New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species
Abstract
:1. Introduction
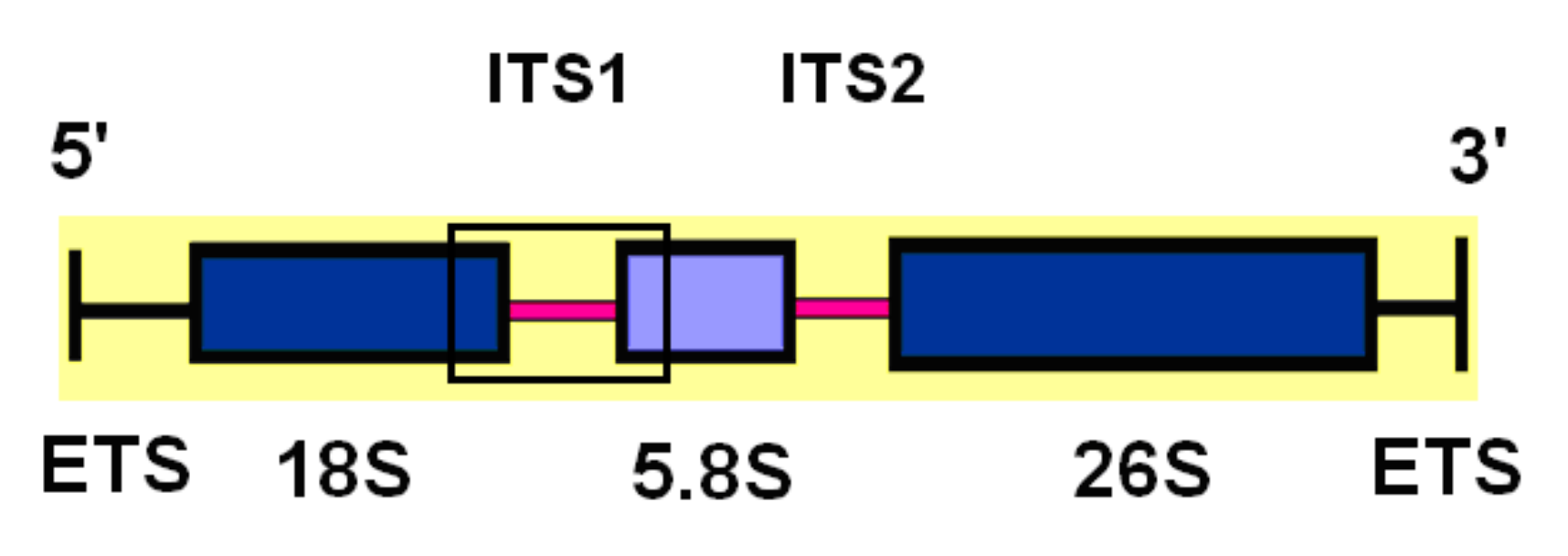
2. Materials and Methods
3. Results
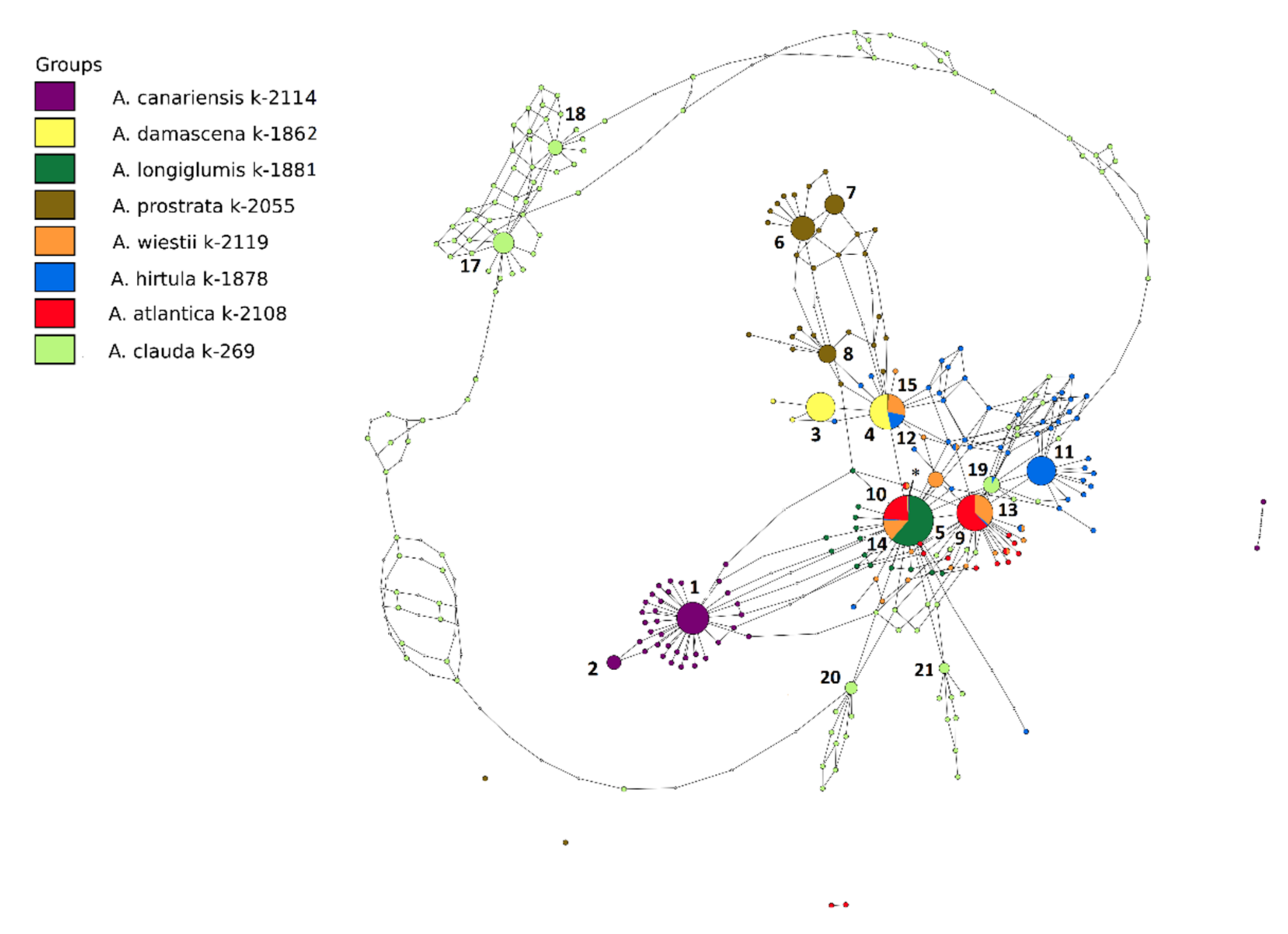
3.1. Sequence Variation and Ribotypes Network
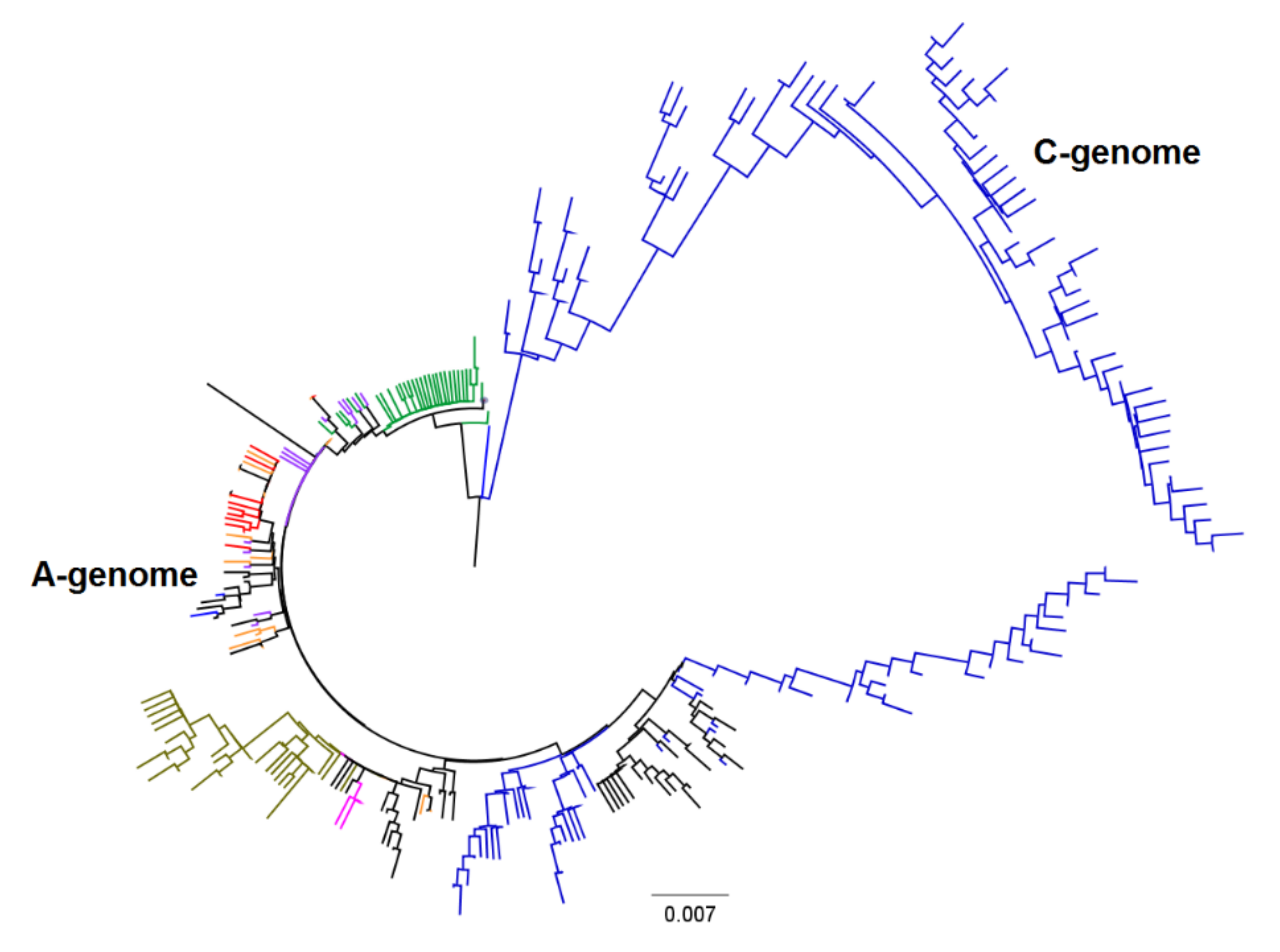
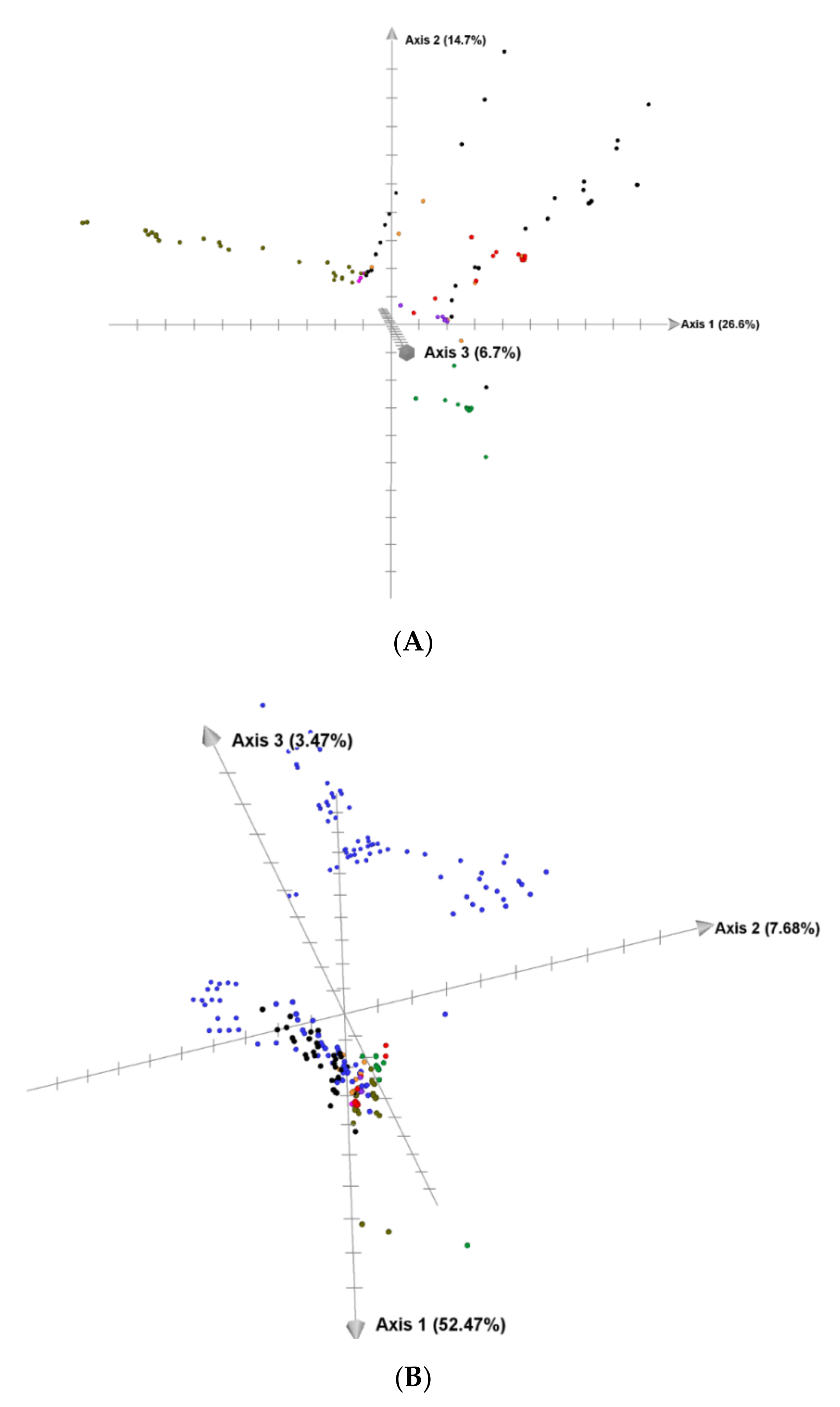
3.2. Genetic Diversity and Relationship
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Judziewicz, E.J.; Zuloaga, F.O.; Filgueiras, T.S.; Morrone, O. A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 2015, 53, 117–137. [Google Scholar] [CrossRef]
- Saarela, J.M.; Bull, R.D.; Paradis, M.J.; Ebata, S.N.; Peterson, P.M.; Soreng, R.J.; Paszko, B. Molecular phylogenetics of cool-season grasses in the subtribes Agrostidinae, Anthoxanthinae, Aveninae, Brizinae, Calothecinae, Koeleriinae and Phalaridinae (Poaceae, Pooideae, Poeae, Poeae chloroplast group 1). PhytoKeys 2017, 87, 1–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loskutov, I.G.; Rines, H.W. Avena. In Wild Crop Relatives: Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Chapter 3; pp. 109–183. [Google Scholar]
- Rajhathy, T.; Thomas, H. Cytogenetics of Oats (Avena L.) Miscellaneous Publications of the Genetics Society of Canada 2; Ontario (Canada) Genetics Society of Canada: Ottawa, ON, Canada, 1974; 90p. [Google Scholar]
- Fominaya, A.; Vega, C.; Ferrer, E. Giemsa C-banded karyotypes of Avena species. Genome 1988, 30, 627–632. [Google Scholar] [CrossRef]
- Drossou, A.; Katsiotis, A.; Leggett, J.M.; Loukas, M.; Tsakas, S. Genome and species relationships in genus Avena based on RAPD and AFLP molecular markers. Theor. Appl. Gen. 2004, 109, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Shelukhina, O.Y.; Diederichsen, A.; Loskutov, I.G.; Pukhalskiy, V.A. Comparative cytogenetic analysis of Avena macrostachya and diploid C-genome Avena species. Genome 2010, 53, 125–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badaeva, E.D.; Shelukhina, O.Y.; Goryunova, S.V.; Loskutov, I.G.; Pukhalskiy, V.A. Phylogenetic relationships of tetraploid AB genome Avena species evaluated by means of cytogenetic (C-banding and FISH) and RAPD analyses. J. Bot. 2010, 2010, 742307. [Google Scholar] [CrossRef] [Green Version]
- Chew, P.; Meade, K.; Hayes, A.; Harjes, C.; Bao, Y.; Beattie, A.D.; Puddephat, I.; Gusmini, G.; Tanksley, S.D. A study on the genetic relationships of Avena taxa and the origins of hexaploid oat. Theor. Appl. Gen. 2016, 129, 1405–1415. [Google Scholar] [CrossRef]
- Latta, R.G.; Bekele, W.A.; Wight, C.P.; Tinker, N.A. Comparative linkage mapping of diploid, tetraploid, and hexaploid Avena species suggests extensive chromosome rearrangement in ancestral diploids. Sci. Rep. 2019, 9, 12298. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Martin, S.L.; Bekele, W.A.; Latta, R.G.; Diederichsen, A.; Peng, Y.; Tinker, N.A. Genome size variation in the genus Avena. Genome 2016, 59, 209–220. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Ren, Z.; Deng, D.; Yang, K. New evidence confirming the CD genomic constitutions of the tetraploid Avena species in the section Pachycarpa Baum. PLoS ONE 2021, 16, e0240703. [Google Scholar] [CrossRef]
- Maughan, P.J.; Lee, R.; Walstead, R.; Vickerstaff, R.J.; Fogarty, M.C.; Brouwer, C.R.; Reid, R.R.; Jay, J.J.; Bekele, W.A.; Jackson, E.W.; et al. Genomic insights from the first chromosome-scale assemblies of oat (Avena spp.) diploid species. BMC Biol. 2019, 17, 92. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Gonzalez, J.J.; Garvin, D.F. Subgenome-specific assembly of vitamin E biosynthesis genes and expression patterns during seed development provide insight into the evolution of oat genome. Plant. Biotechnol. J. 2016, 14, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.B. Oat evolution revealed in the maternal lineages of 25 Avena species. Sci. Rep. 2018, 8, 4252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.Y.; Wei, Y.M.; Baum, B.R.; Zheng, Y.L. Molecular diversity of the 5S rRNA gene and genomic relationships in the genus Avena (Poaceae: Aveneae). Genome 2008, 51, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.; Strömberg, C.A.E.; Leaché, A.D.; Samant, B.; Patnaik, R.; Tang, L.; Mohabey, D.M.; Ge, S.; Sahni, A. Late Cretaceous origin of the rice tribe provides evidence for early diversification in Poaceae. Nat. Commun. 2011, 2, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christin, P.A.; Spriggs, E.; Osborne, C.P.; Strömberg, C.A.E.; Salamin, N.; Edwards, E.J. Molecular dating, evolutionary rates, and the age of the grasses. Syst. Biol. 2014, 63, 153–165. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Marcussen, T.; Meseguer, A.S.; Fjellheim, S. The grass subfamily Pooideae: Cretaceous-Palaeocene origin and climate-driven Cenozoic diversification. Glob. Ecol. Biogeogr. 2019, 28, 1168–1182. [Google Scholar] [CrossRef]
- Thomas, H.; Jones, M.L. Chromosomal differentiation in diploid species of Avena. Can. J. Genet. Cytol. 1965, 7, 108–111. [Google Scholar] [CrossRef]
- Nishiyama, I.; Yabuno, T. Meiotic chromosome pairing in two interspecific hybrids and a criticism of the evolutionary relationship of diploid Avena. Jap. J. Genet. 1975, 50, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Loskutov, I.G. Interspecific crosses in the genus Avena L. Russ. J. Genet. 2001, 37, 467–475. [Google Scholar] [CrossRef]
- Ladizinsky, G. Studies in Oats Evolution: A Man’s Life with Avena; Springer-Verlag: Berlin/Heidelberg, Germany, 2012; 87p. [Google Scholar]
- Nocelli, E.; Giovannini, T.; Bioni, M.; Alicchio, R. RFLP-and RAPD-based genetic relationships of seven diploid species of Avena with the A genome. Genome 1999, 42, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Matyášek, R.; Renny-Byfield, S.; Fulneček, J.; Macas, J.; Grandbastien, M.A.; Nichols, R.; Leitch, A.; Kovařík, A. Next generation sequencing analysis reveals a relationship between rDNA unit diversity and locus number in Nicotiana diploids. BMC Genom. 2012, 13, 722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belyakov, E.A.; Machs, E.M.; Mikhailova, Y.V.; Rodionov, A.V. The study of hybridization processes within genus Sparganium L. subgenus Xanthosparganium Holmb. Based on data of next generation sequencing (NGS). Ecol. Genet. 2019, 17, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Tang, Y.W.; Xu, Y.; Yonezawa, T.; Shao, Y.; Wang, Y.G.; Song, Z.-P.; Yang, J.; Zhang, W.J. Concerted and birth-and-death evolution of 26S ribosomal DNA in Camellia L. Ann. Bot. 2021, 127, 63–73. [Google Scholar] [CrossRef]
- Seitz, U.; Seitz, U. The molecular weight of rRNA precursor molecules and their processing in higher plant cells. Z. Nat. C. Biosci. 1979, 34, 253–258. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Schlessinger, D. Structure and organization of ribosomal DNA. Biochimie 1991, 73, 631–638. [Google Scholar] [CrossRef]
- Garcia, S.; Kovařík, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the Plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [Green Version]
- Rogers, S.O.; Bendich, A.J. Ribosomal RNA genes in plants: Variability in copy number and in intergenic spacer. Plant Mol. Biol. 1987, 9, 509–520. [Google Scholar] [CrossRef]
- Linares, C.; González, J.; Ferrer, E.; Fominaya, A. The use of double fluorescence in situ hybridization to physically map the positions of 5S rDNA genes in relation to the chromosomal location of 18S-5.8S-26S rDNA and a C genome specific DNA sequence in the genus Avena. Genome 1996, 39, 535–542. [Google Scholar] [CrossRef]
- Liu, Q.; Li, X.; Zhou, X.; Li, M.; Zhang, F.; Schwarzacher, T.; Heslop-Harrison, J.S. The repetitive DNA landscape in Avena (Poaceae): Chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads. BMC Plant Biol. 2019, 19, 226. [Google Scholar] [CrossRef] [Green Version]
- Rodionov, A.V.; Amosova, A.V.; Krainova, L.M.; Machs, E.M.; Mikhailova, Y.V.; Gnutikov, A.A.; Muravenko, O.V.; Loskutov, I.G. Phenomenon of Multiple Mutations in the 35S rRNA Genes of the C Subgenome of Polyploid Avena L. Rus. J. Gen. 2020, 56, 674–683. [Google Scholar] [CrossRef]
- Clark, J.W.; Donoghue, P.C.J. Whole-Genome Duplication and Plant Macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, J.T.; Larson, S.R.; Johnson, P.G. Genome relationships in polyploid Poa pratensis and other Poa species inferred from phylogenetic analysis of nuclear and chloroplast DNA sequences. Genome 2005, 48, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Brassac, J.; Blattner, F.R. Species-Level Phylogeny and Polyploid Relationships in Hordeum (Poaceae) Inferred by Next-Generation Sequencing and In Silico Cloning of Multiple Nuclear Loci. Syst. Biol. 2015, 64, 792–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodionov, A.V.; Gnutikov, A.A.; Nosov, N.N.; Machs, E.M.; Mikhaylova, Y.V.; Shneyer, V.S.; Punina, E.O. Intragenomic Polymorphism of the ITS 1 Region of 35S rRNA Gene in the Group of Grasses with Two-Chromosome Species: Different Genome Composition in Closely Related Zingeria Species. Plants 2020, 9, 1647. [Google Scholar] [CrossRef]
- Wang, X.-C.; Liu, C.; Huang, L.; Bengtsson-Palme, J.; Chen, H.; Zhang, J.-H.; Cai, D.; Li, J.-Q. ITS1: A DNA barcode better than ITS2 in eukaryotes? Mol. Ecol. Res. 2015, 15, 573–586. [Google Scholar] [CrossRef]
- Schultz, J.; Maisel, S.; Gerlach, D.; Müller, T.; Wolf, M. A common core of secondary structure of 33 the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [Green Version]
- Coleman, A.W. Nuclear rRNA transcript processing versus internal transcribed spacer 29 secondary structure. Trends Genet. 2015, 31, 157–163. [Google Scholar] [CrossRef]
- Zhang, X.; Cao, Y.; Zhang, W.; Simmons, M.P. Adenine· cytosine substitutions are an alternative 13 pathway of compensatory mutation in angiosperm ITS2. RNA 2020, 26, 209–217. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Machs, E.M.; Blinova, E.V.; Probatova, N.S.; Langdon, T.; Rodionov, A.V. New insights into the genomic structure of the oats (Avena L., Poaceae): Intragenomic polymorphism of ITS1 sequences of rare endemic species Avena bruhnsiana Gruner and its relationship to other species with C-genomes. Euphytica 2022, 218, 3. [Google Scholar] [CrossRef]
- Ridgway, K.P.; Duck, J.M.; Young, J.P.W. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol. 2003, 3, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the ugene team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aronesty, E. Comparison of sequencing utility program. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [Green Version]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef] [Green Version]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/darwin (accessed on 20 January 2022).
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows [electronic resource]. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Volkov, R.A.; Komarova, N.Y.; Hemleben, V. Ribosomal DNA in plant hybrids: Inheritance, rearrangement, expression. Syst. Biodiv. 2007, 5, 261–276. [Google Scholar] [CrossRef]
- Zozomová-Lihová, J.; Mandáková, T.; Kovaříková, A.; Mühlhausen, A.; Mummenhoff, K.; Lysak, M.A.; Kovařík, A. When fathers are instant losers: Homogenization of rDNA loci in recently formed Cardamine× schulzii trigenomic allopolyploid. New Phytol. 2014, 203, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Schanzer, I.A.; Fedorova, A.V.; Galkina, M.A.; Chubar, E.A.; Rodionov, A.V.; Kotseruba, V.V. Is Rosa × archipelagica (Rosaceae, Rosoideae) really a spontaneous intersectional hybrid between R. rugosa and R. maximowicziana? Molecular data confirmation and evidence of paternal leakage. Phytotaxa 2020, 428, 93–103. [Google Scholar] [CrossRef]
- Bashir, T.; Sailer, C.; Gerber, F.; Loganathan, N.; Bhoopalan, H.; Eichenberger, C.; Grossniklaus, U.; Baskar, R. Hybridization Alters Spontaneous Mutation Rates in a Parent-of-Origin-Dependent Fashion in Arabidopsis. Plant Physiol. 2014, 165, 424–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runemark, A.; Vallejo-Marin, M.; Meier, J.I. Eukaryote hybrid genomes. PLoS Genet. 2019, 15, p.e1008404. [Google Scholar] [CrossRef] [PubMed]
- Borisjuk, N.V.; Momot, V.P.; Gleba, Y. Novel class of rDNA repeat units in somatic hybrids between Nicotiana and Atropa. Theor. Appl. Genet. 1988, 76, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Fominaya, A.; Loarce, Y.; Montes, A.; Ferrer, E. Chromosomal distribution patterns of the (AC) 10 microsatellite and other repetitive sequences, and their use in chromosome rearrangement analysis of species of the genus Avena. Genome 2017, 60, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Schlötterer, C.; Tautz, D. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 1994, 4, 777–783. [Google Scholar] [CrossRef]
- Dai, F.; Chen, Z.H.; Wang, X.; Li, Z.; Jin, G.; Wu, D.; Cai, S.; Wang, N.; Wu, F.; Nevo, E.; et al. Transcriptome profiling reveals mosaic genomic origins of modern cultivated barley. Proc. Natl. Acad. Sci. USA 2014, 111, 13403–13408. [Google Scholar] [CrossRef] [Green Version]
- Badaeva, E.D.; Loskutov, I.G.; Shelukhina, O.Y.; Pukhalskiy, V.A. Cytogenetic analysis of diploid Avena L. species containing the as genome. Rus. J. Genet. 2005, 41, 1428–1433. [Google Scholar] [CrossRef]
- Rajhathy, T. Chromosomal differentiation and speciation in diploid Avena. Can. J. Genet. Cytol. 1961, 3, 372–377. [Google Scholar] [CrossRef]
- Ladizinsky, G. Genome relationships in the diploid oats. Chromosoma 1974, 47, 109–117. [Google Scholar] [CrossRef]
- Ladizinsky, G. The cytogenetic position of Avena prostrata among the diploid oats. Can. J. Genet. Cytol. 1973, 15, 443–450. [Google Scholar] [CrossRef]
- Leggett, J.M. Chromosome relationships and morphological comparisons between the diploid oats Avena prostrata, A. canariensis and the tetraploid A. maroccana. Can. J. Genet. Cytol. 1980, 22, 287–294. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations: Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978; 590p. [Google Scholar]
- Jellen, E.N.; Phillips, R.L.; Rines, H.W. C-banded karyotypes and polymorphisms in hexaploid oat accessions (Avena spp.) using Wright’s stain. Genome 1993, 36, 1129–1137. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Tyupa, N.B.; Kim, E.S.; Machs, E.M.; Loskutov, I.G. Genomic configuration of the autotetraploid oat species Avena macrostachya inferred from comparative analysis of ITS1 and ITS2 sequences: On the oat karyotype evolution during the early events of the Avena species divergence. Rus. J. Genet. 2005, 41, 518–528. [Google Scholar] [CrossRef]
- Nikoloudakis, N.; Katsiotis, A. The origin of the C-genome and cytoplasm of Avena polyploids. Theor. Appl. Genet. 2008, 117, 273–281. [Google Scholar] [CrossRef]
- Kamelin, R.V. The Peculiarities of Flowering Plants Speciation; Prilozhenie No. 1; Trudy Zoologicheskogo Instituta RAN: St.-Petersburg, Russia, 2009; pp. 141–149. (In Russian) [Google Scholar]


| Species | Sample ID | Country of Origin | Accession Number | Number of Accessions | Genebank | Genome | Number of NORs in Haploid Genome | Total Number of Reads | Ribotype Number | Ribotype Symbol | Number of Reads | % From the Total Number of the Reads |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avena canariensis | k-2114 | Spain | From OM004567 to OM004604 | 38 | A. Diederichsen | Ac | 2 [1–3] | 20606 | 1 | Ac1 | 10,050 | 49 |
| 2 | Ac2 | 1908 | 9 | |||||||||
| Avena damascena | k-1862 | Syria | From OM004732 to OM004735 | 4 | Ad | 2-3 [1–3] | 12296 | 3 | Ad1 | 4910 | 40 | |
| 4 | Ad2/As3 | 3936 | 32 | |||||||||
| Avena longiglumis | k-1881 | USA | From OM004605 to OM004619 | 15 | Al | 2 [1–3] | 12736 | 5 | Al1/As2 | 9412 | 74 | |
| Avena prostrata | k-2055 | Spain | From OM004620 to OM004650 | 31 | Ap | 2 [1] | 17690 | 6 | Ap1 | 4933 | 28 | |
| 7 | Ap2 | 3188 | 18 | |||||||||
| 8 | Ap3 | 2559 | 15 | |||||||||
| Avena atlantica | k-2108 | Morocco | From OM004717 to OM004731 | 15 | A. Diederichsen | As | 2 [1] | 16241 | 9 | As1 | 6353 | 39 |
| 10 | Al1/As2 | 4571 | 28 | |||||||||
| Avena hirtula | k-1878 | Spain | From OM004668 to OM004716 | 49 | As | 2 [4] | 16538 | 11 | As5 | 12,605 | 40 | |
| 12 | Ad2/As3 | 3587 | 11 | |||||||||
| Avena wiestii | k-2119 | Israel | From OM004651 to OM004667 | 17 | A. Diederichsen | As | 2 [1,4] | 16725 | 13 | As1 | 4005 | 24 |
| 14 | Al1/As2 | 2901 | 17 | |||||||||
| 15 | Ad2/As3 | 2598 | 16 | |||||||||
| 16 | As4 | 1974 | 12 | |||||||||
| Avena clauda | k-269 | Azerbaijan | From OK273905 to OK274031 | 127 | V. N. Soldatov | Cc | 2-4 [3–5] | 40168 | 17 | Ccc1A | 5320 | 13 |
| 18 | Cc1B | 2695 | 7 | |||||||||
| 19 | Cc2A | 3195 | 8 | |||||||||
| 20 | Cc2B | 1777 | 4 | |||||||||
| 21 | Cc2C | 1305 | 3 |
| Samples | N | S | Ps | Θ | π | D |
|---|---|---|---|---|---|---|
| All | 296 | 109 | 0.318 | 0.051 | 0.031 | −2.291 |
| A-genomes | 169 | 92 | 0.269 | 0.047 | 0.013 | −2.291 |
| As-genomes | 81 | 38 | 0.111 | 0.022 | 0.009 | −1.870 |
| Avena canariensis [Ac] | 38 | 40 | 0.117 | 0.028 | 0.008 | −2.476 |
| Avena longiglumis [Al] | 15 | 11 | 0.032 | 0.010 | 0.005 | −1.901 |
| Avena prostrata [Ap] | 31 | 34 | 0.099 | 0.025 | 0.014 | −1.561 |
| Avena wiestii [As] | 17 | 11 | 0.032 | 0.010 | 0.007 | −1.145 |
| Avena hirtula [As] | 49 | 23 | 0.067 | 0.015 | 0.010 | −1.040 |
| Avena atlantica [As] | 15 | 11 | 0.032 | 0.010 | 0.005 | −1.955 |
| Avena damascena [Ad] | 4 | 3 | 0.009 | 0.005 | 0.004 | −0.754 |
| Avena clauda [Cc] | 127 | 34 | 0.099 | 0.018 | 0.037 | 3.160 |
| Species | No. of Accessions | A. canariensis | A. longiglumis | A. prostrata | A. wiestii | A. hirtula | A. atlantica | A. damascena |
|---|---|---|---|---|---|---|---|---|
| A. canariensis | 38 | |||||||
| A. longiglumis | 15 | 0.010 | ||||||
| A. prostrata | 31 | 0.020 | 0.015 | |||||
| A. wiestii | 17 | 0.012 | 0.007 | 0.016 | ||||
| A. hirtula | 49 | 0.015 | 0.010 | 0.018 | 0.010 | |||
| A. atlantica | 15 | 0.012 | 0.007 | 0.017 | 0.006 | 0.010 | ||
| A. damascena | 4 | 0.014 | 0.009 | 0.014 | 0.010 | 0.012 | 0.011 | |
| A. clauda | 127 | 0.051 | 0.043 | 0.051 | 0.044 | 0.045 | 0.045 | 0.046 |
| Source of Variation | Degree of Freedom | Sum of Squares | Variance Components | Percentage Variation | FST |
|---|---|---|---|---|---|
| Among species | 6 | 125.46 | 1.00 | 27.24 | 0.27 |
| Within species | 162 | 438.91 | 2.68 | 72.76 | |
| Total | 168 | 564.36 | 3.68 | ||
| Among genomes | 4 | 125.46 | 0.80 | 21.94 | 0.29 |
| Among species within As-genome | 2 | 16.58 | 0.25 | 6.89 | |
| Within species | 162 | 422.33 | 2.61 | 71.17 | |
| Total | 168 | 564.36 | 3.66 |
| Species | A. canariensis | A. damascena | A. longiglumis | A. prostrata | A. atlantica | A. hirtula | A. wiestii |
|---|---|---|---|---|---|---|---|
| A. canariensis | |||||||
| A. damascena | 0.193 | ||||||
| A. longiglumis | 0.167 | 0.452 | |||||
| A. prostrata | 0.310 | 0.186 | 0.295 | ||||
| A. atlantica | 0.208 | 0.273 | 0.183 | 0.326 | |||
| A. hirtula | 0.289 | 0.278 | 0.199 | 0.323 | 0.172 | ||
| A. wiestii | 0.218 | 0.414 | 0.178 | 0.315 | 0.065 | 0.111 | |
| A. clauda | 0.360 | 0.311 | 0.334 | 0.376 | 0.354 | 0.366 | 0.340 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Blinova, E.V.; Shneyer, V.S.; Probatova, N.S.; Rodionov, A.V. New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species. Plants 2022, 11, 1103. https://doi.org/10.3390/plants11091103
Gnutikov AA, Nosov NN, Loskutov IG, Blinova EV, Shneyer VS, Probatova NS, Rodionov AV. New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species. Plants. 2022; 11(9):1103. https://doi.org/10.3390/plants11091103
Chicago/Turabian StyleGnutikov, Alexander A., Nikolai N. Nosov, Igor G. Loskutov, Elena V. Blinova, Viktoria S. Shneyer, Nina S. Probatova, and Alexander V. Rodionov. 2022. "New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species" Plants 11, no. 9: 1103. https://doi.org/10.3390/plants11091103
APA StyleGnutikov, A. A., Nosov, N. N., Loskutov, I. G., Blinova, E. V., Shneyer, V. S., Probatova, N. S., & Rodionov, A. V. (2022). New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species. Plants, 11(9), 1103. https://doi.org/10.3390/plants11091103








