Optimization of the Transformation Protocol for Increased Efficiency of Genetic Transformation in Hevea brasiliensis
Abstract
:1. Introduction
2. Results
2.1. Kanamycin Sensitivity Analysis
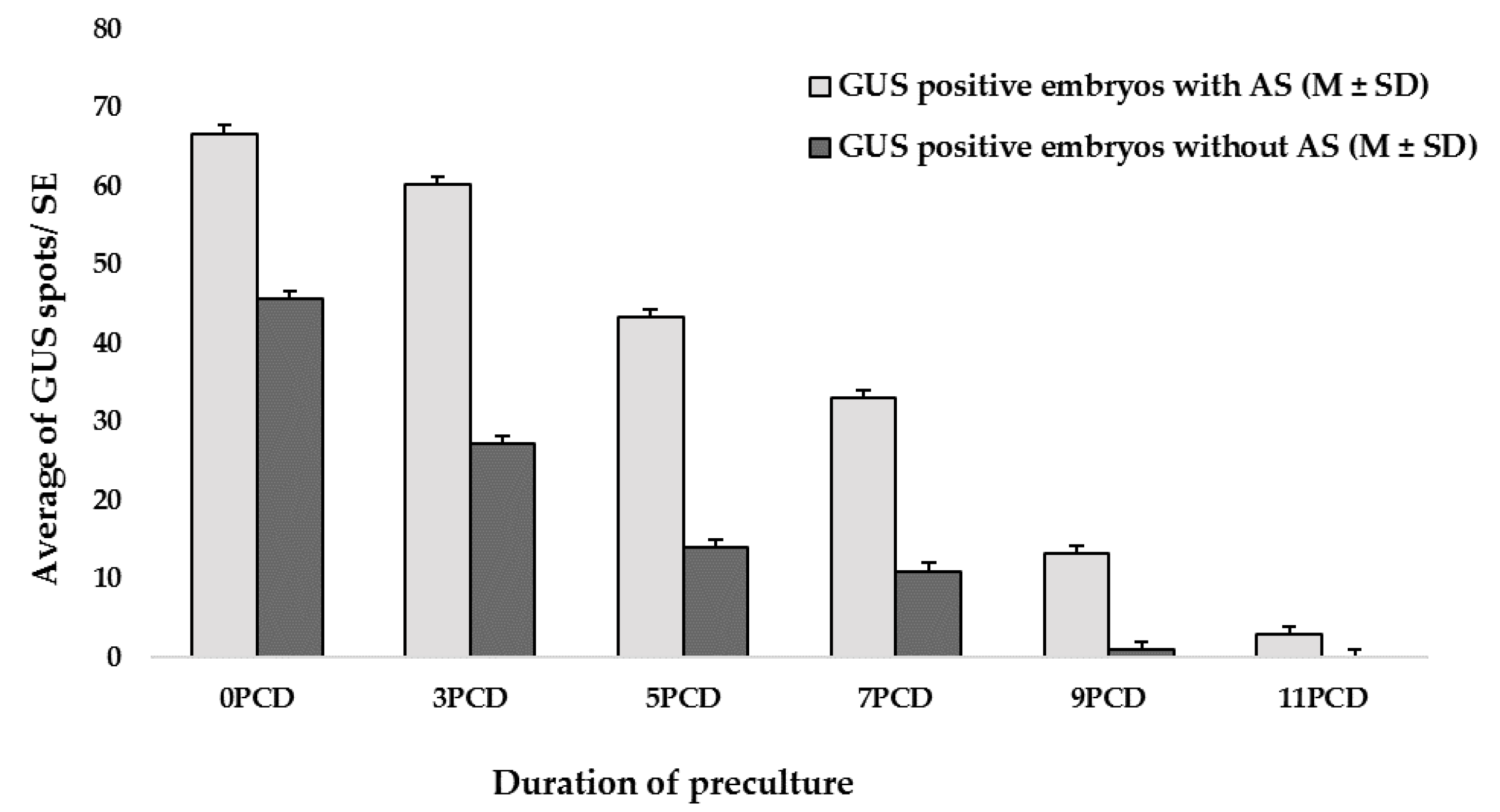
2.2. Effect of the Preculture Period on Transformation Efficiency
2.3. Effect of Agrobacterial Concentration on Transient GUS Expression in Explants
2.4. Sonication-Assisted, Agrobacterium-Mediated Transformation
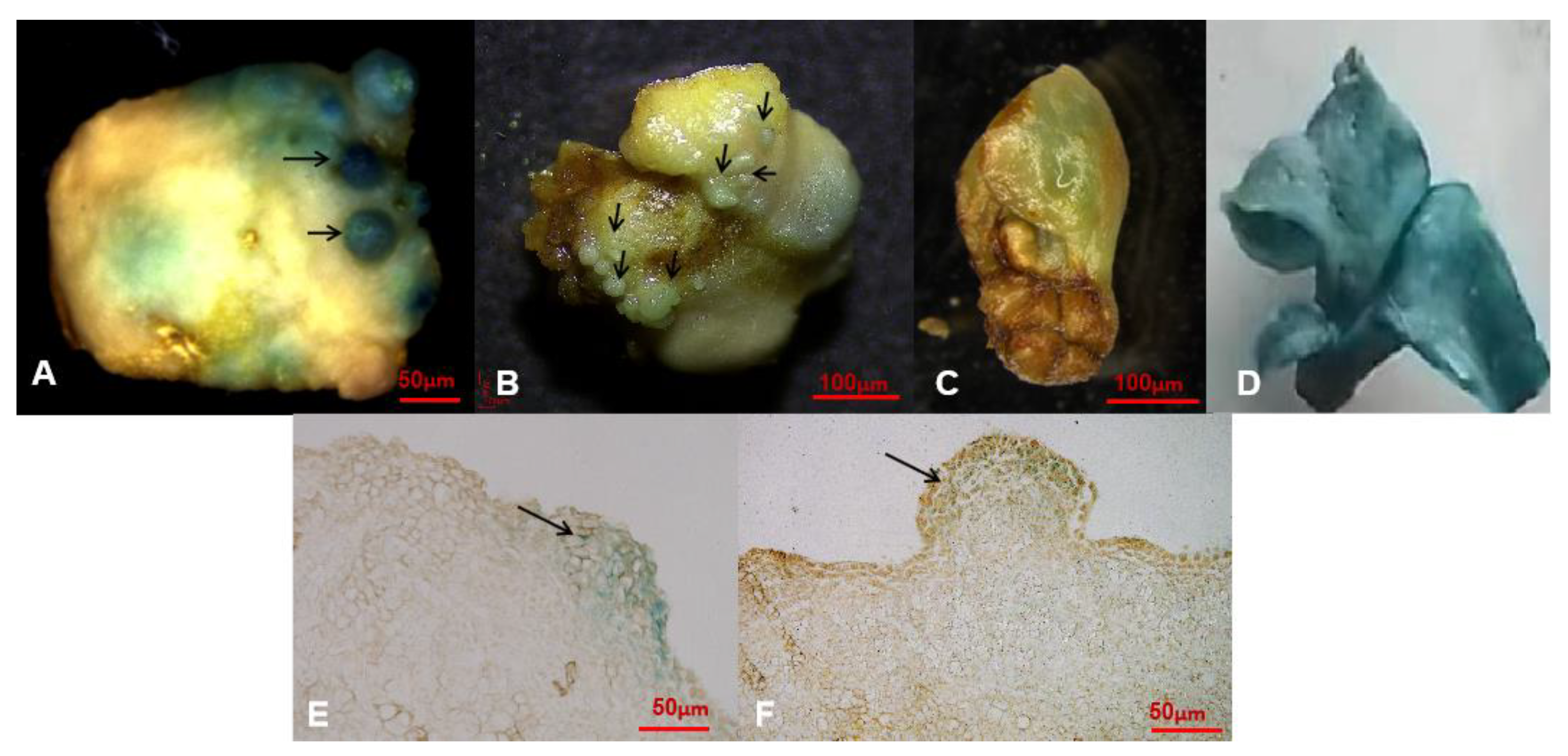
2.5. Screening of Cocultivation Condition
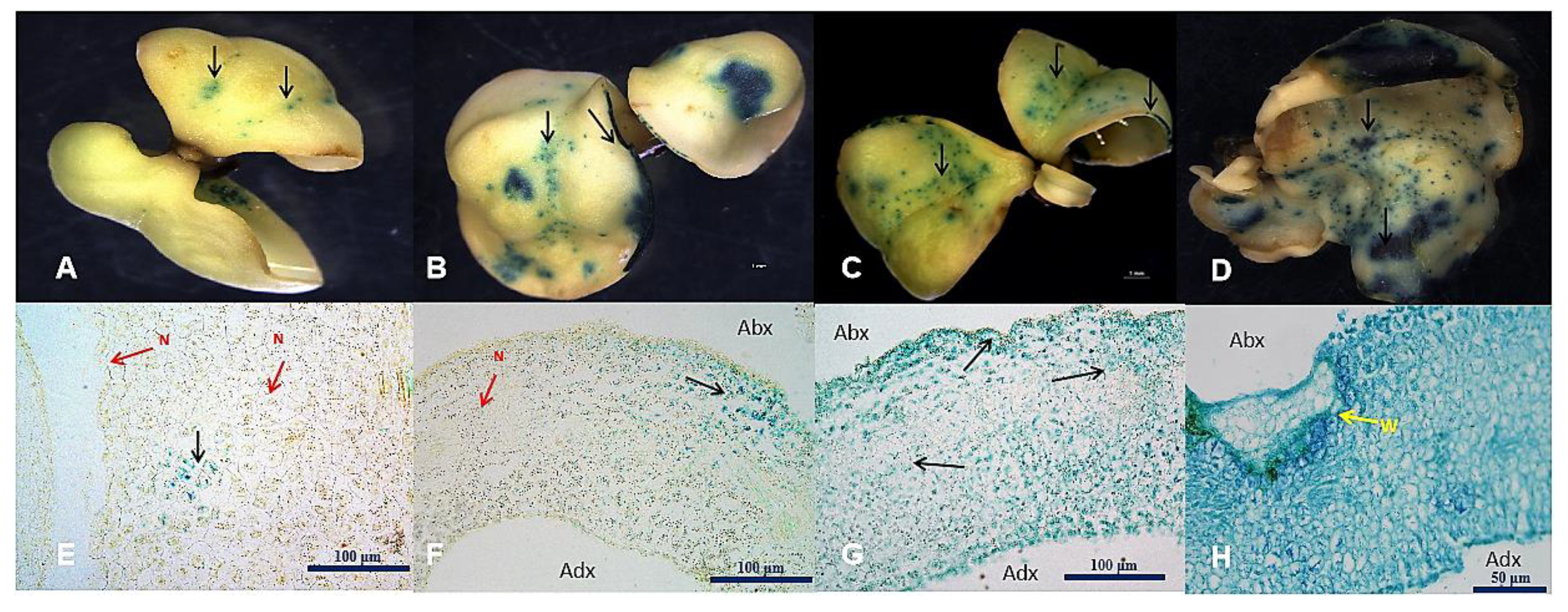
2.6. GUS Histological Observation
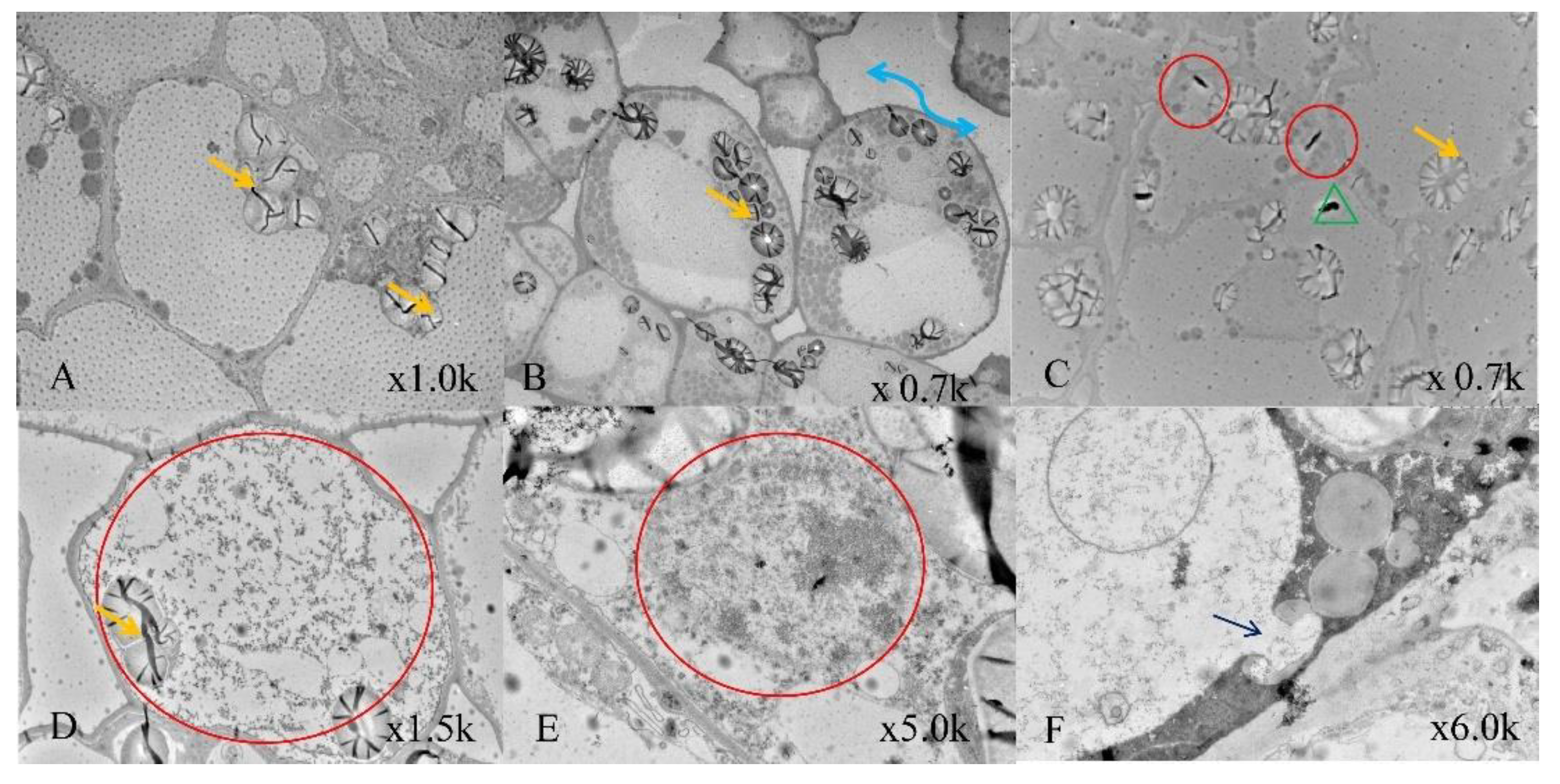
2.7. Transmission Electron Microscopy
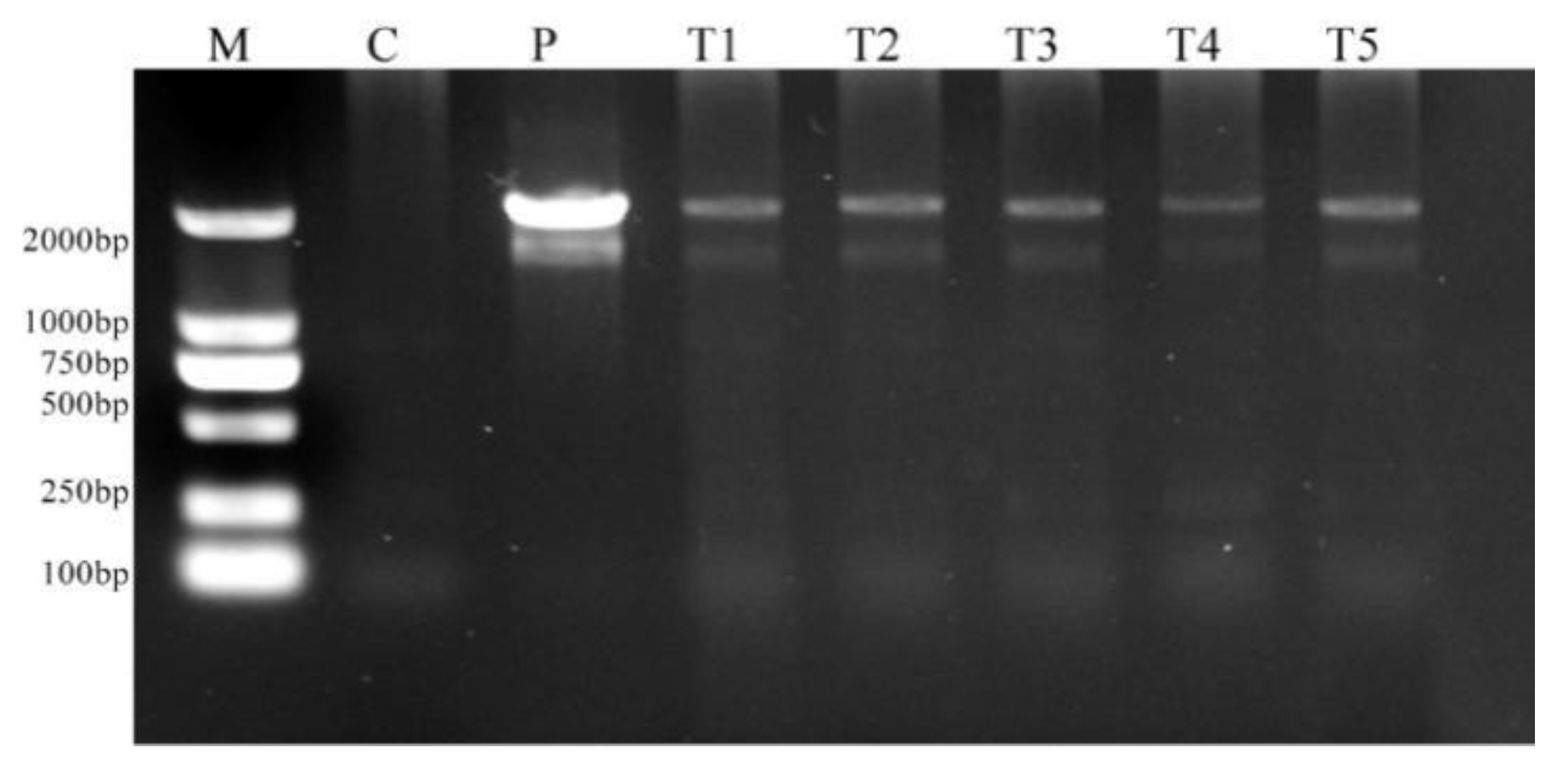
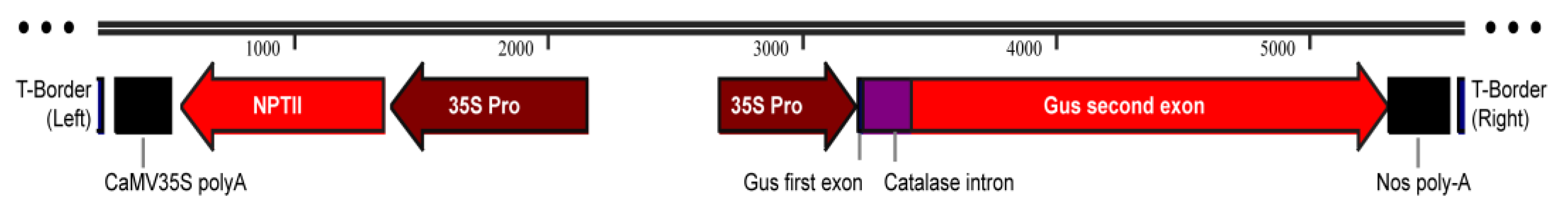
2.8. Molecular Confirmation of Genetic Transformation
3. Discussion
4. Materials and Methods
4.1. Selection of Explant
4.2. Kanamycin Sensitivity Analysis
4.3. Screening Effect of Preculture Days (PCD)
4.4. Preparation of Agrobacterium Strain
4.5. Sonication-Assisted, Agrobacterium-Mediated Transformation (SAAT)
4.6. Screening of Cocultivation Condition
4.7. Histochemical GUS Enzyme Assay
4.8. Transmission Electron Microscopy
4.9. Selection of Transgenic-Resistant Secondary Somatic Embryos
4.10. Molecular Confirmation of Transgenic Gene
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S.; Peng, S.; Huang, G.; Wu, K.; Fu, X.; Chen, Z. Association of decreased expression of a Myb transcription factor with the TPD (tapping panel dryness) syndrome in Hevea brasiliensis. Plant Mol. Biol. 2003, 51, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Begho, E.R. Nursery diseases of Hevea brasiliensis in Nigeria and their control. In Proceedings of a National Workshop on Fruit/Tree Crop Seedling Production; NEARLS: Zaria, Nigeria, 1990; pp. 103–106. [Google Scholar]
- Nandris, D.; Nicole, M.; Geiger, J.P. Root rot diseases of rubber trees. Plant Dis. 1987, 71, 298–306. [Google Scholar] [CrossRef]
- Hua, Y.W.; Huang, T.D.; Huang, H.S. Micropropagation of self-rooting juvenile clones by secondary somatic embryogenesis in Hevea brasiliensis. Plant Breed. 2010, 129, 202–207. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Kumari Jayasree, P.; Sushmakumari, S.; Jayashree, R.; Rekha, K.; Sobha, S.; Priya, P.; Kala, R.G.; Thulaseedharan, A. Current perspectives on application of biotechnology to assist the genetic improvement of rubber tree (Hevea brasiliensis Muell. Arg.): An Overview. Funct. Plant Sci. Biotechnol. 2007, 1, 1–17. [Google Scholar]
- Kala, R.G.; Gimisha, G.C.; Jayasree, P.K.; Sushamakumari, S.; Sobha, S.; Jayasree, R.; Rekha, K.; Thulaseedharan, A. Somatic embryogenesis in leaf cultures of Hevea brasiliensis: Effect of source plant. Nat. Rubber Res. 2009, 22, 117–126. [Google Scholar]
- Carron, M.P.; Etienne, H.; Lardet, L.; Campagna, S.; Perrin, Y.; Leconte, A.; Chaine, C. Somatic embryogenesis in rubber (Hevea brasiliensis Müll. Arg.). In Somatic Embryogenesis in Woody Plants; Jain, S.M., Gupta, P.K., Newton, R.J., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; Volume 2, pp. 117–136. [Google Scholar]
- Carron, M.P.; Le Roux, Y.; Tison, J.; Dea, B.G.; Caussanel, V.; Clair, J.; Keli, J. Compared root system architectures in seedlings and in vitro plantlets of Hevea brasiliensis, in the initial years of growth in the field. Plant Soil 2000, 223, 75–88. [Google Scholar] [CrossRef]
- Shi, X.; Dai, X.; Liu, G.; Zhang, J.; Ning, G.; Bao, M. Cyclic secondary somatic embryogenesis and efficient plant regeneration in camphor tree (Cinnamomum camphora L.). Vitr. Cell. Dev. Biol. Plant 2010, 46, 117–125. [Google Scholar] [CrossRef]
- Sood, A.; Bhattacharya, A.; Sharma, M.; Sharma, R.K.; Harleen, K.N.; Sood, P.; Mehta, R.; Devinder, K.; Jasmine, B.; Ahuja, P.S. Somatic embryogenesis and Agrobacterium mediated genetic transformation in bamboos. In Somatic Embryogenesis and Genetic Transformation in Plant; Aslam, J., Srivastava, P.S., Sharma, M.P., Eds.; Narosa Book Distributors Pvt Ltd.: New Delhi, India, 2013; pp. 168–178. [Google Scholar]
- Wang, T.D.; Huang, T.D.; Huang, H.S.; Hua, T. Origin of secondary somatic embryos and genetic stability of the regenerated plants in Hevea brasiliensis. J. Rubber Res. 2017, 20, 101–116. [Google Scholar] [CrossRef]
- Thulaseedharan, A.; Venkatachalam, P.; Kala, R.G.; Thanseem, I.; Priya, P.; Saleena, A.; Kumari Jayasree, P. Biotechnology research in rubber: Present status and future prospects. J. Plant. Crops 2004, 32, 104–116. [Google Scholar]
- Arokiaraj, P. Genetic transformation of Hevea brasiliensis (rubber tree) and its applications towards crop improvement and production of recombinant proteins of commercial value. In Molecular Biology of Woody Plants; Jain, S.M., Minocha, S.C., Eds.; Springer: Dordrecht, The Netherlands, 2000; Volume 66, pp. 305–325. [Google Scholar]
- Jayashree, R.; Rekha, K.; Venkatachalam, P.; Uratsu, S.L.; Dandekar, A.M.; Jayasree, P.K.; Kala, R.G.; Priya, P.; Sushamakumari, S.; Sobha, S.; et al. Genetic transformation and regeneration of rubber tree (Hevea brasiliensis Muell. Arg) transgenic plants with a constitutive version of an anti-oxidative stress superoxide dismutase gene. Plant Cell Rep. 2003, 22, 201–209. [Google Scholar] [CrossRef]
- Blanc, G.; Baptiste, C.; Oliver, G.; Martin, F.; Montoro, P. Efficient Agrobacterium tumefaciens-mediated transformation of embryogenic calli and regeneration of Hevea brasiliensis Mull Arg. Plants. Plant Cell Rep. 2006, 24, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Omo-Ikerodah, E.E.; Omokhafe, K.O.; Akpobome, F.A.; Mokwunye, M.U. An overview of the potentials of natural rubber (Hevea brasiliensis) engineering for the production of valuable proteins. Afr. J. Biotechnol. 2009, 8, 7303–7307. [Google Scholar]
- Kala, R.G.; Abraham, V.; Sobha, S.; Jayasree, P.K.; Suni, A.M.; Thulaseedharan, A. Agrobacterium-mediated genetic transformation and somatic embryogenesis from leaf callus of Hevea brasiliensis: Effect of silver nitrate. In Prospects in Bioscience: Addressing the Issues; Sabu, A., Augustine, A., Eds.; Springer: New Delhi, India, 2013; pp. 303–315. [Google Scholar]
- Huang, T.D.; Li, J.; Li, Y.T.; Huang, H.S.; Hua, Y.W. Somatic embryo an alternate target tissue for Agrobacterium mediated transformation in Hevea brasiliensis. J. Rubber Res. 2015, 18, 171–188. [Google Scholar]
- Kala, R.G.; Reshmi, J.; Sobha, S.; Jayashree, R.; Thulaseedharan, A. Genetic transformation of Hevea brasiliensis Muell. Arg. using intact explants as target tissues for Agrobacterium infection. J. Trop. Agric. 2014, 52, 21–30. [Google Scholar]
- Flores Solis, J.I.; Mlejnek, P.; Studena, K.; Prochazka, S. Application of sonication-assisted Agrobacterium-mediated transformation in Chenopodium rubrum L. Plant Soil Environ. 2003, 49, 255–260. [Google Scholar] [CrossRef] [Green Version]
- Niazian, M.; Sadat Noori, S.A.; Galuszka, P.; Mortazavian, S.M.M. Tissue Culture-based Agrobacterium-mediated and in planta Transformation Methods. Czech J. Genet. Plant Breed. 2017, 53, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Finer, K.R.; Finer, J.J. Use of Agrobacterium expressing green fluorescent protein to evaluate colonization of sonication-assisted Agrobacterium-mediated transformation-treated soybean cotyledons. Lett. Appl. Microbiol. 2000, 30, 406–410. [Google Scholar] [CrossRef]
- Beranova, M.; Rakousky, S.; Vavrov, Z.; Skalicky, T. Sonication assisted Agrobacterium-mediated transformation enhances the transformation efficiency in flax (Linum usitatissimum L.). Plant Cell Tissue Organ Cult. 2008, 94, 253–259. [Google Scholar] [CrossRef]
- Arokiaraj, P.; Ruker, F.; Obermayr, E.; Shamsul Bahri, A.R.; Hafsah, J.; Carter, D.C.; Yeang, H.Y. Expression of human serum albumin in transgenic Hevea brasiliensis. J. Rubber Res. 2002, 5, 157–166. [Google Scholar]
- Arokiaraj, P.; Jones, H.; Jaafar, H.; Coomber, S.; Charlwood, B.V. Agrobacterium-mediated transformation of Hevea anther callus and their regeneration into plantlets. J. Nat. Rubber Res. 1997, 11, 77–87. [Google Scholar]
- Lardet, L.; Leclercq, J.; Benistan, E.; Dessailly, F.; Oliver, G.; Martin, F.; Montoro, P. Variation in GUS activity in vegetatively propagated Hevea brasiliensis transgenic plants. Plant Cell Rep. 2011, 30, 1847–1856. [Google Scholar] [CrossRef]
- Rojo, F.P.; Seth, S.; Erskine, W.; Kaur, P. An improved protocol for Agrobacterium-mediated transformation in subterranean Clover (Trifolium subterraneum L.). Int. J. Mol. Sci. 2021, 22, 4181. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, C.; Tang, M.; Tao, Y.B.; Pan, B.Z.; Zhang, L.; Niu, L.; He, H.; Wang, X.; Xu, Z.F. An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing Kanamycin concentration and duration of delayed selection. Plant Biotechnol. Rep. 2015, 9, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Fillatti, J.J.; Kiser, J.; Rose, R.; Comai, L. Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 1987, 5, 726–730. [Google Scholar] [CrossRef]
- Sangwan, R.S.; Ducrocq, C.; Sangwan-Norreel, B.S. Effect of culture conditions on Agrobacterium-mediated transformation in datura. Plant Cell Rep. 1991, 10, 90–93. [Google Scholar] [CrossRef]
- Janssen, B.; Gardner, R.C. The use of transient GUS expression to develop an Agrobacterium-mediated gene transfer system for kiwifruit. Plant Cell Rep. 1993, 13, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Cauduro, Y.; Gomes Adamuchio, L.; Carlos Bespalhok Filho, J.; Gerhardt, I.R.; Degenhardt-Goldbach, J.; Bernardes, M.; Quoirin, M. Optimization of factors affecting the Agrobacterium tumefaciens-mediated transformation of Eucalyptus saligna. Rev. Árvore 2017, 41, e410315. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Cong, Y.; Liu, Y.; Wang, T.; Shuai, Q.; Chen, N.; Gai, J.; Li, Y. Optimization of Agrobacterium-mediated transformation in soybean. Front. Plant Sci. 2017, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Pathak, M.R.; Hamzah, R.Y. An effective method of Sonication assisted Agrobacterium-mediated transformation of chickpeas. Plant Cell Tissue Organ Cult. 2008, 93, 65–71. [Google Scholar] [CrossRef]
- Trick, H.N.; Finer, J.J. SAAT: Sonication-assisted Agrobacterium-mediated transformation. Transgenic Res. 1997, 6, 329–336. [Google Scholar] [CrossRef]
- Yenchon, S.; Te-chato, S. Effect of bacteria density, inoculation and co-cultivation period on Agrobacterium-mediated transformation of oil palm embryogenic callus. Int. J. Agric. Technol. 2012, 8, 1485–1496. [Google Scholar]
- Manickavasagam, M.; Subramanyam, K.; Ishwarya, R.; Elayaraja, D.; Ganapathi, A. Assessment of factors influencing the tissue culture-independent Agrobacterium-mediated in planta genetic transformation of okra [Abelmoschus esculentus (L.) Moench]. Plant Cell Tissue Organ Cult. 2015, 123, 309–320. [Google Scholar] [CrossRef]
- Utami, E.S.W.; Hariyanto, S.; Manuhara, Y.S.W. Agrobacterium tumefaciens-mediated transformation of Dendrobium lasianthera J.J.Sm: An important medicinal orchid. J. Genet. Eng. Biotechnol. 2018, 16, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Hasegawa, H.; Suzuki, M. Transformation and regeneration of garlic (Allium sativum L.) by Agrobacterium-mediated gene transfer. Plant Cell Rep. 2000, 19, 989–993. [Google Scholar] [CrossRef]
- Sunilkumar, G.; Rathore, K.S. Transgenic cotton: Factors influencing Agrobacterium-mediated transformation and regeneration. Mol. Breed. 2001, 8, 37–52. [Google Scholar] [CrossRef]
- Liu, S.J.; Wei, Z.M.; Huang, J.Q. The effect of co-cultivation and selection parameters on Agrobacterium-mediated transformation of Chinese soybean varieties. Plant Cell Rep. 2008, 27, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Asande, L.K.; Omwoyo, R.O.; Oduor, R.O.; Nyaboga, E.N. A simple and fast Agrobacterium-mediated transformation system for passion fruit KPF4 (Passiflora edulis f. edulis × Passiflora edulis f. flavicarpa). Plant Methods 2020, 16, 141. [Google Scholar] [CrossRef]
- Baron, C.; Domke, N.; Beinhofer, M.; Hapfelmeier, S. Elevated temperature differentially affects virulence, VirB protein accumulation and T-Pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol. 2001, 183, 6852–6861. [Google Scholar] [CrossRef] [Green Version]
- Borji, M.; Bouamama-Gzara, B.; Chibani, F.; Teyssier, C.; Ben-Ammar, A.; Mliki, A.; Zekri, S.; Ghorbel, A. Micromorphology, structural and ultrastructural changes during somatic embryogenesis of a Tunisian oat variety (Avena sativa L. var ‘Meliane’). Plant Cell Tissue Organ Cult. 2017, 132, 329–342. [Google Scholar] [CrossRef]
- Mikula, A.; Tomaszewicz, W.; Dziurka, M.; Kazmierczak, A.; Grzyb, M.; Sobczak, M.; Zdankowski, P.; Rybczynski, J. The origin of the Cyathea delgadii Sternb. Somatic embryos is determined by the developmental state of donor tissue and mutual balance of selected metabolites. Cells 2021, 10, 1388. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Hwang, O.J.; Cho, M.A.; Han, Y.J.; Kim, Y.M.; Lim, S.H.; Kim, D.S.; Hwang, I.; Kim, J.I. Agrobacterium-mediated genetic transformation of Miscanthus sinensis. Plant Cell Tissue Organ Cult. 2014, 117, 51–63. [Google Scholar] [CrossRef] [Green Version]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]

| Treatment | Explant | Chlorosis and Bleaching | SSE |
|---|---|---|---|
| Control | 30 | 0 | 110 ± 6.43 |
| 25 | 30 | 0 ± 0.58 | 97 ± 7.00 |
| 50 | 30 | 16 ± 1.00 | 74 ± 6.23 |
| 75 | 30 | 22 ± 3.21 | 65 ± 3.51 |
| 100 | 30 | 39 ± 2.51 | 42 ± 4.72 |
| 125 | 30 | 67 ± 0.05 | 14 ± 3.05 |
| 150 | 30 | 78 ± 1.15 | 1 ± 1.00 |
| Temperature | Duration | Number of Replicates | Average of GUS-Positive Embryos | Average GUS Spots/Embryo |
|---|---|---|---|---|
| 22 °C | 4 days | 3 | 0.53 ± 0.03 a | 24.90 ± 1.45 a |
| 25 °C | 4 days | 3 | 0.43 ± 0.03 b | 10.96 ± 1.78 b |
| 28 °C | 4 days | 3 | 0.43 ± 0.03 b | 7.93 ± 2.49 b |
| Time (h) | Temperature | No. of Replicates | GUS-Expressed SEs | GUS Spots/Embryos |
|---|---|---|---|---|
| 72 | 22 °C | 3 | 0.33 ± 0.03 c | 27.33 ± 4.63 c |
| 78 | 22 °C | 3 | 0.53 ± 0.03 b | 60.67 ± 6.33 b |
| 84 | 22 °C | 3 | 0.6 ± 0.03 a | 88.67 ± 3.28 a |
| 90 | 22 °C | 3 | 0.73 ± 0.03 a | 79.67 ± 2.33 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Udayabhanu, J.; Huang, T.; Xin, S.; Cheng, J.; Hua, Y.; Huang, H. Optimization of the Transformation Protocol for Increased Efficiency of Genetic Transformation in Hevea brasiliensis. Plants 2022, 11, 1067. https://doi.org/10.3390/plants11081067
Udayabhanu J, Huang T, Xin S, Cheng J, Hua Y, Huang H. Optimization of the Transformation Protocol for Increased Efficiency of Genetic Transformation in Hevea brasiliensis. Plants. 2022; 11(8):1067. https://doi.org/10.3390/plants11081067
Chicago/Turabian StyleUdayabhanu, Jinu, Tiandai Huang, Shichao Xin, Jing Cheng, Yuwei Hua, and Huasun Huang. 2022. "Optimization of the Transformation Protocol for Increased Efficiency of Genetic Transformation in Hevea brasiliensis" Plants 11, no. 8: 1067. https://doi.org/10.3390/plants11081067
APA StyleUdayabhanu, J., Huang, T., Xin, S., Cheng, J., Hua, Y., & Huang, H. (2022). Optimization of the Transformation Protocol for Increased Efficiency of Genetic Transformation in Hevea brasiliensis. Plants, 11(8), 1067. https://doi.org/10.3390/plants11081067





