Germacrene A Synthases for Sesquiterpene Lactone Biosynthesis Are Expressed in Vascular Parenchyma Cells Neighboring Laticifers in Lettuce
Abstract
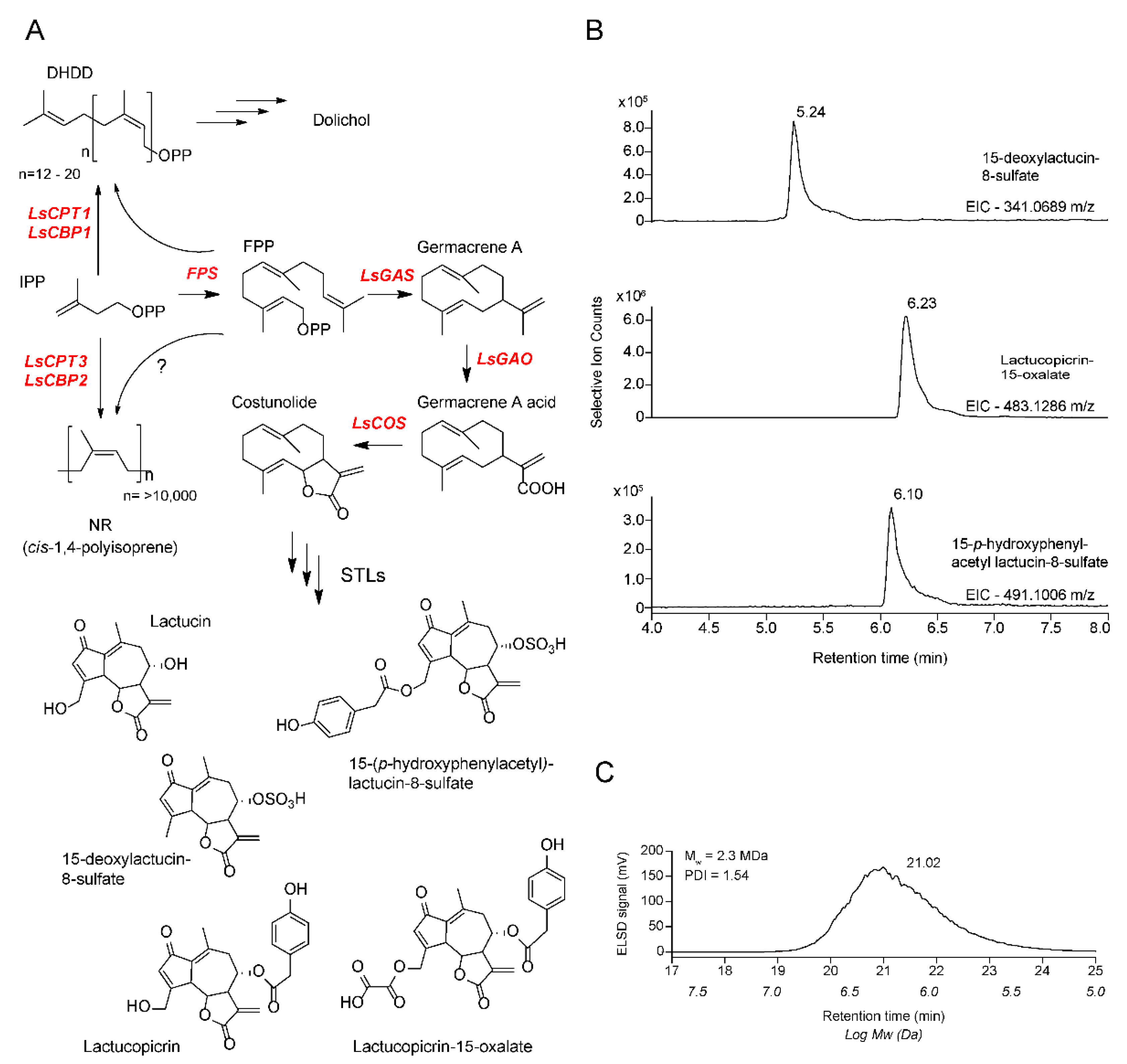
:1. Introduction
2. Results
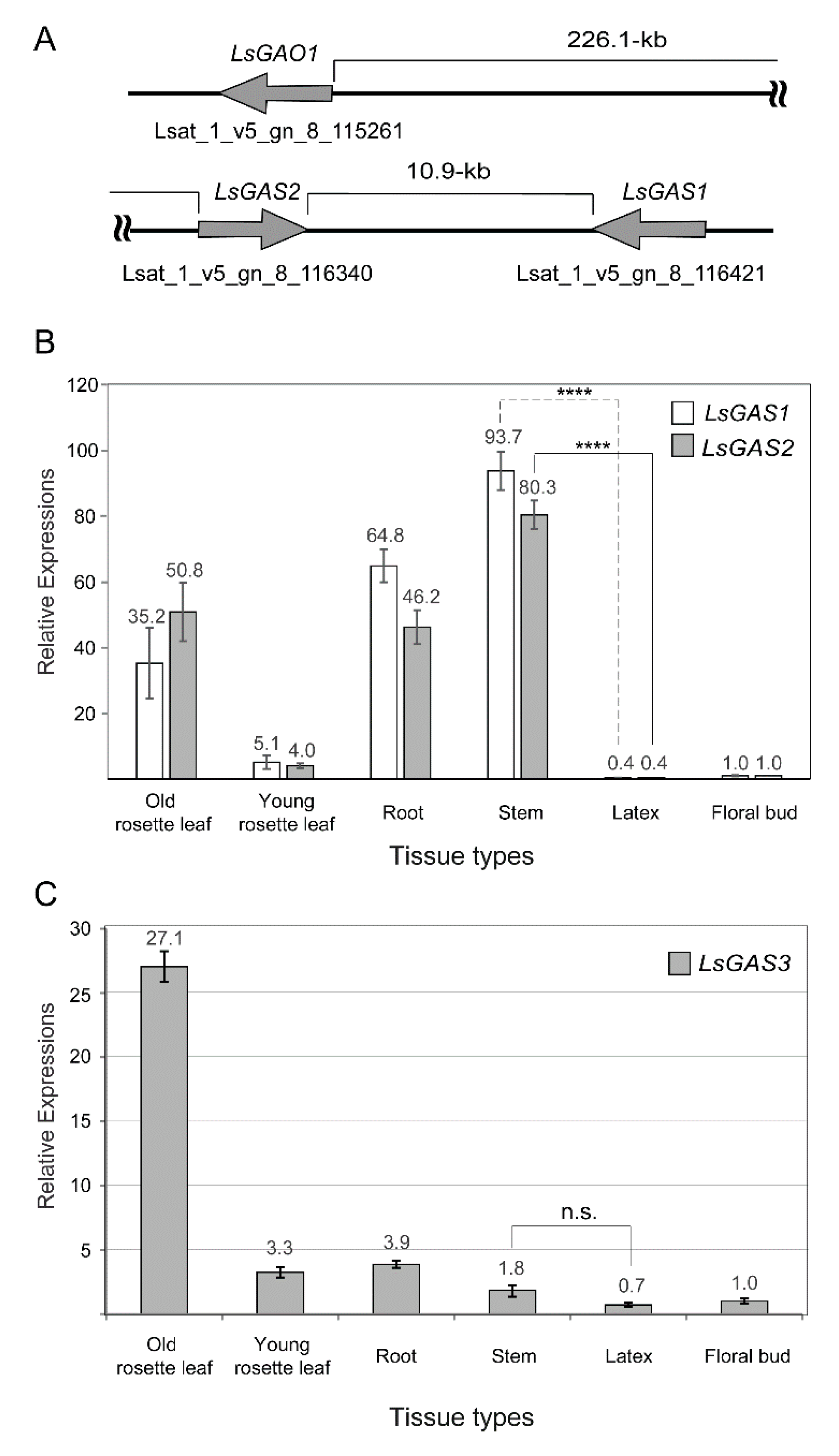
2.1. Assessing Relative LsGAS Transcripts by qPCR Analysis
2.2. Assessing LsGAS Transcription with Targeted RNA-Seq Analysis
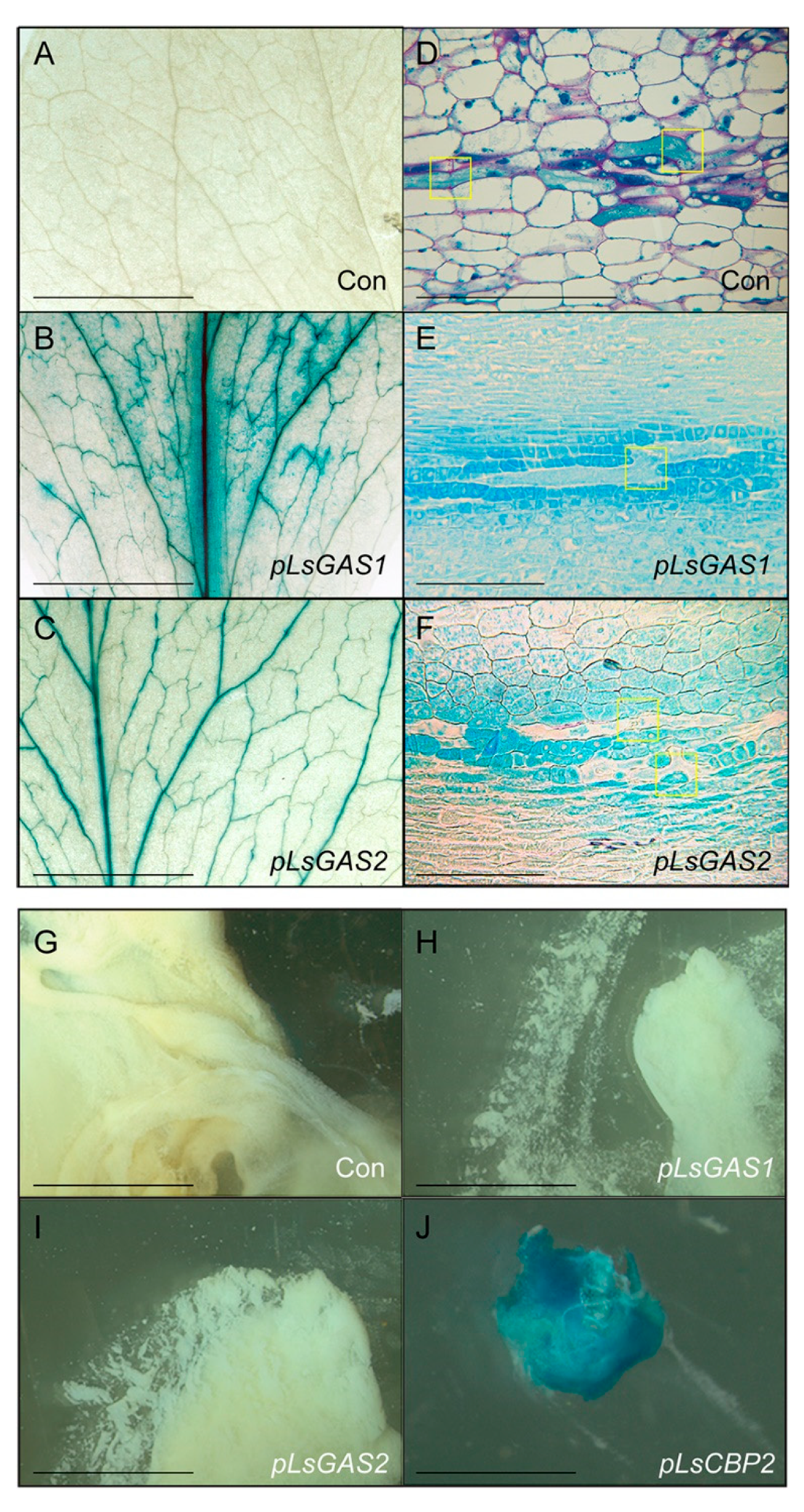
2.3. Characterizing GAS1/2 Promoters with GUS-Fusion Constructs
3. Discussion
4. Materials and Methods
4.1. Plant Growth and DNA Isolation
4.2. Quantitative Real-Time PCR
4.3. RNA-Seq Analysis
4.4. Plasmid Construction
4.5. Generating Transgenic Plants, Histochemical GUS Staining, and Microsections
4.6. Metabolite Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- United Nations, Food and Agriculture Organization of the United Nations. 2020. Available online: http://www.fao.org/faostat/en/#data/QCL (accessed on 10 February 2021).
- Sessa, R.A. Metabolite profiling of sesquiterpene lactones from Lactuca species: Major latexcomponents are novel oxalate and sulfate conjugates of lactucin and its derivatives. J. Biol. Chem. 2000, 275, 26877–26884. [Google Scholar] [CrossRef]
- Bushman, B.S.; Scholte, A.A.; Cornish, K.; Scott, D.J.; Brichta, J.L.; Vederas, J.C.; Ochoa, O.; Michelmore, R.W.; Shintani, D.K.; Knapp, S.J. Identification and comparison of natural rubber from two Lactuca species. Phytochemistry 2006, 67, 2590–2596. [Google Scholar] [CrossRef] [PubMed]
- Beilen, J.B.V.; Poirier, Y. Establishment of new crops for the production of natural rubber. Trends Biotechnol. 2007, 25, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Hehner, S.P.; Hofmann, T.G.; Dröge, W.; Schmitz, M.L. The antiinflammatory sesquiterpene lactone parthenolide inhibits NF-kappa B by targeting the I kappa B kinase complex. J. Immunol. 1999, 163, 5617–5623. [Google Scholar] [PubMed]
- Hagel, J.M.; Yeung, E.C.; Facchini, P.J. Got milk? The secret life of laticifers. Trends Plant Sci. 2008, 13, 631–639. [Google Scholar] [CrossRef]
- Cornish, K. Alternative Natural Rubber Crops: Why Should We Care? Technol. Innov. 2017, 18, 244–255. [Google Scholar] [CrossRef]
- Qu, Y.; Chakrabarty, R.; Tran, H.T.; Kwon, E.-J.G.; Kwon, M.; Nguyen, T.-D.; Ro, D.-K. A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 2015, 290, 1898–1914. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarty, R.; Qu, Y.; Ro, D.-K. Silencing the lettuce homologs of small rubber particle protein does not influence natural rubber biosynthesis in lettuce (Lactuca sativa) This contribution is in honor of Professor Vincenzo de Luca’s 60th birthday. Phytochemistry 2015, 113, 121–129. [Google Scholar] [CrossRef]
- Reyes-Chin-Wo, S.; Wang, Z.; Yang, X.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.; Froenicke, L.; Lavelle, D.O.; Truco, M.-J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef]
- Epping, J.; Deenen, N.V.; Niephaus, E.; Stolze, A.; Fricke, J.; Huber, C.; Eisenreich, W.; Twyman, R.M.; Prüfer, D.; Schulze, C. Gronover, A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants 2015, 1, 15048. [Google Scholar] [CrossRef]
- Lakusta, A.M.; Kwon, M.; Kwon, E.-J.G.; Stonebloom, S.; Scheller, H.V.; Ro, D.-K. Molecular studies of the protein complexes involving cis-prenyltransferase in guayule (Parthenium argentatum), an alternative rubber-producing plant. Front. Plant Sci. 2019, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Barnes, E.K.; Kwon, M.; Hodgins, C.L.; Qu, Y.; Kim, S.-W.; Yeung, E.C.; Ro, D.-K. The promoter sequences of lettuce cis-prenyltransferase and its binding protein specify gene expression in laticifers. Planta 2021, 253, 51. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.D.; Park, E.J.; Gao, N.; Kuo, A.; Rush, J.S.; Waechter, C.J.; Lehrman, M.A.; Sessa, W.C. Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. EMBO J. 2011, 30, 2490–2500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasher, M.I.; Surmacz, L.; Leong, B.; Pitcher, J.; Swiezewska, E.; Pichersky, E.; Akhtar, T.A. A two-component enzyme complex is required for dolichol biosynthesis in tomato. Plant J. 2015, 82, 903–914. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.; Kwon, E.-J.G.; Ro, D.-K. cis-Prenyltransferase Polymer Analysis from a Natural Rubber Perspective. Methods Enzymol. 2016, 576, 121–145. [Google Scholar]
- Sato, M.; Fujisaki, S.; Sato, K.; Nishimura, Y.; Nakano, A. Yeast Saccharomyces cerevisiae has two cis-prenyltransferases with different properties and localizations. Implication for their distinct physiological roles in dolichol synthesis. Genes Cells 2001, 6, 495–506. [Google Scholar] [CrossRef]
- Kato, J.J.; Fujisaki, S.; Nakajima, K.; Nishimura, Y.; Sato, M.; Nakano, A. The Escherichia coli homologue of yeast RER2, a key enzyme of dolichol synthesis, is essential for carrier lipid formation in bacterial cell wall synthesis. J. Bacteriol. 1999, 181, 2733–2738. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.T.; Göpfert, J.C.; Ikezawa, N.; MacNevin, G.; Kathiresan, M.; Conrad, J.; Spring, O.; Ro, D.-K. Biochemical conservation and evolution of germacrene A oxidase in Asteraceae. J. Biol. Chem. 2010, 285, 16588–16598. [Google Scholar] [CrossRef] [Green Version]
- Ikezawa, N.; Göpfert, J.C.; Nguyen, D.T.; Kim, S.-U.; O’Maille, P.E.; Spring, O.; Ro, D.-K. Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio- and stereoselective hydroxylations in sesquiterpene lactone metabolism. J. Biol. Chem. 2011, 286, 21601–21611. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.T.; Kwon, M.; Kim, S.-U.; Fischer, C.; Ro, D.-K. Catalytic Plasticity of Germacrene A Oxidase Underlies Sesquiterpene Lactone Diversification. Plant Physiol. 2019, 181, 945–960. [Google Scholar] [CrossRef] [Green Version]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Yeung, E.C.; Chan, C.K.W. Glycol methacrylate: The art of embedding and serial sectioning. Botany 2015, 93, 1–8. [Google Scholar] [CrossRef]
- Bennett, M.H.; Mansfield, J.W.; Lewis, M.J.; Beale, M.H. Cloning and expression of sesquiterpene synthase genes from lettuce (Lactuca sativa L.). Phytochemistry 2002, 60, 255–261. [Google Scholar] [CrossRef]
- Bouwmeester, H.J.; Kodde, J.; Verstappen, F.W.A.; Altug, I.G.; Kraker, J.-W.D.; Wallaart, T.E. Isolation and characterization of two germacrene A synthase cDNA clones from chicory. Plant Physiol. 2002, 129, 134–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, J.L.; Burke, I.C.; Neff, M.M. Genetic and biochemical evaluation of natural rubber from Eastern Washington prickly lettuce (Lactuca serriola L.). J. Agric. Food Chem. 2015, 63, 593–602. [Google Scholar] [CrossRef]
- Weid, M.; Ziegler, J.; Kutchan, T.M. The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, Papaver somniferum. Proc. Natl. Acad. Sci. USA 2004, 101, 13957–13962. [Google Scholar] [CrossRef] [Green Version]
- Uchida, H.; Yamashita, H.; Kajikawa, M.; Ohyama, K.; Nakayachi, O.; Sugiyama, R.; Yamato, K.T.; Muranaka, T.; Fukuzawa, H.; Takemura, M.; et al. Cloning and characterization of a squalene synthase gene from a petroleum plant, Euphorbia tirucalli L. Planta 2009, 229, 1243–1252. [Google Scholar] [CrossRef]
| Target Genes | 1 Metabolic Products | Stem— 2 Mapped Read Number | Latex— Mapped Read Number | 4 Fold (Stem/ Latex) |
|---|---|---|---|---|
| LsGAS1 | STL | 5 311.5 ± 95.5 | 2.6 ± 1.1 | ** 119.81 |
| LsGAS2 | STL | 521.7 ± 176.6 | 5.0 ± 1.3 | * 104.34 |
| LsGAS3 | STL | 7.5 ± 8.9 | 0.1 ± 0.2 | 75.00 |
| LsGAO1 | STL | 1375.1 ± 343.3 | 8.5 ± 2.4 | ** 161.78 |
| LsGAO2 | STL | 9.5 ± 4.1 | 92.5 ± 23.4 | ** 0.10 |
| LsCOS | STL | 746.9 ± 124.4 | 72.2 ± 29.4 | * 10.34 |
| LsCPT3 | NR | 312.4 ± 38.7 | 2251.7 ± 259.3 | ** 0.14 |
| LsCBP2 | NR | 337.8 ± 51.5 | 2664.2 ± 423.0 | ** 0.13 |
| LsCPT1 | DHDD | 17.7 ± 1.1 | 12.6 ± 1.1 | ** 1.40 |
| LsCBP1 | DHDD | 9.3 ± 0.8 | 9.7 ± 0.6 | 0.96 |
| 3 LsFPS1 | FPP | 528.8 ± 122.0 | 1537.1 ± 164.4 | ** 0.34 |
| 3 LsFPS2 | FPP | 95.7 ± 25.7 | 1.0 ± 0.4 | ** 95.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, M.; Hodgins, C.L.; Haslam, T.M.; Roth, S.A.; Nguyen, T.-D.; Yeung, E.C.; Ro, D.-K. Germacrene A Synthases for Sesquiterpene Lactone Biosynthesis Are Expressed in Vascular Parenchyma Cells Neighboring Laticifers in Lettuce. Plants 2022, 11, 1192. https://doi.org/10.3390/plants11091192
Kwon M, Hodgins CL, Haslam TM, Roth SA, Nguyen T-D, Yeung EC, Ro D-K. Germacrene A Synthases for Sesquiterpene Lactone Biosynthesis Are Expressed in Vascular Parenchyma Cells Neighboring Laticifers in Lettuce. Plants. 2022; 11(9):1192. https://doi.org/10.3390/plants11091192
Chicago/Turabian StyleKwon, Moonhyuk, Connor L. Hodgins, Tegan M. Haslam, Susan A. Roth, Trinh-Don Nguyen, Edward C. Yeung, and Dae-Kyun Ro. 2022. "Germacrene A Synthases for Sesquiterpene Lactone Biosynthesis Are Expressed in Vascular Parenchyma Cells Neighboring Laticifers in Lettuce" Plants 11, no. 9: 1192. https://doi.org/10.3390/plants11091192








