QTL Analysis Revealed One Major Genetic Factor Inhibiting Lesion Elongation by Bacterial Blight (Xanthomonas oryzae pv. oryzae) from a japonica Cultivar Koshihikari in Rice
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Evaluation of Parental Lines and F2 Population to Progression of Lesion Length to Xoo
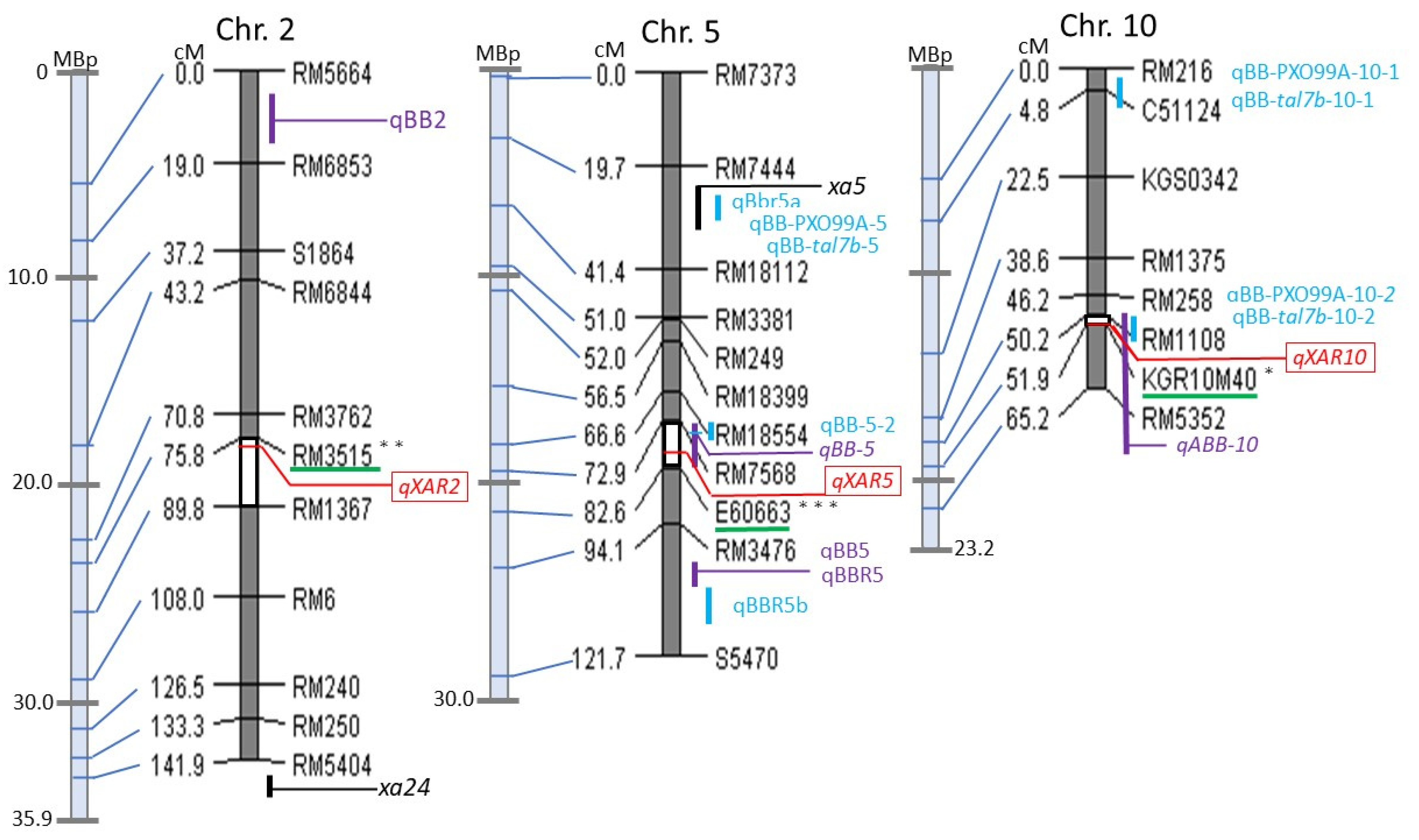
2.2. Putative Quantitative Trait Loci (QTL) Mapping for Resistance to Bacterial Blight (BB)
2.3. Alleles Contributing to Short Lesion Length in the Targeted Regions
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Bacterial Race Preparation
4.2. Genomic DNA Extraction and PCR Analysis Using DNA Markers
4.3. Mapping Putative QTLs against Resistance to BB Disease
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations; Trade and Markets Division. Food Outlook: Biannual Report on Global Food Markets, June 2020; Food and Agriculture Organization: Rome, Italy, 2020; Available online: https://www.fao.org/documents/card/en/c/ca9509en/ (accessed on 9 June 2021).
- Gnanamanickam, S.S. Rice and Its Importance to Human Life. In Biological Control of Rice Diseases; Springer: Dordrecht, The Netherlands, 2009; pp. 1–11. [Google Scholar] [CrossRef]
- Doni, F.; Suhaimi, N.S.M.; Mispan, M.S.; Fathurrahman, F.; Marzuki, B.M.; Kusmoro, J.; Uphoff, N. Microbial Contributions for Rice Production: From Conventional Crop Management to the Use of ‘Omics’ Technologies. Int. J. Mol. Sci. 2022, 23, 737. [Google Scholar] [CrossRef] [PubMed]
- Kottapalli, K.R.; Satoh, K.; Rakwal, R.; Shibato, J.; Doi, K.; Nagata, T.; Kikuchi, S. Combining In Silico Mapping and Arraying: An Approach to Identifying Common Candidate Genes for Submergence Tolerance and Resistance to Bacterial Leaf Blight in Rice. Mol. Cells 2007, 24, 394–408. [Google Scholar] [PubMed]
- Du, X.-X.; Park, J.-R.; Wang, X.-H.; Jan, R.; Lee, G.-S.; Kim, K.-M. Genotype and Phenotype Interaction between OsWKRYq6 and BLB after Xanthomonas oryzae pv. oryzae Inoculation in the Field. Plants 2022, 11, 287. [Google Scholar] [CrossRef] [PubMed]
- Ou, S. Bacterial Diseases. In Rice Diseases; Commonwealth Mycological Institute: Kew, UK, 1985; pp. 61–96. [Google Scholar]
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model Pathogens of a Model Crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef]
- Han, X.; Yang, Y.; Wang, X.; Zhou, J.; Zhang, W.; Yu, C.; Cheng, C.; Cheng, Y.; Yan, C.; Chen, J. Quantitative Trait Loci Mapping for Bacterial Blight Resistance in Rice Using Bulked Segregant Analysis. Int. J. Mol. Sci. 2014, 15, 11847–11861. [Google Scholar] [CrossRef] [Green Version]
- Shobha, B.; Lakshmeesha, T.R.; Ansari, M.A.; Almatroudi, A.; Alzohairy, M.A.; Basavaraju, S.; Alurappa, R.; Niranjana, S.R.; Chowdappa, S. Mycosynthesis of ZnO Nanoparticles Using Trichoderma Spp. Isolated from Rhizosphere Soils and Its Synergistic Antibacterial Effect against Xanthomonas oryzae pv. oryzae. J. Fungi 2020, 6, 181. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, X.-F.; Zhou, M.-G.; Kuang, J.; Zhang, Y.; Shang, Y.; Wang, J.-X. Status of Streptomycin Resistance Development in Xanthomonas oryzae pv. oryzae and Xanthomonas oryzae pv. oryzicola in China and Their Resistance Characters. J. Phytopathol. 2010, 158, 601–608. [Google Scholar] [CrossRef]
- Zhu, X.-F.; Xu, Y.; Peng, D.; Zhang, Y.; Huang, T.-T.; Wang, J.-X.; Zhou, M.-G. Detection and Characterization of Bismerthiazol-Resistance of Xanthomonas oryzae pv. oryzae. Crop Prot. 2013, 47, 24–29. [Google Scholar] [CrossRef]
- Yasmin, S.; Hafeez, F.Y.; Mirza, M.S.; Rasul, M.; Arshad, H.M.I.; Zubair, M.; Iqbal, M. Biocontrol of Bacterial Leaf Blight of Rice and Profiling of Secondary Metabolites Produced by Rhizospheric Pseudomonas aeruginosa BRp3. Front. Microbiol. 2017, 8, 1895. [Google Scholar] [CrossRef] [Green Version]
- Jin, P.; Wang, Y.; Tan, Z.; Liu, W.; Miao, W. Antibacterial Activity and Rice-Induced Resistance, Mediated by C15surfactin A, in Controlling Rice Disease Caused by Xanthomonas oryzae pv. oryzae. Pestic. Biochem. Physiol. 2020, 169, 104669. [Google Scholar] [CrossRef]
- Ji, G.-H.; Wei, L.-F.; He, Y.-Q.; Wu, Y.-P.; Bai, X.-H. Biological Control of Rice Bacterial Blight by Lysobacter antibioticus Strain 13-1. Biol. Control 2008, 45, 288–296. [Google Scholar] [CrossRef]
- Chithrashree; Udayashankar, A.C.; Chandra Nayaka, S.; Reddy, M.S.; Srinivas, C. Plant Growth-Promoting Rhizobacteria Mediate Induced Systemic Resistance in Rice against Bacterial Leaf Blight Caused by Xanthomonas oryzae pv. oryzae. Biol. Control 2011, 59, 114–122. [Google Scholar] [CrossRef]
- Kim, S.-M.; Reinke, R.F. A Novel Resistance Gene for Bacterial Blight in Rice, Xa43(t) Identified by GWAS, Confirmed by QTL Mapping Using a Bi-Parental Population. PLoS ONE 2019, 14, e0211775. [Google Scholar] [CrossRef] [Green Version]
- Bakade, R.; Ingole, K.D.; Deshpande, S.; Pal, G.; Patil, S.S.; Bhattacharjee, S.; Prasannakumar, M.K.; Ramu, V.S. Comparative Transcriptome Analysis of Rice Resistant and Susceptible Genotypes to Xanthomonas oryzae pv. oryzae Identifies Novel Genes to Control Bacterial Leaf Blight. Mol. Biotechnol. 2021, 63, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Cao, Y.; Xu, Z.; Ma, W.; Zakria, M.; Zou, L.; Cheng, Z.; Chen, G. Transcription Activator-Like Effector Tal7 of Xanthomonas oryzae pv. oryzicola Activates Rice Gene Os09g29100 to Suppress Rice Immunity. Sci. Rep. 2017, 7, 5089. [Google Scholar] [CrossRef] [Green Version]
- Busungu, C.; Taura, S.; Sakagami, J.-I.; Anai, T.; Ichitani, K. High-Resolution Mapping and Characterization of xa42, a Resistance Gene against Multiple Xanthomonas oryzae pv. oryzae Races in Rice (Oryza sativa L.). Breed. Sci. 2018, 68, 188–199. [Google Scholar] [CrossRef] [Green Version]
- Acquaah, G. Principles of Plant Genetics and Breeding; Blackwell Pub.: Malden, MA, USA; Oxford, UK, 2007. [Google Scholar]
- Huang, N.; Angeles, E.R.; Domingo, J.; Magpantay, G.; Singh, S.; Zhang, G.; Kumaravadivel, N.; Bennett, J.; Khush, G.S. Pyramiding of Bacterial Blight Resistance Genes in Rice: Marker-Assisted Selection Using RFLP and PCR. Theor. Appl. Genet. 1997, 95, 313–320. [Google Scholar] [CrossRef]
- Wang, S.; Liu, W.; Lu, D.; Lu, Z.; Wang, X.; Xue, J.; He, X. Distribution of Bacterial Blight Resistance Genes in the Main Cultivars and Application of Xa23 in Rice Breeding. Front. Plant Sci. 2020, 11, 555228. [Google Scholar] [CrossRef]
- Mew, T.; Vera Cruz, C.; Medalla, E. Changes in Race Frequency of Xanthomonas oryzae pv. oryzae in Response to Rice Cultivars Planted in the Philippines. Plant Dis. 1992, 76, 1029–1032. [Google Scholar]
- Sahu, A.; Das, A.; Saikia, K.; Barah, P. Temperature Differentially Modulates the Transcriptome Response in Oryza sativa to Xanthomonas oryzae pv. oryzae Infection. Genomics 2020, 112, 4842–4852. [Google Scholar] [CrossRef]
- Webb, K.M.; Oña, I.; Bai, J.; Garrett, K.A.; Mew, T.; Vera Cruz, C.M.; Leach, J.E. A Benefit of High Temperature: Increased Effectiveness of a Rice Bacterial Blight Disease Resistance Gene. New Phytol. 2010, 185, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.-M.; Qiu, D.-Y.; Shen, X.-L.; Li, X.-H.; Wang, S.-P. Isolation and Manipulation of Quantitative Trait Loci for Disease Resistance in Rice Using a Candidate Gene Approach. Mol. Plant 2008, 1, 786–793. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, J.; Vera Cruz, C.M.; Kukreja, K.; Chittoor, J.M.; Wu, J.-L.; Lee, S.W.; Baraoidan, M.; George, M.L.; Cohen, M.B.; Hulbert, S.H.; et al. Candidate Defense Genes from Rice, Barley, and Maize and Their Association with Qualitative and Quantitative Resistance in Rice. Mol. Plant-Microbe Interact. 2003, 16, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.K.; Arif, M.; Zhong, D.B.; Fu, B.Y.; Xu, J.L.; Domingo-Rey, J.; Ali, J.; Vijayakumar, C.H.M.; Yu, S.B.; Khush, G.S. Complex Genetic Networks Underlying the Defensive System of Rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. Proc. Natl. Acad. Sci. USA 2006, 103, 7994–7999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, T.; Ohuchi, A. A New Pathogenic Race of Xanthomonas campestris pv. oryzae and Inheritance of Resistance of Differential Rice Variety, Te-Tep to It. Jpn. J. Phytopathol. 1989, 55, 201–207. [Google Scholar] [CrossRef] [Green Version]
- Busungu, C.; Taura, S.; Sakagami, J.-I.; Ichitani, K. Identification and Linkage Analysis of a New Rice Bacterial Blight Resistance Gene from XM14, a Mutant Line from IR24. Breed. Sci. 2016, 66, 636–645. [Google Scholar] [CrossRef] [Green Version]
- Msami, J.A.; Kawaguchi, Y.; Ichitani, K.; Taura, S. Linkage Analysis of Rice Bacterial Blight Resistance Gene xa20 in XM6, a Mutant Line from IR24. Breed. Sci. 2021, 71, 144–154. [Google Scholar] [CrossRef]
- McCouch, S.R.; Cho, Y.G.; Yano, M.; Paul, E.; Blinstrub, M.; Morishima, H.; Kinosita, T. Report on QTL Nomenclature. Rice Genet. Newsl. 1997, 14, 11–13. [Google Scholar]
- Yamazaki, Y.; Sakaniwa, S.; Tsuchiya, R.; Nonomura, K.-I.; Kurata, N. Oryzabase: An Integrated Information Resource for Rice Science. Breed. Sci. 2010, 60, 544–548. [Google Scholar] [CrossRef] [Green Version]
- Iwata, H.; Ninomiya, S. AntMap: Constructing Genetic Linkage Maps Using an Ant Colony Optimization Algorithm. Breed. Sci. 2006, 56, 371–377. [Google Scholar] [CrossRef] [Green Version]
- Kosambi, D.D. The Estimation of Map Distances from Recombination Values; Kosambi, D.D., Ramaswamy, R., Eds.; Springer: New Delhi, India, 2016. [Google Scholar] [CrossRef]
- Vikal, Y.; Bhatia, D. Genetics and Genomics of Bacterial Blight Resistance in Rice. In Advances in International Rice Research; Li, J., Ed.; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Hasan, M.M.; Oladosu, Y.A.; Magaji, U.G.; Akos, I.; Olalekan, K.K. Bacterial Leaf Blight Resistance in Rice: A Review of Conventional Breeding to Molecular Approach. Mol. Biol. Rep. 2019, 46, 1519–1532. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Hori, K.; Yamamoto, T.; Yano, M. Koshihikari: A Premium Short-Grain Rice Cultivar—Its Expansion and Breeding in Japan. Rice 2018, 11, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broman, K.W.; Sen, S. A Guide to QTL Mapping with R/Qtl; Statistics for Biology and Health; Springer: New York, NY, USA, 2009. [Google Scholar]
- Djedatin, G.; Ndjiondjop, M.-N.; Sanni, A.; Lorieux, M.; Verdier, V.; Ghesquiere, A. Identification of Novel Major and Minor QTLs Associated with Xanthomonas oryzae pv. oryzae (African Strains) Resistance in Rice (Oryza sativa L.). Rice 2016, 9, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta, A.I.; Delorean, E.E.; Bossa-Castro, A.M.; Tonnessen, B.W.; Raghavan, C.; Corral, R.; Pérez-Quintero, Á.L.; Leung, H.; Verdier, V.; Leach, J.E. Resistance and Susceptibility QTL Identified in a Rice MAGIC Population by Screening with a Minor-effect Virulence Factor from Xanthomonas oryzae pv. oryzae. Plant Biotechnol. J. 2021, 19, 51–63. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare Reference Genome Using Next Generation Sequence and Optical Map Data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Blair, M.W.; Garris, A.J.; Iyer, A.S.; Chapman, B.; Kresovich, S.; McCouch, S.R. High Resolution Genetic Mapping and Candidate Gene Identification at the xa5 Locus for Bacterial Blight Resistance in Rice (Oryza sativa L.). Theor. Appl. Genet. 2003, 107, 62–73. [Google Scholar] [CrossRef]
- Wu, X.; Li, X.; Xu, C.; Wang, S. Fine Genetic Mapping of xa24, a Recessive Gene for Resistance against Xanthomonas oryzae pv. oryzae in Rice. Theor. Appl. Genet. 2008, 118, 185–191. [Google Scholar] [CrossRef]
- Temnykh, S.; Park, W.D.; Ayres, N.; Cartinhour, S.; Hauck, N.; Lipovich, L.; Cho, Y.G.; Ishii, T.; McCouch, S.R. Mapping and Genome Organization of Microsatellite Sequences in Rice (Oryza sativa L.). Theor. Appl. Genet. 2000, 100, 697–712. [Google Scholar] [CrossRef]
- Hu, K.; Cao, J.; Zhang, J.; Xia, F.; Ke, Y.; Zhang, H.; Xie, W.; Liu, H.; Cui, Y.; Cao, Y.; et al. Improvement of Multiple Agronomic Traits by a Disease Resistance Gene via Cell Wall Reinforcement. Nat. Plants 2017, 3, 17009. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of Function of a Proline-Containing Protein Confers Durable Disease Resistance in Rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef]
- Fonseca, J.P.; Mysore, K.S. Genes Involved in Nonhost Disease Resistance as a Key to Engineer Durable Resistance in Crops. Plant Sci. 2019, 279, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Taura, S.; Ogawa, T.; Yoshimura, A.; Omura, T. Induction of Mutants Resistant to Bacterial Blight in Rice. Jpn. J. Breed. 1991, 41, 279–288. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, T.; Yamamoto, T.; Khush, G.S.; Mew, T.-W. Breeding of Near-Isogenic Lines of Rice with Single Genes for Resistance to Bacterial Blight Pathogen (Xanthomonas campestris pv. oryzae). Jpn. J. Breed. 1991, 41, 523–529. [Google Scholar] [CrossRef] [Green Version]
- Wakimoto, S. Classification of Strains of Xanthomonas oryzae on the Basis of Their Susceptibility against Bacteriophages. Jpn. J. Phytopathol. 1960, 25, 193–198. [Google Scholar] [CrossRef]
- Kauffman, H. An Improved Technique for Evaluating Resistance of Rice Varieties to Xanthomonas oryzae. Plant Rep. 1973, 57, 537–541. [Google Scholar]
- Kim, S.-M. Identification of Novel Recessive Gene Xa44(t) Conferring Resistance to Bacterial Blight Races in Rice by QTL Linkage Analysis Using an SNP Chip. Theor. Appl. Genet. 2018, 131, 2733–2743. [Google Scholar] [CrossRef] [Green Version]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A Plant DNA Minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar]
- Panaud, O.; Chen, X.; McCouch, S.R. Development of Microsatellite Markers and Characterization of Simple Sequence Length Polymorphism (SSLP) in Rice (Oryza sativa L.). Mol. Gen. Genet. 1996, 252, 597–607. [Google Scholar] [CrossRef]
- Chen, X.; Temnykh, S.; Xu, Y.; Cho, Y.G.; McCouch, S.R. Development of a Microsatellite Framework Map Providing Genome-Wide Coverage in Rice (Oryza sativa L.). Theor. Appl. Genet. 1997, 95, 553–567. [Google Scholar] [CrossRef]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and Mapping of 2240 New SSR Markers for Rice (Oryza sativa L.). DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2002, 9, 199–207. [Google Scholar] [CrossRef]
- Ichitani, K.; Taura, S.; Tezuka, T.; Okiyama, Y.; Kuboyama, T. Chromosomal Location of HWA1 and HWA2, Complementary Hybrid Weakness Genes in Rice. Rice 2011, 4, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Kuboyama, T.; Saito, T.; Matsumoto, T.; Wu, J.; Kanamori, H.; Taura, S.; Sato, M.; Marubashi, W.; Ichitani, K. Fine Mapping of HWC2, a Complementary Hybrid Weakness Gene, and Haplotype Analysis Around the Locus in Rice. Rice 2009, 2, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Toyomoto, D.; Uemura, M.; Taura, S.; Sato, T.; Henry, R.; Ishikawa, R.; Ichitani, K. Segregation Distortion Observed in the Progeny of Crosses Between Oryza sativa and O. meridionalis Caused by Abortion During Seed Development. Plants 2019, 8, 398. [Google Scholar] [CrossRef] [Green Version]
- International Rice Genome Sequencing Project (IRGSP). The map-based sequence of the rice genome. Nature 2005, 436, 793–800. Available online: https://pubmed.ncbi.nlm.nih.gov/16100779/ (accessed on 9 March 2021).
- Rice Genome Research Program. Available online: https://rgp.dna.affrc.go.jp/E/publicdata/caps/index.html (accessed on 4 February 2021).
- Broman, K.W.; Wu, H.; Sen, S.; Churchill, G.A. R/Qtl: QTL Mapping in Experimental Crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [Green Version]





| Lesion Length (cm) | |||||
|---|---|---|---|---|---|
| Rice Accession | Race I T7174 | Race II T7147 | Race III T7133 | Race IV H75373 | Race V H75304 |
| Koshihikari Japan | 14.9 | 13.5 | 13.5 | 10.1 | 17.7 |
| ARC7013 India | 40.5 | 48.5 | 43.3 | 43.3 | 54.0 |
| QTL | Chr. | Nearest Marker | Lod Scores Using (SIM) a & (CIM) b | QTL cM (CI) c | R2% SIM d | Additive Effect e | Dominance Effect f | DPE g |
|---|---|---|---|---|---|---|---|---|
| qXAR2 | 2 | RM3515 | (2.4) a; (3.1) b | 75.8 (75.8–89.8) | 3.9 | −2.8 | 2.5 | KO ** |
| qXAR5 | 5 | E60663 | (11.6) a; (14.0) b | 79.0 (72.9–82.9) | 20.5 | −7.0 | 1.8 | KO *** |
| qXAR10 | 10 | KGR10M40 | (1.4) a; (3.4) b | 51.0 (50.2–51.9) | 2.3 | 2.3 | 0.4 | AR * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, S.; Tsuneyoshi, H.; Ichitani, K.; Taura, S. QTL Analysis Revealed One Major Genetic Factor Inhibiting Lesion Elongation by Bacterial Blight (Xanthomonas oryzae pv. oryzae) from a japonica Cultivar Koshihikari in Rice. Plants 2022, 11, 867. https://doi.org/10.3390/plants11070867
Shah S, Tsuneyoshi H, Ichitani K, Taura S. QTL Analysis Revealed One Major Genetic Factor Inhibiting Lesion Elongation by Bacterial Blight (Xanthomonas oryzae pv. oryzae) from a japonica Cultivar Koshihikari in Rice. Plants. 2022; 11(7):867. https://doi.org/10.3390/plants11070867
Chicago/Turabian StyleShah, Shameel, Hiroaki Tsuneyoshi, Katsuyuki Ichitani, and Satoru Taura. 2022. "QTL Analysis Revealed One Major Genetic Factor Inhibiting Lesion Elongation by Bacterial Blight (Xanthomonas oryzae pv. oryzae) from a japonica Cultivar Koshihikari in Rice" Plants 11, no. 7: 867. https://doi.org/10.3390/plants11070867








