Molecular Characterization, Evolutionary Analysis, and Expression Profiling of BOR Genes in Important Cereals
Abstract
:1. Introduction
2. Results
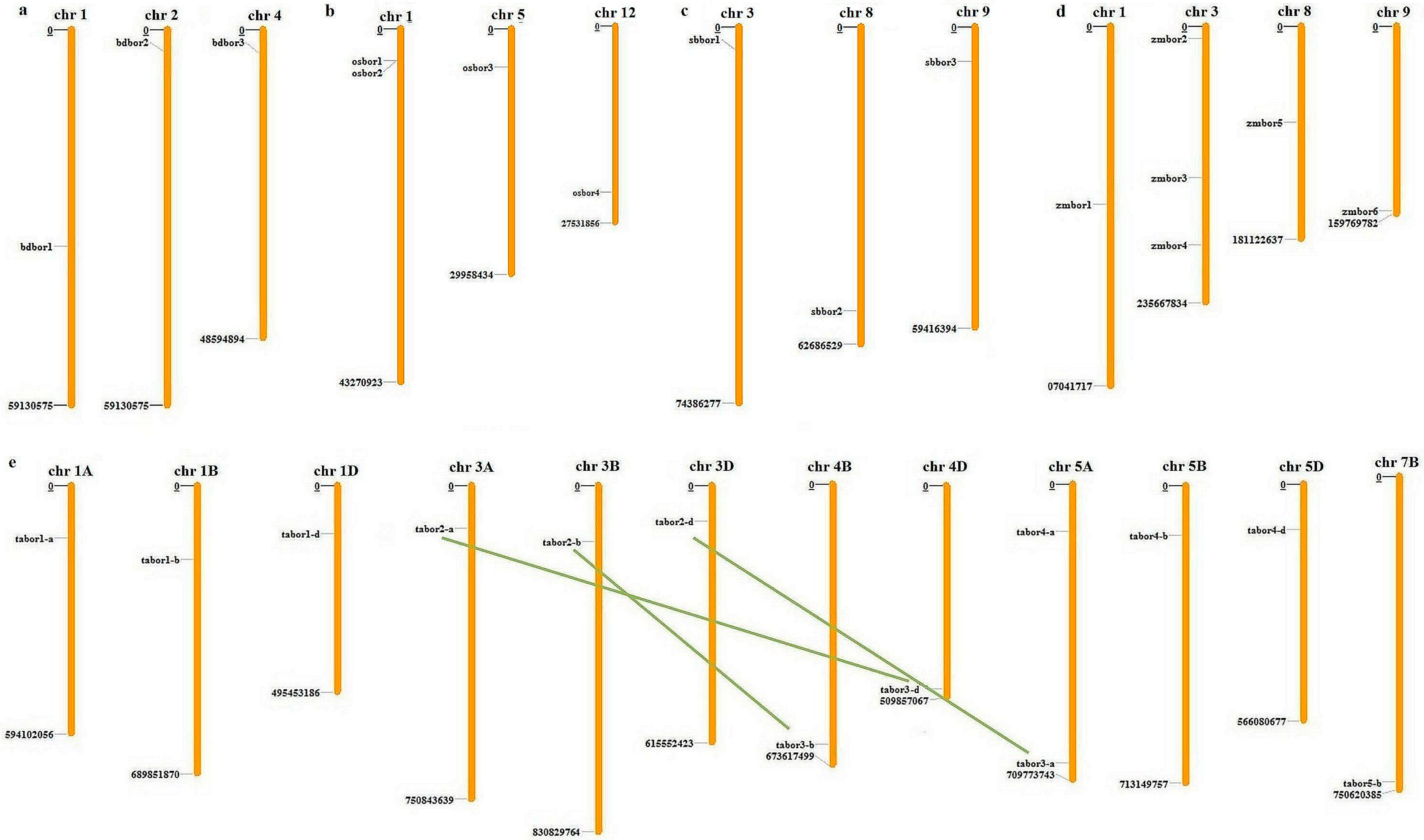
2.1. Identification, Distribution, and Paralogous Analysis of BOR Genes
2.2. Multiple Sequence Alignment and Evolutionary Analysis
2.3. Gene Structure Analysis
2.4. Divergence Analysis
2.5. Protein Characterization
2.6. Cis-Regulatory Elements
2.7. Expression Analysis of BOR Genes during Tissue Developmental Stages
2.8. Expression Analysis of TaBORs in Different Stress Conditions
2.9. Co-Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Identification and Nomenclature of BOR Genes
4.2. Chromosomal Mapping and Duplication Events
4.3. Multiple Sequence Alignment and Evolutionary Analysis
4.4. Ka/Ks Substitutions
4.5. Gene Structure Analysis
4.6. Promoter Analysis
4.7. Protein Characterization
4.8. Expression Profiling Studies
4.9. Co-Expression Analysis
4.10. Plant Growth, Treatments, and qRT-PCR Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reid, R. Understanding the boron transport network in plants. Plant Soil 2014, 385, 1–13. [Google Scholar] [CrossRef]
- Tanaka, M.; Fujiwara, T. Physiological roles and transport mechanisms of boron: Perspectives from plants. Pflugers Arch. Eur. J. Physiol. 2008, 456, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Matoh, T.; Azuma, J.I. Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 1996, 110, 1017–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, M.A.; Eberhard, S.; Albersheim, P.; Darvill, A.G. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for A. thaliana growth. Science 2001, 294, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Rashid, A.; Nadeem, F.; Stuerz, S.; Asch, F.; Bell, R.W.; Siddique, K.H.M. Boron nutrition of rice in different production systems. Agron. Sustain. Dev. 2018, 38, 25. [Google Scholar] [CrossRef] [Green Version]
- Brown, P.H.; Shelp, B.J. Boron mobility in plants. Plant Soil 1997, 193, 85–101. [Google Scholar] [CrossRef]
- Gupta, U.C. Boron nutrition of crops. Adv. Agron. 1980, 31, 273–307. [Google Scholar] [CrossRef]
- Takano, J.; Miwa, K.; Fujiwara, T. Boron transport mechanisms: Collaboration of channels and transporters. Trends Plant Sci. 2008, 13, 451–457. [Google Scholar] [CrossRef]
- Takano, J.; Noguchi, K.; Yasumori, M.; Kobayashi, M.; Gajdos, Z.; Miwa, K.; Hayashi, H.; Yoneyama, T.; Fujiwara, T. A. thaliana boron transporter for xylem loading. Nature 2002, 420, 337–340. [Google Scholar] [CrossRef]
- Takano, J.; Tanaka, M.; Toyoda, A.; Miwa, K.; Kasai, K.; Fuji, K.; Onouchi, H.; Naito, S.; Fujiwara, T. Polar localization and degradation of A. thaliana boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 5220–5225. [Google Scholar] [CrossRef] [Green Version]
- Kasai, K.; Takano, J.; Miwa, K.; Toyoda, A.; Fujiwara, T. High boron-induced ubiquitination regulates vacuolar sorting of the bor1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 2011, 286, 6175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miwa, K.; Takano, J.; Omori, H.; Seki, M.; Shinozaki, K.; Fujiwara, T. Plants tolerant of high boron levels. Science 2007, 318, 1417. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.D.; Boron, W.F. The divergence, actions, roles, and relatives of sodium-coupled bicarbonate transporters. Physiol. Rev. 2013, 93, 803–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, Y.; Hanaoka, H.; Kobayashi, M.; Miyoshi, K.; Miwa, K.; Fujiwara, T. Cell-type specificity of the expression of OsBOR1, a rice efflux boron transporter gene, is regulated in response to boron availability for efficient boron uptake and xylem loading. Plant Cell 2007, 19, 2634–2635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, R. Identification of Boron Transporter Genes Likely to be Responsible for Tolerance to Boron Toxicity in Wheat and Barley. Plant Cell Physiol. 2007, 48, 1673–1678. [Google Scholar] [CrossRef]
- Pérez-Castro, R.; Kasai, K.; Gainza-Cortés, F.; Ruiz-Lara, S.; Casaretto, J.A.; Peña-Cortés, H.; Tapia, J.; Fujiwara, T.; González, E. VvBOR1, the Grapevine Ortholog of AtBOR1, Encodes an Efflux Boron Transporter That is Differentially Expressed Throughout Reproductive Development of Vitis vinifera L. Plant Cell Physiol. 2012, 53, 485–494. [Google Scholar] [CrossRef] [Green Version]
- Cañon, P.; Aquea, F.; Rodríguez-Hoces de la Guardia, A.; Arce-Johnson, P. Functional characterization of Citrus macrophylla BOR1 as a boron transporter. Physiol. Plant 2013, 149, 329–339. [Google Scholar] [CrossRef]
- Leaungthitikanchana, S.; Fujibe, T.; Tanaka, M.; Wang, S.; Sotta, N.; Takano, J.; Fujiwara, T. Differential Expression of Three BOR1 Genes Corresponding to Different Genomes in Response to Boron Conditions in Hexaploid Wheat (Triticum aestivum L.). Plant Cell Physiol. 2013, 54, 1056–1063. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, N.; Uraguchi, S.; Saito, A.; Kajikawa, M.; Kasai, K.; Sato, Y.; Nagamura, Y.; Fujiwara, T. Roles of Pollen-Specific Boron Efflux Transporter, OsBOR4, in the Rice Fertilization Process. Plant Cell Physiol. 2013, 54, 2011–2019. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Zhang, Q.; He, M.; Wang, S.; Shi, L.; Xu, F. Molecular characterization of the genome-wide BOR transporter gene family and genetic analysis of BnaC04.BOR1;1c in Brassica napus. BMC Plant Biol. 2018, 18, 193. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liang, Z.; Wu, M.; Mei, L. Genome-wide identification of BOR genes in poplar and their roles in response to various environmental stimuli. Environ. Exp. Bot. 2019, 164, 101–113. [Google Scholar] [CrossRef]
- Onuh, A.F.; Miwa, O. Regulation, diversity and evolution of boron transporters in plants. Plant Cell Physiol. 2021, 62, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, S.; Zhi, H.; Xing, L.; Zhang, H.; Tang, C.; Wang, E.; Zhao, M.; Jia, G.; Feng, B.; et al. The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet. Crop J. 2021, 10, 342–353. [Google Scholar] [CrossRef]
- Wakuta, S.; Mineta, K.; Amano, T.; Toyoda, A.; Fujiwara, T.; Naito, S.; Takano, J. Evolutionary Divergence of Plant Borate Exporters and Critical Amino Acid Residues for the Polar Localization and Boron-Dependent Vacuolar Sorting of AtBOR1. Plant Cell Physiol. 2015, 56, 852–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 1993, 135, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Ozyigit, I.I.; Filiz, E.; Saracoglu, I.A.; Karadeniz, S. Exploration of two major boron transport genes BOR1 and NIP5;1 in the genomes of different plants. Biotechnol. Biotechnol. Equip. 2020, 34, 455–468. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; International Wheat Genome Sequencing Consortium; Jackobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, A.; Yashvika; Sidhu, S.; Upadhyay, S.K. A glimpse of boron transport in plants. In Cation Transporters in Plants; Upadhyay, S.K., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 281–306. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- IWGSC; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef]
- Lee, S.K.; Hong, W.-J.; Silva, J.; Kim, E.-J.; Park, S.K.; Jung, K.-H.; Kim, Y.-J. Global Identification of ANTH Genes Involved in Rice Pollen Germination and Functional Characterization of a Key Member, OsANTH3. Front. Plant Sci. 2021, 12, 609473. [Google Scholar] [CrossRef]
- Feng, Y.; Cui, R.; Wang, S.; He, M.; Hua, Y.; Shi, L.; Ye, X.; Xu, F. Transcription factor BnaA9.WRKY47 contributes to the adaptation of Brassica napus to low boron stress by up-regulating the boric acid channel gene BnaA3.NIP5;1. Plant Biotechnol. J. 2020, 18, 1241–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, Y.; Feng, Y.; Zhou, T.; Xu, F. Genome-scale mRNA transcriptomic insights into the responses of oilseed rape (Brassica napus L.) to varying boron availabilities. Plant Soil 2017, 416, 205–225. [Google Scholar] [CrossRef]
- Sharma, A.; Tyagi, S.; Alok, A.; Singh, K.; Upadhyay, S.K. Thaumatin-like protein kinases: Molecular characterization and transcriptional profiling in five cereal crops. Plant Sci. 2020, 290, 110317. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Taneja, M.; Upadhyay, S.K. Identification, characterization and expression profiling of cation-proton antiporter superfamily in Triticum aestivum L. and functional analysis of TaNHX4-B. Genomics 2020, 112, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Contreras-Moreira, B.; De Silva, N.; Maslen, G.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Barba, M.; Bolser, D.M.; Cambell, L. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020, 48, D689–D695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Tyagi, S.; Shumayla; Madhu; Singh, K.; Upadhyay, S.K. Molecular characterization revealed the role of catalases under abiotic and arsenic stress in bread wheat (Triticum aestivum L.). J. Hazard. Mater. 2021, 403, 123585. [Google Scholar] [CrossRef]
- Shumayla; Tyagi, S.; Sharma, A.; Singh, K.; Upadhyay, S.K. Genomic dissection and transcriptional profiling of Cysteine-rich receptor-like kinases in five cereals and functional characterization of TaCRK68-A. Int. J. Biol. Macromol. 2019, 134, 316–329. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, H.; Rajput, R.; Pandey, A.; Upadhyay, S.K. Molecular Characterization Revealed the Role of Thaumatin-Like Proteins of Bread Wheat in Stress Response. Front Plant Sci. 2022, 12, 807448. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef] [Green Version]
- Kaur, A.; Taneja, M.; Tyagi, S.; Sharma, A.; Singh, K.; Upadhyay, S. Genome-wide characterization and expression analysis suggested diverse functions of the mechanosensitive channel of small conductance-like (MSL) genes in cereal crops. Sci. Rep. 2020, 10, 16583. [Google Scholar] [CrossRef]
- Gaut, B.S.; Morton, B.R.; McCaig, B.C.; Clegg, M.T. Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 1996, 93, 10274. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef]
- King, B.R.; Guda, C. ngLOC: An n-gram-based Bayesian method for estimating the subcellular proteomes of eukaryotes. Genome Biol. 2007, 8, R68. [Google Scholar] [CrossRef] [Green Version]
- Petersen, T.N.; Brunak, S.; Heijne, G.V.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.M.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E. Structural and Functional Partitioning of Bread Wheat Chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef] [PubMed]
- Pingault, L.; Choulet, F.; Alberti, A.; Glover, N.; Wincker, P.; Feuillet, C.; Paux, E. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 2015, 16, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, Y.A.; Barrera, E.; Bazant, W.; Burke, M.; Füllgrabe, A.; Fuentes, A.M.-P.; George, N.; et al. Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018, 46, D246–D251. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Wang, C.; Liu, M.; Li, H.; Fu, Y.; Wang, Y.; Nie, Y.; Liu, X.; Ji, W. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genom. 2014, 15, 898. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Z.; Khan, A.A.; Lin, Q.; Han, Y.; Mu, P.; Liu, Y.; Zhang, H.; Li, L.; Meng, X.; et al. Expression partitioning of homeologs and tandem duplications contribute to salt tolerance in wheat (Triticum aestivum L.). Sci. Rep. 2016, 6, 21476. [Google Scholar] [CrossRef]
- Pearce, S.; Vazquez-Gross, H.; Herin, S.Y.; Hane, D.; Wang, Y.; Gu, Y.Q.; Dubcovsky, J. WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015, 15, 299. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabhher, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Seo, J.; Gordish-Dressman, H.; Hoffman, E.P. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics 2006, 22, 808–814. [Google Scholar] [CrossRef]
- Nazarov, P.V.; Muller, A.; Khutko, V. Co-expression analysis of large microarray data sets using CoExpress software tool. In Proceedings of the Conference: WCSB, Luxemborg, 16–18 June 2010. [Google Scholar]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks Visualization and Exploration of Large Graphs. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shumayla; Madhu; Singh, K.; Upadhyay, S.K. LysM domain-containing proteins modulate stress response and signalling in Triticum aestivum L. Environ. Exp. Bot. 2021, 189, 104558. [Google Scholar] [CrossRef]
- Madhu; Kaur, A.; Tyagi, S.; Shumayla; Singh, K.; Upadhyay, S.K. Exploration of glutathione reductase for abiotic stress response in bread wheat (Triticum aestivum L.). Plant Cell Rep. 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]






| Gene A | Gene B | Ka | Ks | Ka/Ks | Selection Pressure | T = Ks/2r |
|---|---|---|---|---|---|---|
| TaBOR2-A | TaBOR3-D | 0.0581 | 0.3206 | 0.1814 | Purifying | 24.7 |
| TaBOR2-B | TaBOR3-B | 0.0570 | 0.3275 | 0.1741 | Purifying | 25.2 |
| TaBOR2-D | TaBOR3-A | 0.0614 | 0.3081 | 0.1992 | Purifying | 23.7 |
| Group A | Group B | Out Group | Nt | Na | Nb | χ2 | P |
|---|---|---|---|---|---|---|---|
| TaBOR2-A | TaBOR3-D | TaBOR4-A | 1149 | 64 | 83 | 2.46 | 0.11709 |
| TaBOR2-B | TaBOR3-B | TaBOR4-A | 1156 | 79 | 77 | 0.03 | 0.87278 |
| TaBOR2-D | TaBOR3-A | TaBOR4-A | 1145 | 73 | 77 | 0.11 | 0.74397 |
| B. distachyon | O. sativa | S. bicolor | T. aestivum | Z. mays | |
|---|---|---|---|---|---|
| Average length (aa) | 683 | 683 | 684 | 691 | 717 |
| Average MW (kDa) | 76.6 | 76.7 | 76.5 | 77.2 | 80.2 |
| Signal peptides | 0 | 0 | 0 | 0 | 0 |
| Average pI value | 8 | 7.8 | 8 | 7.6 | 8.2 |
| Average GRAVY | 0.2 | 0.2 | 0.2 | 0.2 | 0.16 |
| Sub-cellular localization | PM | PM | PM | PM | PM |
| Transmemberane regions (TMs) | 9–12 | 9–12 | 8–10 | 9–12 | 5–12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, H.; Sharma, A.; Rajput, R.; Sidhu, S.; Dhillon, H.; Verma, P.C.; Pandey, A.; Upadhyay, S.K. Molecular Characterization, Evolutionary Analysis, and Expression Profiling of BOR Genes in Important Cereals. Plants 2022, 11, 911. https://doi.org/10.3390/plants11070911
Sharma H, Sharma A, Rajput R, Sidhu S, Dhillon H, Verma PC, Pandey A, Upadhyay SK. Molecular Characterization, Evolutionary Analysis, and Expression Profiling of BOR Genes in Important Cereals. Plants. 2022; 11(7):911. https://doi.org/10.3390/plants11070911
Chicago/Turabian StyleSharma, Himanshu, Alok Sharma, Ruchika Rajput, Sukhjeet Sidhu, Harpal Dhillon, Praveen Chandra Verma, Ashutosh Pandey, and Santosh Kumar Upadhyay. 2022. "Molecular Characterization, Evolutionary Analysis, and Expression Profiling of BOR Genes in Important Cereals" Plants 11, no. 7: 911. https://doi.org/10.3390/plants11070911







