Differential Expression of Arabinogalactan in Response to Inclination in Stem of Pinus radiata Seedlings
Abstract
:1. Introduction
2. Results
2.1. Molecular and Biochemical Description of Pine AGP
2.2. Determination of Total AGP and Identification of Differential Epitopes
3. Discussion
4. Materials and Methods
4.1. Sequence Analysis
4.2. Multiple Sequence Alignment, Phylogenetic Analysis and Motif Prediction
4.3. Cryo-Sectioning
4.4. AGP Extraction
4.5. Immunolabelling and Staining
4.6. Confocal Microscopy
4.7. Quantitative PCR Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knoch, E.; Dilokpimol, A.; Geshi, N. Arabinogalactan proteins: Focus on carbohydrate active enzymes. Front. Plant Sci. 2014, 5, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orbović, V.; Göllner, E.M.; Soria, P. The effect of arabinogalactan proteins on regeneration potential of juvenile citrus explants used for genetic transformation by Agrobacterium tumefaciens. Acta Physiol. Plant. 2013, 35, 1409–1419. [Google Scholar] [CrossRef]
- Palacio-López, K.; Tinaz, B.; Holzinger, A.; Domozych, D.S. Arabinogalactan Proteins and the Extracellular Matrix of Charophytes: A Sticky Business. Front. Plant Sci. 2019, 10, 447. [Google Scholar] [CrossRef]
- Zhou, K. Glycosylphosphatidylinositol-Anchored Proteins in Arabidopsis and One of Their Common Roles in Signaling Transduction. Front. Plant Sci. 2019, 10, 1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Zhao, H.; Cheng, Z.; Ke, Y.; Liu, J.; Ma, H. Evolution Analysis of the Fasciclin-Like Arabinogalactan Proteins in Plants Shows Variable Fasciclin-AGP Domain Constitutions. Int. J. Mol. Sci. 2019, 20, 1945. [Google Scholar] [CrossRef] [Green Version]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef]
- Zang, L.; Zheng, T.; Chu, Y.; Ding, C.; Zhang, W.; Huang, Q.; Su, X. Genome-Wide Analysis of the Fasciclin-like Arabinogalactan Protein Gene Family Reveals Differential Expression Patterns, Localization, and Salt Stress Response in Populus. Front. Plant Sci. 2015, 6, 1140. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Showalter, A.M.; Egelund, J.; Hernandez-Sanchez, A.; Doblin, M.S.; Bacic, A.F. Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 2012, 3, 140. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.L.; Jones, B.J.; Bacic, A.; Schultz, C.J. The Fasciclin-Like Arabinogalactan Proteins of Arabidopsis. A Multigene Family of Putative Cell Adhesion Molecules. Plant Physiol. 2003, 133, 1911–1925. [Google Scholar] [CrossRef] [Green Version]
- Hernández Sánchez, A.M.; Capataz Tafur, J.; Rodríguez-Monroy, M.; Sepúlveda-Jiménez, G. Arabinogalactan proteins in plant cell cultures. Interciencia 2009, 34, 170–176. [Google Scholar]
- Iris Pérez-Almeida, N.C.C. Las β-galactosidasas y la dinámica de la pared celular. Interciencia 2006, 31, 10. [Google Scholar]
- Elkins, T.; Zinn, K.; McAllister, L.; HoffMann, F.M.; Goodman, C.S. Genetic analysis of a drosophila neural cell adhesion molecule: Interaction of fasciclin I and abelson tyrosine kinase mutations. Cell 1990, 60, 565–575. [Google Scholar] [CrossRef]
- Seifert, G.J. Fascinating Fasciclins: A Surprisingly Widespread Family of Proteins that Mediate Interactions between the Cell Exterior and the Cell Surface. Int. J. Mol. Sci. 2018, 19, 1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacMillan, C.P.; Mansfield, S.D.; Stachurski, Z.H.; Evans, R.; Southerton, S.G. Fasciclin-like arabinogalactan proteins: Specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 2010, 62, 689–703. [Google Scholar] [CrossRef]
- Bossy, A.; Blaschek, W.; Classen, B. Characterization and immunolocalization of arabinogalactan-proteins in roots of Echinacea purpurea. Planta Med. 2009, 75, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Göllner, E.M.; Gramann, J.C.; Classen, B. Antibodies against Yariv’s reagent for immunolocalization of arabinogalactan-proteins in aerial parts of Echinacea purpurea. Planta Med. 2013, 79, 175–180. [Google Scholar]
- Motose, H.; Sugiyama, M.; Fukuda, H. A proteoglycan mediates inductive interaction during plant vascular development. Nature 2004, 429, 873–878. [Google Scholar] [CrossRef]
- Seifert, G.J.; Roberts, K. The Biology of Arabinogalactan Proteins. Annu. Rev. Plant Biol. 2007, 58, 137–161. [Google Scholar] [CrossRef]
- Yang, J.; Sardar, H.S.; McGovern, K.R.; Zhang, Y.; Showalter, A.M. A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. Plant J. 2007, 49, 629–640. [Google Scholar] [CrossRef]
- Mohammadinejad, R.; Kumar, A.; Ranjbar-Mohammadi, M.; Ashrafizadeh, M.; Han, S.S.; Khang, G.; Roveimiab, Z. Recent Advances in Natural Gum-Based Biomaterials for Tissue Engineering and Regenerative Medicine: A Review. Polymers 2020, 12, 176. [Google Scholar] [CrossRef] [Green Version]
- Makarova, E.N.; Shakhmatov, E.G.; Udoratina, E.V.; Kutchin, A.V. Structural and chemical charactertistics of pectins, arabinogalactans, and arabinogalactan proteins from conifers. Russ. Chem. Bull. 2015, 64, 1302–1318. [Google Scholar] [CrossRef]
- Ca, L.; Jd, P.; Eg, N. Purification and cloning of an arabinogalactan-protein from xylem of loblolly pine. Planta 2000, 210, 686–689. [Google Scholar]
- Yang, S.-H.; Loopstra, C.A. Seasonal variation in gene expression for loblolly pines (Pinus taeda) from different geographical regions. Tree Physiol. 2005, 25, 1063–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-vaz, J.E.; Fernandez, A.; Valenzuela, L.; Torres, M. Madera de compresión en Pinus Radiata D. Don: I, características anatómicas. Maderas Cienc. Tecnol. 2007, 9, 29–43. [Google Scholar] [CrossRef] [Green Version]
- Groover, A. Gravitropisms and reaction woods of forest trees—evolution, functions and mechanisms. New Phytol. 2016, 211, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Lomagno, J.; Rozas, C. Determinación De la madera de compresión en Pinus radiata D.DON. Maderas Cienc. Tecnol. 2001, 3, 63–67. [Google Scholar] [CrossRef]
- Ramos, P.; Provost, G.L.; Gantz, C.; Plomion, C.; Herrera, R. Transcriptional analysis of differentially expressed genes in response to stem inclination in young seedlings of pine. Plant Biol. 2012, 14, 923–933. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, C.; Wang, C.; Yang, Y.; Yang, L.; Gao, X.; Zhang, H. Antisense expression of the fasciclin-like arabinogalactan protein FLA6 gene in Populus inhibits expression of its homologous genes and alters stem biomechanics and cell wall composition in transgenic trees. J. Exp. Bot. 2015, 66, 1291–1302. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Jin, Y.; Wang, C.; Li, B.; Jiang, C.; Sun, Z.; Zhang, Z.; Kong, F.; Zhang, H. Fasciclin-like arabinogalactan proteins, PtFLAs, play important roles in GA-mediated tension wood formation in Populus. Sci. Rep. 2017, 7, 6182. [Google Scholar] [CrossRef]
- Li, X.; Yang, X.; Wu, H.X. Transcriptome profiling of radiata pine branches reveals new insights into reaction wood formation with implications in plant gravitropism. BMC Genom. 2013, 14, 768. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wu, H.X.; Dillon, S.K.; Southerton, S.G. Generation and analysis of expressed sequence tags from six developing xylem libraries in Pinus radiata D. Don. BMC Genom. 2009, 10, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenhaber, B.; Bork, P.; Yuan, Y.; Löffler, G.; Eisenhaber, F. Automated annotation of GPI anchor sites: Case study C. elegans. Trends Biochem. Sci. 2000, 25, 340–341. [Google Scholar] [CrossRef]
- Macquet, A.; Ralet, M.-C.; Kronenberger, J.; Marion-Poll, A.; North, H.M. In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 2007, 48, 984–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Happ, K.; Classen, B. Arabinogalactan-Proteins from the Liverwort Marchantia polymorpha L., a Member of a Basal Land Plant Lineage, Are Structurally Different to Those of Angiosperms. Plants 2019, 8, 460. [Google Scholar] [CrossRef] [Green Version]
- MacMillan, C.P.; Taylor, L.; Bi, Y.; Southerton, S.G.; Evans, R.; Spokevicius, A. The fasciclin-like arabinogalactan protein family of Eucalyptus grandis contains members that impact wood biology and biomechanics. New Phytol. 2015, 206, 1314–1327. [Google Scholar] [CrossRef]
- Ma, H.; Zhao, J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 2647–2668. [Google Scholar] [CrossRef]
- Showalter, A.M.; Keppler, B.D.; Liu, X.; Lichtenberg, J.; Welch, L.R. Bioinformatic Identification and Analysis of Hydroxyproline-Rich Glycoproteins in Populus trichocarpa. BMC Plant Biol. 2016, 16, 229. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Suzuki, Y.; Miyamoto, K.; Ueda, J.; Yamaguchi, I. AtFLA11, a Fasciclin-Like Arabinogalactan-Protein, Specifically Localized in Screlenchyma Cells. Biosci. Biotechnol. Biochem. 2005, 69, 1963–1969. [Google Scholar] [CrossRef]
- Guerriero, G.; Mangeot-Peter, L.; Legay, S.; Behr, M.; Lutts, S.; Siddiqui, K.S.; Hausman, J.-F. Identification of fasciclin-like arabinogalactan proteins in textile hemp (Cannabis sativa L.): In silico analyses and gene expression patterns in different tissues. BMC Genom. 2017, 18, 741. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.; Pereira, A.M.; Pinto, S.C.; Silva, J.; Pereira, L.G.; Coimbra, S. In silico and expression analyses of fasciclin-like arabinogalactan proteins reveal functional conservation during embryo and seed development. Plant Reprod. 2019, 32, 353–370. [Google Scholar] [CrossRef] [Green Version]
- Bygdell, J.; Srivastava, V.; Obudulu, O.; Srivastava, M.K.; Nilsson, R.; Sundberg, B.; Trygg, J.; Mellerowicz, E.J.; Wingsle, G. Protein expression in tension wood formation monitored at high tissue resolution in Populus. J. Exp. Bot. 2017, 68, 3405–3417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, H.; Veit, C.; Abas, L.; Tryfona, T.; Maresch, D.; Ricardi, M.M.; Estevez, J.M.; Strasser, R.; Seifert, G.J. Arabidopsis thaliana FLA4 functions as a glycan-stabilized soluble factor via its carboxy-proximal Fasciclin 1 domain. Plant J. 2017, 91, 613–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Yu, M.; Geng, L.-L.; Zhao, J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 2010, 64, 482–497. [Google Scholar] [CrossRef]
- Yariv, J.; Lis, H.; Katchalski, E. Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem. J. 1967, 105, 1C–2C. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domon, J.-M.; Neutelings, G.; Roger, D.; David, A.; David, H. A Basic Chitinase-like Protein Secreted by Embryogenic Tissues of Pinus caribaea acts on Arabinogalactan Proteins Extracted from the same Cell Lines. J. Plant Physiol. 2000, 156, 33–39. [Google Scholar] [CrossRef]
- Lafarguette, F.; Leplé, J.-C.; Déjardin, A.; Laurans, F.; Costa, G.; Lesage-Descauses, M.-C.; Pilate, G. Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytol. 2004, 164, 107–121. [Google Scholar] [CrossRef]
- Cassab, G.I. Arabinogalactan proteins during the development of soybean root nodules. Planta 1986, 168, 441–446. [Google Scholar] [CrossRef]
- Baumann, A.; Pfeifer, L.; Classen, B. Arabinogalactan-proteins from non-coniferous gymnosperms have unusual structural features. Carbohydr. Polym. 2021, 261, 117831. [Google Scholar] [CrossRef]
- Valenzuela, C.; Ramos, P.; Carrasco, C.; Moya-Leon, M.A.; Herrera, R. Cloning and characterization of a xyloglucan endo-transglycosylase/hydrolase gene expressed in response to inclination in radiata pine seedlings. Tree Genet. Genomes 2014, 10, 1305–1315. [Google Scholar] [CrossRef]
- Mateluna, P.; Valenzuela-Riffo, F.; Morales-Quintana, L.; Herrera, R.; Ramos, P. Transcriptional and computational study of expansins differentially expressed in response to inclination in radiata pine. Plant Physiol. Biochem. 2017, 115, 12–24. [Google Scholar] [CrossRef]
- Cruz, N.; Méndez, T.; Ramos, P.; Urbina, D.; Vega, A.; Gutiérrez, R.A.; Moya-León, M.A.; Herrera, R. Induction of PrMADS10 on the lower side of bent pine tree stems: Potential role in modifying plant cell wall properties and wood anatomy. Sci. Rep. 2019, 9, 18981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Brown, G.; Whetten, R.; Loopstra, C.A.; Neale, D.; Kieliszewski, M.J.; Sederoff, R.R. An arabinogalactan protein associated with secondary cell wall formation in differentiating xylem of loblolly pine. Plant Mol. Biol. 2003, 52, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Szarejko, I. Arabinogalactan proteins are involved in root hair development in barley. J. Exp. Bot. 2014, 66, 1245–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; MacMillan, C.P.; de Vries, L.; Mansfield, S.D.; Hao, P.; Ratcliffe, J.; Bacic, A.; Johnson, K.L. FLA11 and FLA12 glycoproteins fine-tune stem secondary wall properties in response to mechanical stresses. New Phytol. 2022, 233, 1750–1767. [Google Scholar] [CrossRef]
- Ma, Y.; Yan, C.; Li, H.; Wu, W.; Liu, Y.; Wang, Y.; Chen, Q.; Ma, H. Bioinformatics Prediction and Evolution Analysis of Arabinogalactan Proteins in the Plant Kingdom. Front. Plant Sci. 2017, 8, 66. [Google Scholar] [CrossRef] [Green Version]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- de Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Herrera, R.; Krier, C.; Lalanne, C.; Ba, E.H.M.; Stokes, A.; Salin, F.; Fourcaud, T.; Claverol, S.; Plomion, C. (Not) Keeping the stem straight: A proteomic analysis of maritime pine seedlings undergoing phototropism and gravitropism. BMC Plant Biol. 2010, 10, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chabannes, M.; Barakate, A.; Lapierre, C.; Marita, J.M.; Ralph, J.; Pean, M.; Danoun, S.; Halpin, C.; Grima-Pettenati, J.; Boudet, A.M. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001, 28, 257–270. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ding, L.; Zhu, J.-K. A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 1997, 203, 289–294. [Google Scholar] [CrossRef]
- Hall, H.C.; Cheung, J.; Ellis, B.E. Immunoprofiling reveals unique cell-specific patterns of wall epitopes in the expanding Arabidopsis stem. Plant J. 2013, 74, 134–147. [Google Scholar]
- le Provost, G.; Herrera, R.; Paiva, J.A.; Chaumeil, P.; Salin, F.; Plomion, C. A micromethod for high throughput RNA extraction in forest trees. Biol. Res. 2007, 40, 291–297. [Google Scholar] [CrossRef] [Green Version]
- Ramos, P.; Valenzuela, C.; le Provost, G.; Plomion, C.; Gantz, C.; Moya-León, M.A.; Herrera, R. ACC Oxidase and ACC Synthase Expression Profiles after Leaning of Young Radiata (P. radiata D. Don) and Maritime Pine (P. pinaster Ait.) Seedlings. J. Plant Growth Regul. 2012, 31, 382–391. [Google Scholar] [CrossRef]

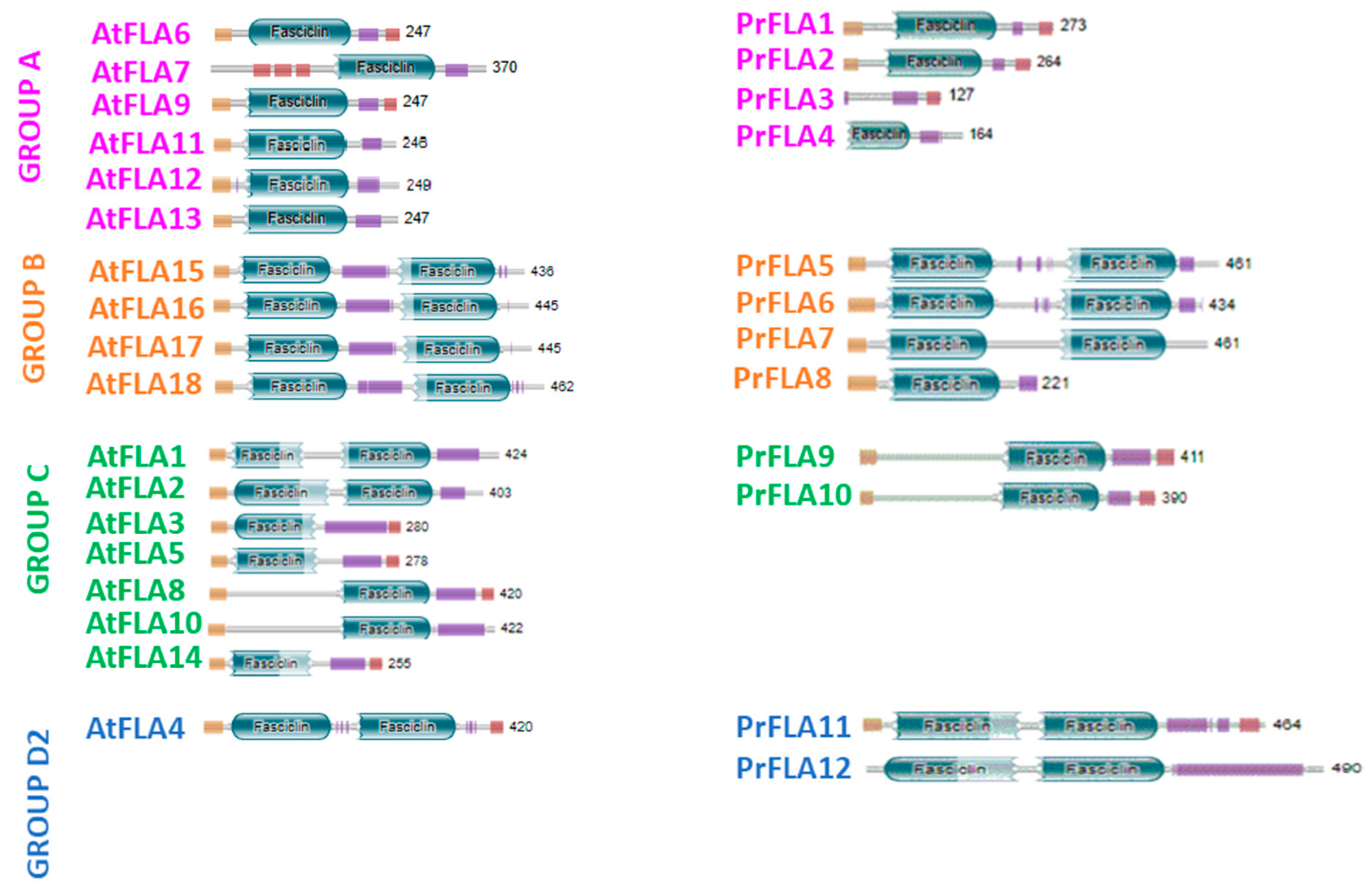
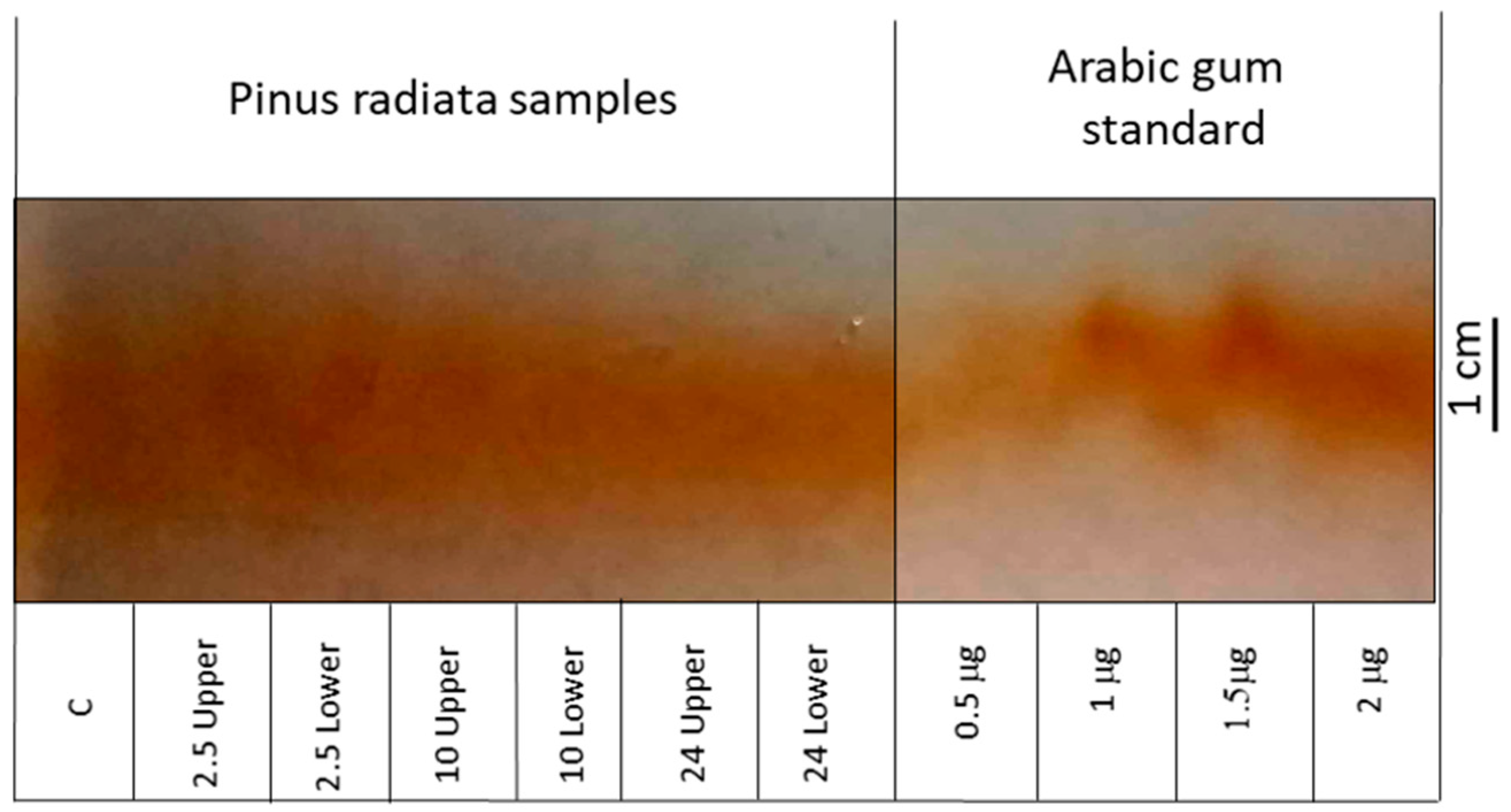

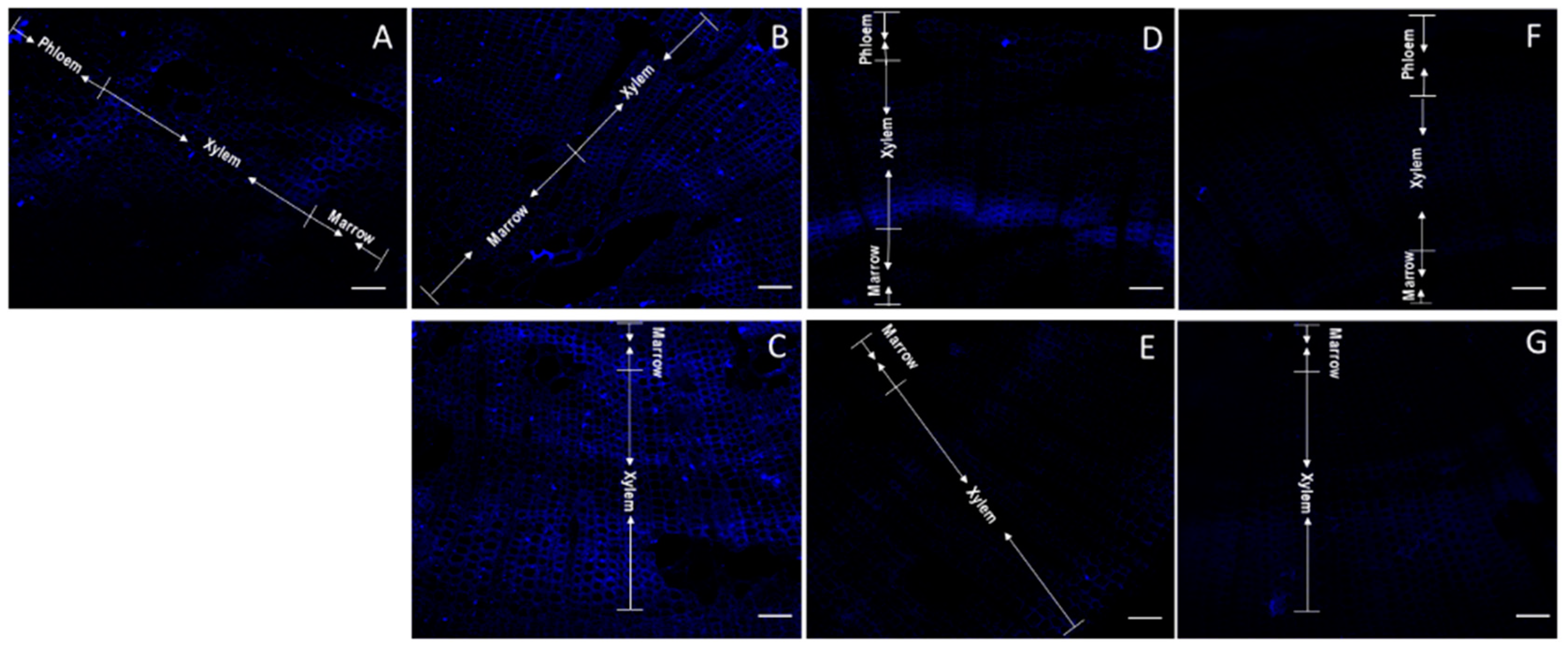
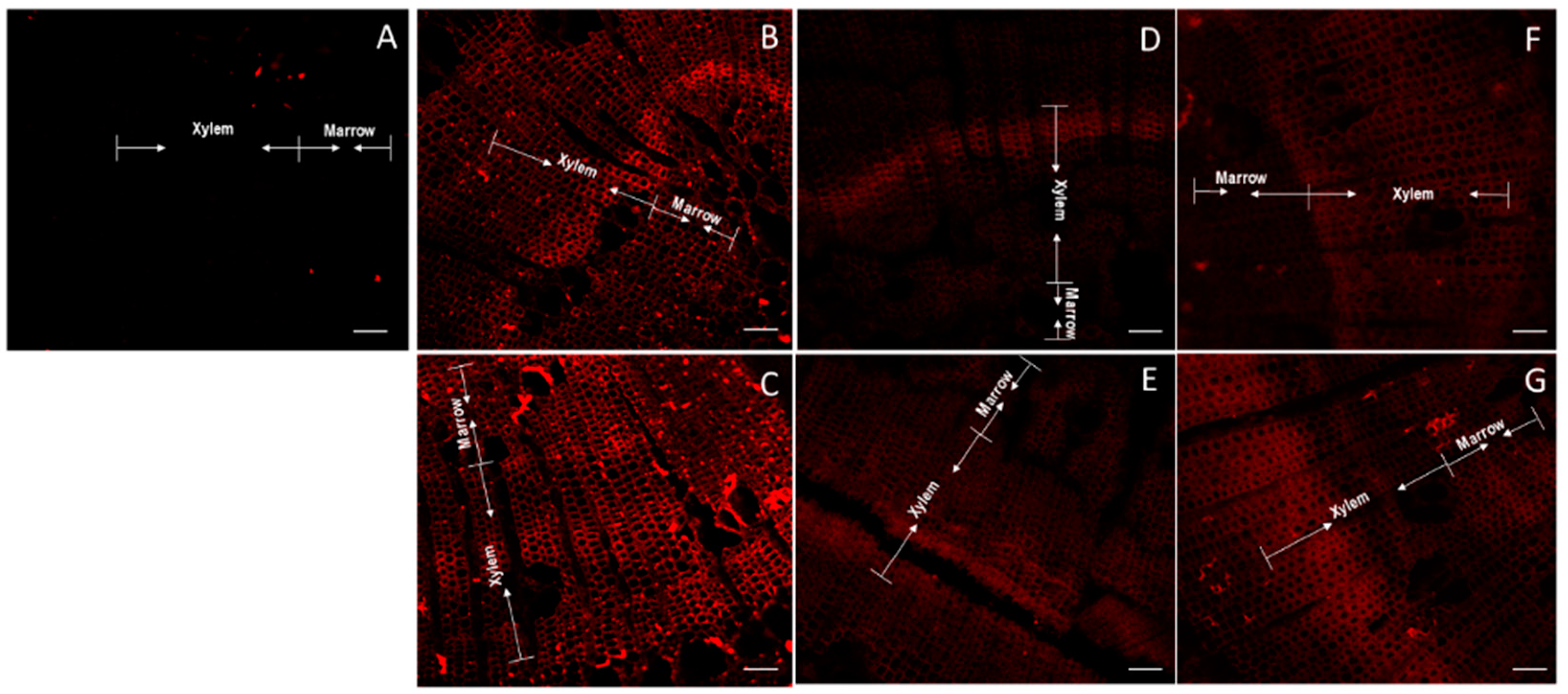

| Gene | Group Length (aa) | AP/PA/SP/TP/GP/VP Repeats in AGP Region | Total PAST% | Part PAST% | Part Length (aa) | Fasciclin Domain | Predicted Signal Peptide | Predicted GPI Anchor | A. thaliana FLA BLASTP Hits | Intraspecies FLA BLASTP Hits | Hmmscan |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PrFLA1 | A/273 | 6-5-2-1-0-1 | 0.38 | 0.08 | 19 and 8 | 1 | Yes | No | AtFLA11 | FLA1(Pinus taeda) | SP-FAS-Dis-TM |
| PrFLA2 | A/264 | 3-4-1-2-0-0 | 0.41 | 0.09 | 16 and 8 | 1 | Yes | Yes | AtFLA6 | p14A9 (Pinus taeda)/arabinogalactan-like protein (Pinus armandii) | SP-FAS-Dis-TM |
| PrFLA3 | A/127 | 2-2-3-0-0-0 | 0.48 | 0.086 | 11 | 1 | None | Yes | AtFLA6 | FLA7(Pinus taeda) | Dis-Dis-TM |
| PrFLA4 | A/164 | 4-4-1-0-0-1 | 0.45 | 0.08 | 15 | 1 | None | Yes | AtFLA6 | FLA7-11(Pinus taeda)-FLA7 (Cinnamomum micranthum f. kanehirae)-AGP-like (Pinus densata) | FAS-Dis |
| PrFLA5 | B/461 | 4-2-0-0-2-0 | 0.24 | 0.04 | 26 | 2 | Yes | None | AtFLA17 | FLA16(Amborella trichopoda)-FLA17(Elaeis guineensis)-FLA17(Elaeis guineensis)-FLA17(Populus trichocarpa) | SP-FAS-Dis-Dis-FAS-Dis |
| PrFLA6 | B/434 | 4-2-0-0-2-0 | 0.28 | 0.25 | 14 | 2 | None | None | AtFLA17 | FLA16(Amborella trichopoda)-FLA16-17(Dendrobium catenatum)-FLA17(Carica papaya) | SP-FAS-Dis-Dis-FAS-Dis |
| PrFLA7 | B/411 | 9-7-3-2-1-0 | 0.38 | 0.09 | 8 and 42 | 2 | Yes | Yes | AtFLA17 | FLA8(Pinus taeda)-FLA10(Hevea brasiliensis)-FLA10(Citrus clementina) | SP-FAS-FAS |
| PrFLA8 | B/221 | 0-0-0-0-0-0 | 0.27 | 0 | 0 | 1 | None | None | AtFLA17 | FLA7-11(Pinus taeda)-FLA7 (Cinnamomum micranthum f. kanehirae)-AGP-like (Pinus densata) | SP-FAS-Dis |
| PrFLA9 | C/845 | 13-9-3-2-3-0 | 0.33 | 0.06 | 8, 41 and 14 | 3 | None | None | AtFLA8 | FLA8(Pinus taeda)-FLA16(Amborella trichopoda)-FLA16(Dendrobium catenatum) | SP-FAS-Dis-TM |
| PrFLA10 | C/390 | 5-6-1-2-0-1 | 0.36 | 0.09 | 44 | 2 | Yes | None | AtFLA8 | FLA8(Pinus taeda)-FLA10(Populus alba)-FLA10(Populus trichocarpa)-FLA8(Prunus avium) | SP-FAS-Dis-TM |
| PrFLA11 | D2/464 | 4-2-1-0-0-1 | 0.34 | 0.03 | 4, 5 and 8 | 2 | Yes | None | AtFLA4 | FLA4 isoform X1(Amborella trichopoda)-FLA4(Nymphaea colorata)-FLA4(Glycine soja) | SP-FAS-FAS-Dis-Dis-TM |
| PrFLA12 | D2/415 | 3-4-5-1-0-0 | 0.36 | 0.05 | 7 and 14 | 2 | None | None | AtFLA4 | FLA4 isoform X1(Amborella trichopoda)-FLA4(Nymphaea colorata)-FLA4(Elaeis guineensis) | FAS-FAS-Dis |
| Gene | Fordward | Reverse |
|---|---|---|
| PrFLA1 | GATAGGATTTATTGGGGCAATTCAC | CTGCCCCTATAAATCAGAATTCCAT |
| PrFLA2 | TAGCGCCCGTCGTTTAAATG | CTGACGCAACGATTACTTACCAAAT |
| PrFLA3 | AGCACCAGCACCAGCACCAGTCTT | GGGAAATGCAATGGGCCAA |
| PrFLA4 | AGCCAATCTGACACAACTGCTATCA | CAGGATCAGATGGTGCAAATATTGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méndez, T.; Stappung, Y.; Moya-León, M.A.; Herrera, R. Differential Expression of Arabinogalactan in Response to Inclination in Stem of Pinus radiata Seedlings. Plants 2022, 11, 1190. https://doi.org/10.3390/plants11091190
Méndez T, Stappung Y, Moya-León MA, Herrera R. Differential Expression of Arabinogalactan in Response to Inclination in Stem of Pinus radiata Seedlings. Plants. 2022; 11(9):1190. https://doi.org/10.3390/plants11091190
Chicago/Turabian StyleMéndez, Tamara, Yazmina Stappung, María A. Moya-León, and Raúl Herrera. 2022. "Differential Expression of Arabinogalactan in Response to Inclination in Stem of Pinus radiata Seedlings" Plants 11, no. 9: 1190. https://doi.org/10.3390/plants11091190








