY c | pI d | No. of TMD e | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AhIRT1.1 | arahy.T4CX6H | 8962 | 1095 | 38.94 | 364 | 33.45 | 109.29 | 0.55 | 6.3 | 8/out-out | PM f |
| AhIRT1.2 | arahy.B3ZT22 | 2373 | 456 | 16.55 | 151 | 47.1 | 109.07 | 0.654 | 5.28 | 3/out-in | PM |
| AhIRT1.3 | arahy.234RNS | 4415 | 1089 | 39.25 | 362 | 36.8 | 111.99 | 0.573 | 6.86 | 8/out-out | PM |
| AhIRT1.4 | arahy.8VMZ7D | 4094 | 1101 | 39.56 | 366 | 37.5 | 108.63 | 0.53 | 6.34 | 8/out-out | PM |
| AhZIP1.1 | arahy.XJH13Y | 5732 | 1140 | 40.51 | 379 | 41.77 | 104.01 | 0.477 | 6.29 | 8/out-out | PM |
| AhZIP1.2 | arahy.ZLZ7ZM | 1608 | 714 | 25.74 | 237 | 44.01 | 86.03 | 0.167 | 6.53 | 3/out-in | EMS g |
| AhZIP2.1 | arahy.VX1J70 | 2322 | 1062 | 38.77 | 353 | 26.98 | 107.48 | 0.495 | 6.52 | 8/in-out | PM |
| AhZIP2.2 | arahy.1Q0IUD | 2309 | 1059 | 38.71 | 352 | 27.91 | 103.35 | 0.472 | 6.62 | 8/in-out | PM |
| AhZIP3.1 | arahy.CK2LDM | 4766 | 894 | 32.38 | 297 | 39.12 | 107.71 | 0.254 | 6.03 | 5/out-in | PM |
| AhZIP3.2 | arahy.E7VKLQ | 1747 | 831 | 30.12 | 276 | 38.03 | 110.62 | 0.288 | 6.62 | 5/out-in | EMS |
| AhZIP3.3 | arahy.ZVRF07 | 1554 | 783 | 28.26 | 260 | 33.11 | 107.19 | 0.423 | 6.08 | 5/out-in | PM |
| AhZIP3.4 | arahy.30BR38 | 1211 | 699 | 25.09 | 232 | 33.22 | 109.66 | 0.568 | 5.28 | 5/out-in | PM |
| AhZIP3.5 | arahy.WQ3KQR | 2618 | 888 | 32.26 | 295 | 37.42 | 99.49 | 0.212 | 6.45 | 3/out-in | EMS |
| AhZIP3.6 | arahy.05ZQZB | 937 | 747 | 26.99 | 248 | 31.92 | 117.14 | 0.524 | 5.91 | 5/out-in | PM |
| AhZIP4.1 | arahy.KIJ6L7 | 2325 | 1227 | 43.82 | 408 | 42.93 | 95.22 | 0.324 | 6.02 | 8/out-out | PM |
| AhZIP4.2 | arahy.2FF2JF | 2561 | 1101 | 38.83 | 366 | 41.96 | 95.25 | 0.34 | 6.02 | 8/out-out | PM |
| AhZIP6.1 | arahy.58GIJL | 2185 | 999 | 35.60 | 332 | 35.92 | 106.99 | 0.631 | 5.77 | 8/out-out | PM |
| AhZIP6.2 | arahy.78T540 | 2812 | 999 | 35.54 | 332 | 35.89 | 107.29 | 0.647 | 5.9 | 8/out-out | PM |
| AhZIP6.3 | arahy.0DI5A2 | 3196 | 1035 | 36.29 | 344 | 37.88 | 106.1 | 0.629 | 6.29 | 8/out-out | PM |
| AhZIP6.4 | arahy.E6QUMC | 3026 | 1035 | 36.31 | 344 | 37.66 | 106.37 | 0.630 | 6.29 | 8/out-out | PM |
| AhZIP7.1 | arahy.BTX8K3 | 1585 | 819 | 29.67 | 272 | 45.6 | 103.2 | 0.400 | 6.10 | 5/out-in | PM |
| AhZIP7.2 | arahy.092K8V | 1514 | 819 | 29.61 | 272 | 44.17 | 104.3 | 0.413 | 6.58 | 5/out-in | PM |
| AhZIP7.3 | arahy.RP74H9 | 1398 | 810 | 29.45 | 269 | 45.52 | 109.44 | 0.412 | 6.04 | 5/out-in | PM |
| AhZIP7.4 | arahy.FX0GN0 | 1151 | 819 | 29.88 | 272 | 44.84 | 103.57 | 0.347 | 6.62 | 5/out-in | PM |
| AhZIP7.5 | arahy.1C0EWX | 1510 | 822 | 29.68 | 273 | 44.23 | 106.78 | 0.429 | 6.62 | 5/out-in | PM |
| AhZIP7.6 | arahy.QZI7QE | 1207 | 843 | 30.33 | 280 | 46.91 | 104.11 | 0.399 | 6.62 | 5/out-in | PM |
| AhZIP7.7 | arahy.0E1GBK | 1397 | 822 | 29.88 | 273 | 45.75 | 104.62 | 0.408 | 6.1 | 5/out-in | PM |
| AhZIP7.8 | arahy.NIU36G | 3869 | 480 | 17.69 | 159 | 46.83 | 99.25 | 0.432 | 6.57 | 4/out-out | EMS |
| AhZIP11.1 | arahy.HSP4SF | 3404 | 1038 | 36.62 | 345 | 30.58 | 112.87 | 0.767 | 5.73 | 8/out-out | PM |
| AhZIP11.2 | arahy.W56MR2 | 2492 | 1047 | 36.94 | 348 | 32.71 | 113.05 | 0.749 | 5.49 | 8/out-out | PM |
| Gene Pairs | Duplicate Type | Ka a | Ks b | Ka/Ks c | Positive Selection |
|---|---|---|---|---|---|
| AhZIP6.1/6.4 | Segmental | 0.1912 | 1.3731 | 0.1393 | No |
| AhZIP6.1/6.3 | Segmental | 0.1884 | 1.3068 | 0.1442 | No |
| AhZIP6.2/6.4 | Segmental | 0.1887 | 1.3548 | 0.1393 | No |
| AhZIP6.3/6.2 | Segmental | 0.1859 | 1.2902 | 0.1441 | No |
| AhIRT1.2/1.4 | Whole-genome | 0.0463 | 0.0956 | 0.4843 | No |
| AhZIP1.1/1.2 | Whole-genome | 0.0338 | 0.0667 | 0.5068 | No |
| AhZIP2.1/2.2 | Whole-genome | 0.0112 | 0.0373 | 0.3006 | No |
| AhZIP3.1/3.2 | Whole-genome | 0.0393 | 0.0474 | 0.8291 | No |
| AhZIP3.3/3.4 | Whole-genome | 0.0208 | 0.0861 | 0.2415 | No |
| AhZIP4.1/4.2 | Whole-genome | 0.0024 | 0.0460 | 0.0525 | No |
| AhZIP6.3/6.4 | Whole-genome | 0.0026 | 0.0485 | 0.0532 | No |
| AhZIP6.1/6.2 | Whole-genome | 0.0013 | 0.0477 | 0.0277 | No |
| AhZIP7.3/7.4 | Whole-genome | 0.0180 | 0.0848 | 0.2119 | No |
| AhZIP7.1/7.7 | Whole-genome | 0.0227 | 0.0835 | 0.2717 | No |
| AhZIP7.5/7.6 | Whole-genome | 0.0113 | 0.0698 | 0.1620 | No |
| AhZIP11.1/11.2 | Whole-genome | 0.0077 | 0.0282 | 0.2733 | No |
| Gene Expression a | [Fe]root b | [Fe]shoot c | Total Fe in Plants | % of Fe in Shoots | [Zn]root d | [Zn]shoot e | Total Zn in Plants | [Mn]shoot f | Total Mn in Plants | % of Mn in Shoots |
|---|---|---|---|---|---|---|---|---|---|---|
| AhIRT1.1 | −0.07 | −0.08 | −0.52 ** | 0.65 ** | 0.21 | 0.60 ** | 0.16 | 1.19 *** | 0.05 | 0.95 *** |
| AhIRT1.2 | −0.39 * | 0.02 | 0.10 | 0.16 | 0.30 | 0.32 | 0.28 | 0.06 | 0.44 * | −0.34 * |
| AhZIP1.1 | 0.10 | 0.11 | −0.14 | −0.07 | −0.21 | −0.17 | −0.20 | −0.06 | −0.27 | 0.15 |
| AhZIP1.2 | −0.01 | 0.12 | −0.24 | −0.05 | 0.13 | 0.03 | −0.12 | −0.05 | −0.26 | 0.10 |
| AhZIP2.1 | 0.01 | 0.21 | −0.30 | 0.07 | 0.18 | 0.16 | −0.12 | −0.04 | −0.11 | 0.01 |
| AhZIP3.5 | 0.06 | 0.27 | −0.14 | 0.09 | 0.26 | 0.28 | 0.09 | −0.12 | −0.05 | −0.14 |
| AhZIP3.6 | −0.12 | −0.15 | −0.32 | 0.11 | 0.04 | 0.10 | −0.26 | 0.57 ** | −0.25 | 0.70 *** |
| AhZIP4.1 | 0.00 | 0.13 | −0.35 | −0.11 | −0.06 | −0.02 | −0.17 | −0.18 | −0.22 | 0.05 |
| AhZIP6.1 | −0.60 ** | −0.42 * | 0.02 | 0.06 | 0.02 | −0.04 | 0.10 | 0.13 | 0.37 | −0.15 |
| AhZIP7.2 | 0.10 | −0.04 | 0.15 | 0.09 | 0.49 * | 0.06 | 0.45 * | 0.07 | 0.28 | −0.10 |
| AhZIP7.8 | 0.01 | 0.13 | −0.27 | −0.08 | 0.21 | 0.13 | −0.10 | −0.03 | −0.18 | −0.01 |
| AhZIP11.1 | 0.11 | 0.23 | 0.19 | 0.26 | −0.05 | 0.43 | −0.03 | 0.56 * | 0.20 | −0.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Chen, N.; Zhang, Z.; Shi, G. Genome-Wide Identification and Expression Profile Reveal Potential Roles of Peanut ZIP Family Genes in Zinc/Iron-Deficiency Tolerance. Plants 2022, 11, 786. https://doi.org/10.3390/plants11060786
Zhang Z, Chen N, Zhang Z, Shi G. Genome-Wide Identification and Expression Profile Reveal Potential Roles of Peanut ZIP Family Genes in Zinc/Iron-Deficiency Tolerance. Plants. 2022; 11(6):786. https://doi.org/10.3390/plants11060786
Chicago/Turabian StyleZhang, Zhen, Nannan Chen, Zheng Zhang, and Gangrong Shi. 2022. "Genome-Wide Identification and Expression Profile Reveal Potential Roles of Peanut ZIP Family Genes in Zinc/Iron-Deficiency Tolerance" Plants 11, no. 6: 786. https://doi.org/10.3390/plants11060786
APA StyleZhang, Z., Chen, N., Zhang, Z., & Shi, G. (2022). Genome-Wide Identification and Expression Profile Reveal Potential Roles of Peanut ZIP Family Genes in Zinc/Iron-Deficiency Tolerance. Plants, 11(6), 786. https://doi.org/10.3390/plants11060786






