Advances in Understanding the Genetic Basis of Fatty Acids Biosynthesis in Perilla: An Update
Abstract
1. Introduction
2. Earlier Identification and Cloning of Fatty Acid Encoding Gene in Perilla
3. Transcriptomics Sheds Lights into Key Master Player Enzymes of Perilla Fatty Acid Biosynthesis
4. Whole-Genome-Driven Fatty Acid Genes Discovery
5. Concluding Remarks and Outlook
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nitta, M.; Lee, J.K.; Kang, C.W.; Katsuta, M.; Yasumoto, S.; Liu, D.; Nagamine, T.; Ohnishi, O. The Distribution of Perilla Species. Genet. Resour. Crop Evol. 2005, 52, 797–804. [Google Scholar] [CrossRef]
- Nitta, M.; Lee, J.K.; Ohnishi, O. Asian Perilla crops and their weedy forms: Their cultivation, utilization and genetic relationships. Econ. Bot. 2003, 57, 245–253. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, Q.; Leng, L.; Zhang, D.; Chen, S.; Shi, Y.; Ning, Z.; Chen, S. Incipient diploidization of the medicinal plant Perilla within 10,000 years. Nat. Commun. 2021, 12, 5508. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.M. Ethnomedicinal, phytochemical and pharmacological investigations of Perilla frutescens (L.) Britt. Molecules 2019, 24, 102. [Google Scholar] [CrossRef]
- Banno, N.; Akihisa, T.; Tokuda, H.; Yasukawa, K.; Higashihara, H.; Ukiya, M.; Watanabe, K.; Kimura, Y.; Hasegawa, J.I.; Nishino, H. Triterpene acids from the leaves of Perilla frutescens and their anti-inflammatory and antitumor-promoting effects. Biosci. Biotechnol. Biochem. 2004, 68, 85–90. [Google Scholar] [CrossRef]
- Narisawa, T.; Takahashi, M.; Kotanagi, H.; Kusaka, H.; Yamazaki, Y.; Koyama, H.; Fukaura, Y.; Nishizawa, Y.; Kotsugai, M.; Isoda, Y.; et al. Inhibitory Effect of Dietary Perilla Oil Rich in the n-3 Polyunsaturated Fatty Acid α-Linolenic Acid on Colon Carcinogenesis in Rats. Jpn. J. Cancer Res. 1991, 82, 1089–1096. [Google Scholar] [CrossRef]
- Lin, C.S.; Kuo, C.L.; Wang, J.P.; Cheng, J.S.; Huang, Z.W.; Chen, C.F. Growth inhibitory and apoptosis inducing effect of Perilla frutescens extract on human hepatoma HepG2 cells. J. Ethnopharmacol. 2007, 112, 557–567. [Google Scholar] [CrossRef]
- Wijendran, V.; Hayes, K.C. Dietary n-6 and n-3 fatty acid balance and cardiovascular health. Annu. Rev. Nutr. 2004, 24, 597–615. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, H.; Hu, M.; Wang, J.; Xia, H.; Yang, X.; Yang, L.; Sun, G. Perilla Oil Supplementation Improves Hypertriglyceridemia and Gut Dysbiosis in Diabetic KKAy Mice. Mol. Nutr. Food Res. 2018, 62, 1800299. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, S.; Li, W.; Ma, L.; Ding, M.; Li, R.; Liu, Y. High-fat diet from perilla oil induces insulin resistance despite lower serum lipids and increases hepatic fatty acid oxidation in rats. Lipids Health Dis. 2014, 13, 15. [Google Scholar] [CrossRef]
- Paradee, N.; Utama-ang, N.; Uthaipibull, C.; Porter, J.B.; Garbowski, M.W.; Srichairatanakool, S. Extracts of Thai Perilla frutescens nutlets attenuate tumour necrosis factor-α-activated generation of microparticles, ICAM-1 and IL-6 in human endothelial cells. Biosci. Rep. 2020, 40, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, T.; Yasuda, T.; Ueda, J.; Ohsawa, K. Antidepressant-like effects of apigenin and 2,4,5-trimethoxycinnamic acid from Perilla frutescens in the forced swimming test. Biol. Pharm. Bull. 2003, 26, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Tsuji, M.; Inazu, M.; Egashira, T.; Matsumiya, T. Rosmarinic acid and caffeic acid produce antidepressive-like effect in the forced swimming test in mice. Eur. J. Pharmacol. 2002, 449, 261–267. [Google Scholar] [CrossRef]
- Takeda, H.; Tsuji, M.; Miyamoto, J.; Matsumiya, T. Rosmarinic acid and caffeic acid reduce the defensive freezing behavior of mice exposed to conditioned fear stress. Psychopharmacology 2002, 164, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Kamalashiran, C.; Pattaraarchachai, J.; Muengtaweepongsa, S. Feasibility and Safety of Perilla Seed Oil as an Additional Antioxidative Therapy in Patients with Mild to Moderate Dementia. J. Aging Res. 2018, 2018, 5302105. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Gou, Z.; Fan, Q.; Li, L.; Lin, X.; Wang, Y.; Jiang, S.; Jiang, Z. Effects of dietary perilla seed oil supplementation on lipid metabolism, meat quality, and fatty acid profiles in Yellow-feathered chickens. Poult. Sci. 2019, 98, 5714–5723. [Google Scholar] [CrossRef] [PubMed]
- Peiretti, P.G.; Gasco, L.; Brugiapaglia, A.; Gai, F. Effects of perilla (Perilla frutescens L.) seeds supplementation on performance, carcass characteristics, meat quality and fatty acid composition of rabbits. Livest. Sci. 2011, 138, 118–124. [Google Scholar] [CrossRef]
- Chung, C.-H.; Kim, J.-L.; Lee, Y.-C.; Choi, Y.-L. Cloning and Characterization of a Seed-Specific -3 Fatty Acid Desaturase cDNA from Perilla frutescens. Plant Cell Physiol. 1999, 40, 114–118. [Google Scholar] [CrossRef][Green Version]
- Hwang, S.K.; Hwang, Y.S. Molecular cloning and functional expression of Perilla frutescens 3-ketoacyl-(acyl carrier protein) synthase III. Mol. Cells 2000, 10, 375–381. [Google Scholar] [CrossRef]
- Hwang, S.K.; Kim, K.H.; Hwang, Y.S. Molecular cloning and expression analysis of 3-ketoacyl-ACP synthases in the immature seeds of Perilla frutescens. Mol. Cells 2000, 10, 533–539. [Google Scholar] [CrossRef]
- Lee, K.-R.; Lee, Y.; Kim, E.-H.; Lee, S.-B.; Roh, K.H.; Kim, J.-B.; Kang, H.-C.; Kim, H.U. Functional identification of oleate 12-desaturase and ω-3 fatty acid desaturase genes from Perilla frutescens var. frutescens. Plant Cell Rep. 2016, 35, 2523–2537. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Chen, B.; Win, A.N.; Fu, C.; Lian, J.; Liu, X.; Wang, R.; Zhang, X.; Chai, Y. Omega-3 fatty acid desaturase gene family from two ω-3 sources, Salvia hispanica and Perilla frutescens: Cloning, characterization and expression. PLoS ONE 2018, 13, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.N.; Hao, Y.J.; Lu, J.X.; Bai, H.Y.; Guan, L.; Zhang, T. Transcriptomic analysis of Perilla frutescens seed to insight into the biosynthesis and metabolic of unsaturated fatty acids. BMC Genom. 2018, 19, 213. [Google Scholar] [CrossRef]
- Zhang, T.; Song, C.; Song, L.; Shang, Z.; Yang, S.; Zhang, D.; Sun, W.; Shen, Q.; Zhao, D. RNA sequencing and coexpression analysis reveal key genes involved in α-linolenic acid biosynthesis in Perilla frutescens seed. Int. J. Mol. Sci. 2017, 18, 2433. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Lee, K.R.; Shim, D.; Lee, J.H.; Chen, G.Q.; Hwang, S. Transcriptome analysis and identification of genes associated with ω-3 fatty acid biosynthesis in Perilla frutescens (L.) var. frutescens. BMC Genom. 2016, 17, 474. [Google Scholar] [CrossRef]
- Fukushima, A.; Nakamura, M.; Suzuki, H.; Saito, K.; Yamazaki, M. High-throughput sequencing and de novo assembly of red and green forms of the Perilla frutescens var. crispa transcriptome. PLoS ONE 2015, 10, e0129154. [Google Scholar] [CrossRef]
- Duan, W.; Shi-Mei, Y.; Zhi-Wei, S.; Jing, X.; De-Gang, Z.; Hong-Bin, W.; Qi, S. Genome-Wide Analysis of the Fatty Acid Desaturase Gene Family Reveals the Key Role of PfFAD3 in α-Linolenic Acid Biosynthesis in Perilla Seeds. Front. Genet. 2021, 12, 735862. [Google Scholar] [CrossRef]
- Liping, W.; Shen, W.; Kazachkov, M.; Chen, G.; Chen, Q.; Carlsson, A.S.; Stymne, S.; Weselake, R.J.; Zou, J. Metabolic interactions between the lands cycle and the kennedy pathway of glycerolipid synthesis in arabidopsis developing seeds. Plant Cell 2012, 24, 4652–4669. [Google Scholar] [CrossRef]
- Bates, P.D.; Fatihi, A.; Snapp, A.R.; Carlsson, A.S.; Browse, J.; Lu, C. Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol. 2012, 160, 1530–1539. [Google Scholar] [CrossRef]
- Konishi, T.; Shinohara, K.; Yamada, K.; Sasaki, Y. Acetyl-CoA Carboxylase in Higher Plants: Most Plants Other Than Gramineae Have Both the Prokaryotic and the Eukaryotic Forms of This Enzyme. Plant Cell Physiol. 1996, 37, 117–122. [Google Scholar] [CrossRef]
- Jung, S.H.; Kim, R.J.; Kim, K.J.; Lee, D.H.; Suh, M.C. Plastidial and mitochondrial malonyl CoA-ACP malonyltransferase is essential for cell division and its overexpression increases storage oil content. Plant Cell Physiol. 2019, 60, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Munz, J.; Cass, C.; Zienkiewicz, A.; Kong, Q.; Ma, W.; Sanjaya, S.; Sedbrook, J.C.; Benning, C. Ectopic expression of WRI1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol. 2015, 169, 1836–1847. [Google Scholar] [CrossRef]
- An, D.; Kim, H.; Ju, S.; Go, Y.S.; Kim, H.U.; Suh, M.C. Expression of Camelina WRINKLED1 Isoforms Rescue the Seed Phenotype of the Arabidopsis wri1 Mutant and Increase the Triacylglycerol Content in Tobacco Leaves. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Grimberg, Å.; Carlsson, A.S.; Marttila, S.; Bhalerao, R.; Hofvander, P. Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 2015, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Ye, R.; Gao, L.; Zhang, L.; Wang, R.; Mao, T.; Zheng, Y.; Li, D.; Lin, Y. Characterization and Ectopic Expression of CoWRI1, an AP2/EREBP Domain-Containing Transcription Factor from Coconut (Cocos nucifera L.) Endosperm, Changes the Seeds Oil Content in Transgenic Arabidopsis thaliana and Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Arondel, V.; Kilaru, A.; Bates, P.D.; Thrower, N.A.; Benning, C.; Ohlrogge, J.B. WRINKLED1, A Ubiquitous Regulator in Oil Accumulating Tissues from Arabidopsis Embryos to Oil Palm Mesocarp. PLoS ONE 2013, 8, e68887. [Google Scholar] [CrossRef]
- Yang, H.; Yu, C.; Yan, J.; Wang, X.; Chen, F.; Zhao, Y.; Wei, W. Overexpression of the Jatropha curcas JcERF1 gene coding an AP2/ERF-Type transcription factor increases tolerance to salt in transgenic tobacco. Biochemistry 2014, 79, 1226–1236. [Google Scholar] [CrossRef]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoët, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef]
- Fukuda, N.; Ikawa, Y.; Aoyagi, T.; Kozaki, A. Expression of the genes coding for plastidic acetyl-CoA carboxylase subunits is regulated by a location-sensitive transcription factor binding site. Plant Mol. Biol. 2013, 82, 473–483. [Google Scholar] [CrossRef]
- Kazaz, S.; Barthole, G.; Domergue, F.; Ettaki, H.; To, A.; Vasselon, D.; de Vos, D.; Belcram, K.; Lepiniec, L.; Baud, S. Differential activation of partially redundant Δ9 stearoyl-ACP desaturase genes is critical for omega-9 monounsaturated fatty acid biosynthesis during seed development in arabidopsis. Plant Cell 2020, 32, 3613–3637. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shao, J.; Tang, S.; Shen, Q.; Wang, T.; Chen, W.; Hong, Y. Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front. Plant Sci. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhai, Z.; Kuczynski, K.; Keereetaweep, J.; Schwender, J.; Shanklin, J. Wrinkled1 regulates biotin attachment domain-containing proteins that inhibit fatty acid synthesis. Plant Physiol. 2019, 181, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Maeo, K.; Tokuda, T.; Ayame, A.; Mitsui, N.; Kawai, T.; Tsukagoshi, H.; Ishiguro, S.; Nakamura, K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009, 60, 476–487. [Google Scholar] [CrossRef]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.; Py, C.; Gendrot, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef]
- Ruuska, S.A.; Girke, T.; Benning, C.; Ohlrogge, J.B. Contrapuntal networks of gene expression during Arabidopsis seed fillingW. Plant Cell 2002, 14, 1191–1206. [Google Scholar] [CrossRef]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef]
- Browse, J.; McConn, M.; James, D.; Miquel, M. Mutants of Arabidopsis deficient in the synthesis of α-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 1993, 268, 16345–16351. [Google Scholar] [CrossRef]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [CrossRef]
- Speerling, P.; Heinz, E. Isomeric sn-1-octadecenyl and sn-2-octadecenyl analogues of lysophosphatidylcholine as substrates for acylation and desaturation by plant microsomal membranes. Eur. J. Biochem. 1993, 213, 965–971. [Google Scholar] [CrossRef]
- Chen, J.; Tan, R.K.; Guo, X.J.; Fu, Z.L.; Wang, Z.; Zhang, Z.Y.; Tan, X.L. Transcriptome analysis comparison of lipid biosynthesis in the leaves and developing seeds of Brassica napus. PLoS ONE 2015, 10, e0130067. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kazachkov, M.; Shen, W.; Bai, M.; Wu, H.; Zou, J. Deciphering the roles of Arabidopsis LPCAT and PAH in phosphatidylcholine homeostasis and pathway coordination for chloroplast lipid synthesis. Plant J. 2014, 80, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pérez, A.J.; Santos-Pereira, J.M.; Martins-Noguerol, R.; DeAndrés-Gil, C.; Troncoso-Ponce, M.A.; Venegas-Calerón, M.; Sánchez, R.; Garcés, R.; Salas, J.J.; Tena, J.J.; et al. Genome-Wide Mapping of Histone H3 Lysine 4 Trimethylation (H3K4me3) and Its Involvement in Fatty Acid Biosynthesis in Sunflower Developing Seeds. Plants 2021, 10, 706. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, C.; Liu, X.; Cheng, J.; Li, S.; Zhu, J.K.; Gong, Z. Peroxisomal β-oxidation regulates histone acetylation and DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 10576–10585. [Google Scholar] [CrossRef]
- Yamazaki, M.; Kobayashi, M.; Saito, K. Transformation of Perill frutescens var. crispa Using an Agrobacterium-Ri Binary Vector System. Plant Biotechnol. 1997, 14, 169–173. [Google Scholar] [CrossRef][Green Version]
- Kim, K.-H.; Lee, Y.-H.; Kim, D.; Park, Y.-H.; Lee, J.-Y.; Hwang, Y.-S.; Kim, Y.-H. Agrobacterium-mediated genetic transformation of Perilla frutescens. Plant Cell Rep. 2004, 23, 386–390. [Google Scholar] [CrossRef]
- Dolgin, E. T-cell vaccines could top up immunity to COVID, as variants loom large. Nat. Biotechnol. 2022, 40, 3–4. [Google Scholar] [CrossRef]
- Schwartz, C.; Lenderts, B.; Feigenbutz, L.; Barone, P.; Llaca, V.; Fengler, K.; Svitashev, S. CRISPR–Cas9-mediated 75.5-Mb inversion in maize. Nat. Plants 2020, 6, 1427–1431. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Mark Cigan, A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. [Google Scholar] [CrossRef]
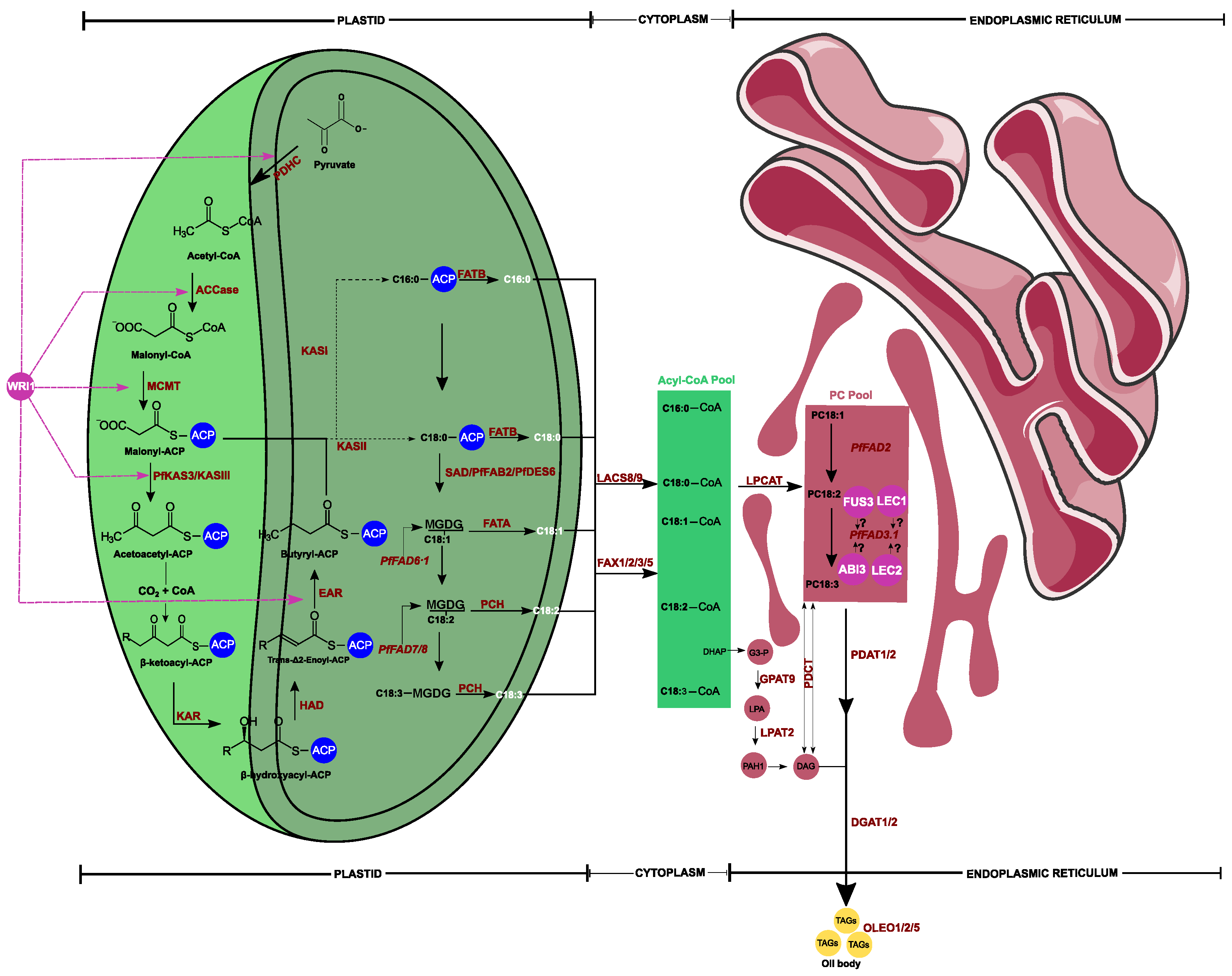
| Enzyme ID | Enzyme Name | GeneID | Homologous | Pathways Involved | Field of Study | References | ||
|---|---|---|---|---|---|---|---|---|
| PF40 * | Dayudeulkkae ** | PC *** | A. Thaliana | |||||
| PDH(E1α) | Pyruvate Dehydrogenase E1 Subunit Alpha 1 | Locus_2112 | AT1G01090.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| PDH(E1ß) | Pyruvate Dehydrogenase E1 Subunit beta 1 | Locus_25208 | AT2G34590.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| EMB3003(E2) | Pyruvate dehydrogenase e2 component (dihydrolipoamide acetyltransferase) | Locus_33306 | AT1G34430.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| LTA2 (E2) | Plastid E2 Subunit of Pyruvate Decarboxylase, PLE2 | Locus_5104 | AT3G25860.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| LPD1 (E3) | Lipoamide dehydrogenase | Locus_7407 | AT3G16950.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| α-CTa | Alpha-carboxyltransferase Isoform a | Locus_8492 | AT2G38040.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| α-CTb | Apha-carboxyltransferase Isoform b | Locus_2178 | AT2G38040.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| ß-CT | Beta-carboxyltransferase | Locus_53041 | ATCG00500.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| BC | Biotin carboxylase | Locus_22078 | AT5G35360.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| BCCP1 | Biotin carboxyl carrier protein of acetyl-CoA carboxylase 1 | Locus_29162 | AT5G16390.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| BCCP2 | Biotin carboxyl carrier protein of acetyl-CoA carboxylase 2 | Locus_17340 | AT5G15530.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| MCMT | Malonyl-CoA ACP transacylase | Locus_14579 | AT2G30200.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| KASIII | 3-Ketoacyl-ACP synthase | Locus_10821 | AT1G62640.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| KAR | 3-ketoacyl-ACP reductase | Locus_1445 | AT1G24360.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| HAD | 3-hydroxyacyl-ACP dyhydratase | Locus_19332 | AT5G10160.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| EAR | 2-enoyl-ACP reductase | Locus_25443 | AT2G05990.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| FATA | Fatty acyl-ACP thioesterase A | Locus_29919 | AT3G25110.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| FATB | Fatty acyl-ACP thioesterase B | Locus_6603 | AT1G08510.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| FAB2 | Fatty acid biosynthesis2 | Locus_13564 | AT2G43710.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| DES6 | Stearoyl-acyl carrier protein desaturase | Locus_9486 | AT1G43800.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| KASI | Ketoacyl-ACP Synthase I | Locus_26341 | AT5G46290.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| KASII | Ketoacyl-ACP Synthase II | Locus_1373 | AT1G74960.1 | FA de novo biosynthesis and export from plastid | Transcriptomics | [25] | ||
| LACS8 | Long-chain acyl-CoA synthetase 8 | chr07_36292788_36299197 chr19_22302145_22308533 | Locus_3838 | chr06_37084362_37090768 | AT2G04350.1 | FA de novo biosynthesis and export from plastid | Genome Assembly, Transcriptomics | [3,25] |
| LACS9 | Long-chain acyl-CoA synthetase 9 | chr03_70622879_70627324 chr09_58852417_58856892 chr01_02424545_02428997 | Locus_23636 | chr01_02424545_02428997 | AT1G77590.1 | FA de novo biosynthesis and export from plastid | Genome Assembly, Transcriptomics | [3,25] |
| FAX1 | Fatty acid export 1 | chr05_24282740_24284950 chr01_71691539_71693779 | chr02_42552603_42554830 | FA de novo biosynthesis and export from plastid | [25] | |||
| FAX2 | Fatty acid export 2 | chr07_10626150_10628000 | chr06_11381976_11383822 | FA de novo biosynthesis and export from plastid | [25] | |||
| FAX3 | Fatty acid export 3 | chr04_00857340_00859552 | chr03_67540865_67543081 | FA de novo biosynthesis and export from plastid | [25] | |||
| FAX5 | Fatty acid export 5 | chr04_65527957_65529911 chr07_22534802_22537586 chr06_00746938_00748860 chr19_10735560_10738363 | chr03_02347871_02349825 chr06_23562111_23564893 | FA de novo biosynthesis and export from plastid | [25] | |||
| FAD2 | Omega-6 fatty acid desaturase | chr12_56933298_56934446 chr11_05592060_05593208 chr11_05575254_05576393 | Locus_733 | chr08_55538081_55539229 | AT3G12120.1 | Acyl editing of phospatidylcholine | Genome Assembly, Transcriptomics | [3,25] |
| chr12_56948107_56949167 | chr08_55558209_55559348 | |||||||
| FAD3 | Omega-3 fatty acid desaturase | chr12_04645208_04647776 chr11_54194712_54197265 | Locus_22029 | chr08_04030082_04032640 | AT2G29980.1 | Acyl editing of phospatidylcholine | Genome Assembly, Transcriptomics | [3,25] |
| FAD8 | Omega-8 fatty acid desaturase | Locus_5107 | AT5G05580.2 | Acyl editing of phospatidylcholine | Transcriptomics | [25] | ||
| GPAT9 | Glycerol-3-phosphate acyltransferase 9 | chr12_33733527_33737891 chr11_26255533_26259881 | Locus_10180 | chr08_33038421_33042132 | AT5G60620.1 | Acyl-CoA-dependent TAG synthesis in Kennedy pathway | Genome Assembly, Transcriptomics | [3,25] |
| LPAT2 | 1-acyl-sn-glycerol-3-phosphate acyltransferase 2 | chr05_23583386_23588593 chr05_34400913_34404444 chr01_72114246_72119454 | Locus_6587 | chr02_43313059_43318262 chr02_32585727_32589258 | AT3G57650.1 | Acyl-CoA-dependent TAG synthesis in Kennedy pathway | Genome Assembly, Transcriptomics | [3,25] |
| PAH1 | Phenylalanine hydrolase 1 | chr01_61567423_61570965 chr14_08597119_08602056 chr15_37103964_37108907 chr03_61656532_61661875 chr18_09154357_09159306 chr17_34575710_34580664 chr09_50343045_50349360 | chr10_43830659_43835596 chr01_11516392_11522733 | Acyl-CoA-dependent TAG synthesis in Kennedy pathway | ||||
| DGAT1 | Diacylglycerol O-acyltransferase 1 | chr01_09730655_09741367 chr01_48275733_48286173 | Locus_14696 | chr05_08797620_08808333 | AT2G19450.1 | Acyl-CoA-dependent TAG synthesis in Kennedy pathway | Genome Assembly, Transcriptomics | [3,25] |
| DGAT2 | Diacylglycerol O-acyltransferase 2 | chr14_26782964_26787941 chr18_25811826_25816791 | Locus_12629 | chr10_25785382_25790335 | AT3G51520.1 | Acyl-CoA-dependent TAG synthesis in Kennedy pathway | Genome Assembly, Transcriptomics | [3,25] |
| DGAT3 | Diacylglycerol O-acyltransferase 3 | Locus_1560 | AT1G48300.1 | Acyl-CoA-dependent TAG synthesis in Kennedy pathway | Transcriptomics | [25] | ||
| LPCAT | Lysophosphatidylcholine acyltransferase | chr01_06996630_07001595 chr05_56678891_56685081 chr01_03079195_03084058 chr07_53028425_53034567 chr01_43224061_43229071 chr02_66141068_66147271 chr02_04634020_04638876 chr19_35211932_35217537 | Locus_43749 | PC00000058_00436672_00441634 chr02_10454190_10460391 chr05_03185967_03190829 chr06_54113419_54119561 | AT1G12640.1 | PC-mediated TAG synthesis | Transcriptomics | [3,25] |
| CPT1 | Diacylglycerol cholinephosphotransferase | Locus_7821 | AT1G13560.1 | PC-mediated TAG synthesis | Transcriptomics | [25] | ||
| CPT2 | Diacylglycerol cholinephosphotransferase | Locus_22567 | AT3G25585.1 | PC-mediated TAG synthesis | Transcriptomics | [25] | ||
| PDAT1 | Phospholipid:diacylglycerol acyltransferase 1 | chr05_44104376_44108847 | Locus_7255 | chr02_22969948_22974420 | AT5G13640.1 | Acyl-CoA independent pathway | Transcriptomics | [3,25] |
| chr03_00447151_00451507 | PC00002899_00154872_00159184 | |||||||
| chr02_52135886_52140327 | ||||||||
| chr09_00376677_00380564 | ||||||||
| PDAT2 | Phospholipid:diacylglycerol acyltransferase 2 | chr05_38922115_38924735 | Locus_29208 | chr02_28050267_28052887 | AT3G44830.1 | Acyl-CoA independent pathway | Transcriptomics | [3,25] |
| chr02_45992086_45994691 | ||||||||
| PDCT | Phosphatidylcholine:diacylglycerol cholinephosphotransferase | chr03_46291224_46293449 chr09_37050943_37053194 | Locus_15867 | chr01_27228085_27230144 | AT3G15820.1 | Acyl-CoA independent pathway | Genome Assembly, Transcriptomics | [3,25] |
| OLEO2 | Oleosin2 | chr15_52133834_52134256 | Locus_31790 | AT5G40420.1 | TAG assembly | Transcriptomics | [3,25] | |
| chr17_50355018_50355440 | chr09_02008310_02008732 | |||||||
| OLEO | Oleosin | chr14_08347244_08347714 | Locus_31788 | chr10_44101965_44102435 | AT3G18570.1 | TAG assembly | Transcriptomics | [3,25] |
| chr18_08871500_08871970 | ||||||||
| OLEO1 | Oleosin1 | chr05_05196095_05196523 | Locus_29266 | chr02_64426568_64426996 | AT4G25140.1 | TAG assembly | Transcriptomics | [3,25] |
| chr01_30156121_30156549 | ||||||||
| OLEO5 | Oleosin5 | chr05_59989345_59989911 | Locus_29276 | AT3G01570.1 | TAG assembly | Transcriptomics | [3,25] | |
| chr05_59997449_59997976 | chr02_07157257_07157823 | |||||||
| chr02_69562819_69563393 | chr02_07149192_07149719 | |||||||
| chr02_69577662_69578195 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, S.-H.; Zoclanclounon, Y.A.B.; Kumar, T.S.; Oh, J.-H.; Lee, J.; Kim, T.-H.; Park, K.Y. Advances in Understanding the Genetic Basis of Fatty Acids Biosynthesis in Perilla: An Update. Plants 2022, 11, 1207. https://doi.org/10.3390/plants11091207
Bae S-H, Zoclanclounon YAB, Kumar TS, Oh J-H, Lee J, Kim T-H, Park KY. Advances in Understanding the Genetic Basis of Fatty Acids Biosynthesis in Perilla: An Update. Plants. 2022; 11(9):1207. https://doi.org/10.3390/plants11091207
Chicago/Turabian StyleBae, Seon-Hwa, Yedomon Ange Bovys Zoclanclounon, Thamilarasan Senthil Kumar, Jae-Hyeon Oh, Jundae Lee, Tae-Ho Kim, and Ki Young Park. 2022. "Advances in Understanding the Genetic Basis of Fatty Acids Biosynthesis in Perilla: An Update" Plants 11, no. 9: 1207. https://doi.org/10.3390/plants11091207
APA StyleBae, S.-H., Zoclanclounon, Y. A. B., Kumar, T. S., Oh, J.-H., Lee, J., Kim, T.-H., & Park, K. Y. (2022). Advances in Understanding the Genetic Basis of Fatty Acids Biosynthesis in Perilla: An Update. Plants, 11(9), 1207. https://doi.org/10.3390/plants11091207









