Adaptive Divergence without Distinct Species Relationships Indicate Early Stage Ecological Speciation in Species of the Rhododendronpseudochrysanthum Complex Endemic to Taiwan
Abstract
:1. Introduction
2. Results
2.1. Genetic Diversity Based on the Total AFLP Variation
2.2. Environmental Heterogeneity
2.3. Genetic Relationships and Clustering Based on the Total AFLP Variation
2.4. Potential Genetic and Epigenetic Outliers Associated with Environmental Variables and the Most Important Environmental Variables Explaining Outlier Variation
2.5. Relative Contribution of IBD and IBE Explaining Outlier Genetic and Epigenetic Variations
3. Discussion
4. Materials and Methods
4.1. Sampling, Genotyping, and Epigenotyping
4.2. Genetic Diversity Based on the Total AFLP Variation
4.3. Environmental Heterogeneity
4.4. AFLP Genetic Clustering and Relationships
4.5. Test for AFLP and MSAP FST Outliers
4.6. AFLP Genetic Differentiation
4.7. Associations of Genetic and Epigenetic Loci with Environmental Variables
4.8. AFLP and MSAP Isolation-by-Environment and Isolation-by-Distance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schluter, D. Evidence for ecological speciation and its alternative. Science 2009, 323, 737–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jump, A.S.; Marchant, R.; Peñuelas, J. Environmental change and the option value of genetic diversity. Trends Plant Sci. 2009, 14, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.J.; Akçakaya, H.R.; Araújo, M.B.; Fordham, D.A.; Martinez-Meyer, E.; Thuiller, W.; Brook, B.W. Dynamics of range margins for metapopulations under climate change. Proc. R. Soc. B 2009, 276, 1415–1420. [Google Scholar] [CrossRef] [Green Version]
- Hargreaves, A.L.; Bailey, S.F.; Laird, R.A. Fitness declines towards range limits and local adaptation to climate affect dispersal evolution during climate-induced range shifts. J. Evol. Biol. 2015, 28, 1489–1501. [Google Scholar] [CrossRef]
- Lenoir, J.; Gégout, J.C.; Marquet, P.A.; de Ruffray, P.; Brisse, H.A. Significant upward shift in plant species optimum elevation during the 20th century. Science 2008, 320, 1768–1771. [Google Scholar] [CrossRef]
- Jump, A.S.; Huang, T.-J.; Chou, C.-H. Rapid altitudinal migration of mountain plants in Taiwan and its implications for high altitude biodiversity. Ecography 2012, 35, 204–210. [Google Scholar] [CrossRef]
- Ackerly, D.D. Community assembly, niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 2003, 164, S165–S184. [Google Scholar] [CrossRef]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-Y.; Liang, B.-K.; Chung, J.-D.; Chang, C.-T.; Hsieh, Y.-C.; Lin, T.-C.; Hwang, S.-Y. Demography of the upward-shifting temperate woody species of the Rhododendron pseudochrysanthum complex and ecologically relevant adaptive divergence in its trailing edge populations. Tree Genet. Genom. 2014, 10, 11–126. [Google Scholar] [CrossRef]
- Dirnböck, T.; Essl, F.; Rabitsch, W. Disproportional risk for habitat loss of high-altitude endemic species under climate change. Glob. Chang. Biol. 2011, 17, 990–996. [Google Scholar] [CrossRef]
- Bennett, S.; Duarte, C.M.; Marbà, N.; Wernberg, T. Integrating within-species variation in thermal physiology into climate change ecology. Phil. Trans. R. Soc. B 2019, 374, 20180550. [Google Scholar] [CrossRef] [Green Version]
- Leimu, R.; Fischer, M. A meta-analysis of local adaptation in plants. PLoS ONE 2008, 3, e4010. [Google Scholar] [CrossRef] [Green Version]
- Holderegger, R.; Herrmann, D.; Poncet, B.; Gugerli, F.; Thuiller, W.; Taberlet, P.; Gielly, L.; Rioux, D.; Brodbeck, S.; Aubert, S.; et al. Land ahead: Using genome scans to identify molecular markers of adaptive relevance. Plant Ecol. Div. 2008, 1, 273–283. [Google Scholar] [CrossRef]
- Richards, C.L.; Bossdorf, O.; Verhoeven, K.J. Understanding natural epigenetic variation. New Phytol. 2010, 187, 562–564. [Google Scholar] [CrossRef]
- Richards, C.L.; Alonso, C.; Becker, C.; Bossdorf, O.; Bucher, E.; Colomé-Tatché, M.; Durka, W.; Engelhardt, J.; Gaspar, B.; Gogol-Döring, A.; et al. Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecol. Lett. 2017, 20, 1576–1590. [Google Scholar] [CrossRef] [Green Version]
- Shih, K.-M.; Chang, C.-T.; Chung, J.-D.; Chiang, Y.-C.; Hwang, S.-Y. Adaptive genetic divergence despite significant isolation-by-distance in populations of Taiwan Cow-tail fir (Keteleeria davidiana var. formosana). Front. Plant Sci. 2018, 9, 92. [Google Scholar] [CrossRef] [Green Version]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van der Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucl. Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Chien, W.-M.; Chang, C.-T.; Chiang, Y.-C.; Hwang, S.-Y. Ecological factors generally not altitude related played main roles in driving potential adaptive evolution at elevational range margin populations of Taiwan incense cedar (Calocedrus formosana). Front. Genet. 2020, 11, 580630. [Google Scholar] [CrossRef]
- Lister, R.; Ecker, J.R. Finding the fifth base: Genome-wide sequencing of cytosine methylation. Genome Res. 2009, 19, 959–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, L.Z.; Xu, C.G.; Saghai Maroof, M.A.; Zhang, Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 1999, 261, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Bazaga, P. Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis. New Phytol. 2010, 187, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-L.; Chen, J.-H.; Tsang, M.-H.; Chung, J.-D.; Chang, C.-T.; Hwang, S.-Y. Influences of environmental and spatial factors on genetic and epigenetic variations in Rhododendron oldhamii (Ericaceae). Tree Genet. Genom. 2015, 11, 823. [Google Scholar] [CrossRef]
- Li, H.-L.; Lu, S.-Y.; Yang, Y.-P.; Tseng, Y.-H. Ericaceae. In Flora of Taiwan, 2nd ed.; Committee of the Flora of Taiwan; Epoch Publishing Co. Ltd.: Taipei, Taiwan, 1998; Volume 4, pp. 17–39. [Google Scholar]
- Chung, J.-D.; Lin, T.-P.; Chen, Y.-L.; Cheng, Y.-P.; Hwang, S.-Y. Phylogeographic study reveals the origin and evolutionary history of a Rhododendron species complex in Taiwan. Mol. Phylogenet. Evol. 2007, 42, 14–24. [Google Scholar] [CrossRef]
- Wang, I.J.; Glor, R.E.; Losos, J.B. Quantifying the roles of ecology and geography in spatial genetic divergence. Ecol. Lett. 2013, 16, 175–182. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Frichot, E.; François, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.; Moulton, V. Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 23, 341–369. [Google Scholar] [CrossRef]
- Foll, M.; Gaggiotti, O. A genome scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef] [Green Version]
- Beaumont, M.A.; Nichols, R.A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B Biol. Sci. 1996, 263, 1619–1626. [Google Scholar]
- Frichot, E.; Schoville, S.D.; Bouchard, G.; François, O. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol. Biol. Evol. 2013, 30, 1687–1699. [Google Scholar] [CrossRef] [Green Version]
- Stucki, S.; Orozco-terWengel, P.; Bruford, M.W.; Colli, L.; Masembe, C.; Negrini, R.; Landguth, E.; Jones, M.R.; Nextgen, C.; Bruford, M.W.; et al. High performance computation of landscape genomic models integrating local indices of spatial association. Mol. Ecol. Resour. 2017, 17, 1072–1089. [Google Scholar] [CrossRef] [Green Version]
- Bürkner, P.-C. Brms: Visualization of Regression Models Using visreg. J. Stat. Softw. 2017, 80, 1–27. [Google Scholar]
- Bürkner, P.-C. Advanced Bayesian Multilevel Modeling with the R Package brms. R J. 2018, 10, 395–411. [Google Scholar] [CrossRef]
- Blanchet, F.G.; Legendre, P.; Borcard, D. Forward selection of explanatory variables. Ecology 2008, 89, 2623–2632. [Google Scholar] [CrossRef]
- Wang, I.J. Examining the full effects of landscape heterogeneity on spatial genetic variation: A multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution 2013, 67, 3403–3411. [Google Scholar] [CrossRef]
- Zhao, B.; Yin, Z.-F.; Xu, M.; Wang, Q.-C. AFLP analysis of genetic variation in wild populations of five Rhododendron species in Qinling Mountain in China. Biochem. System. Ecol. 2012, 45, 198–205. [Google Scholar] [CrossRef]
- Wu, F.Q.; Shen, S.K.; Zhang, Z.J.; Wang, Y.H.; Sun, W.B. Genetic diversity and population structure of an extremely endangered species: The world’s largest Rhododendron. AoB Plants 2014, 7, plu082. [Google Scholar] [CrossRef] [Green Version]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2009; ISBN 9780511808999. [Google Scholar]
- Hirao, A.S.; Kameyama, Y.; Ohara, M.; Isagi, Y.; Kudo, G. Seasonal changes in pollinator activity influence pollen dispersal and seed production of the alpine shrub Rhododendron aureum (Ericaceae). Mol. Ecol. 2006, 15, 1165–1173. [Google Scholar] [CrossRef]
- Ono, A.; Dohzono, I.; Sugawara, T. Bumblebee pollination and reproductive biology of Rhododendron semibarbatum (Ericaceae). J. Plant Res. 2008, 121, 319–327. [Google Scholar] [CrossRef]
- Hirao, A.S. Kinship between parents reduces offspring fitness in a natural population of Rhododendron brachycarpum. Ann. Bot. 2010, 105, 637–646. [Google Scholar] [CrossRef] [Green Version]
- Liew, P.-M.; Chung, N.-J. Vertical migration of forests during the last glacial period in subtropical Taiwan. West Pac. Earth Sci. 2001, 1, 405–414. [Google Scholar]
- Aguilar, R.; Quesada, M.; Ashworth, L.; Herrerias-Diego, Y.; Lobo, J. Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol. Ecol. 2008, 17, 5177–5188. [Google Scholar] [CrossRef] [PubMed]
- Vranckx, G.; Jacquemyn, H.; Muys, B.; Honnay, O. Meta-analysis of susceptibility of woody plants to loss of genetic diversity through habitat fragmentation. Conserv. Biol. 2012, 26, 228–237. [Google Scholar] [CrossRef]
- Charlesworth, B.; Bartolomé, C.; Noël, V. The detection of shared and ancestral polymorphisms. Genet. Res. 2005, 86, 149–157. [Google Scholar] [CrossRef]
- Lu, S.-Y.; Yang, Y.-P. A revision of Rhododendron (Ericaceae) of Taiwan. Bull. Taiwan For. Res. Inst. 1989, 4, 155–166. [Google Scholar]
- Tsai, C.-C.; Huang, S.-C.; Chen, C.-H.; Tseng, Y.-H.; Huang, P.-L.; Tsai, S.-H.; Chou, C.-H. Genetic relationships of Rhododendron (Ericaceae) in Taiwan based on the sequence of the internal transcribed spacer of ribosomal DNA. J. Hort. Sci. Biotech. 2003, 78, 234–240. [Google Scholar] [CrossRef]
- Petit, R.J.; Excoffier, L. Gene flow and species delimitation. Trends Ecol. Evol. 2009, 24, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.R. Biological flora of the British Isles. Rhododendron ponticum L. J. Ecol. 1975, 63, 345–364. [Google Scholar]
- Ng, S.C.; Corlett, R.T. Comparative reproductive biology of the six species of Rhododendron (Ericaceae) in Hong Kong, South China. Can. J. Bot. 2000, 78, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, J.; Lai, L.; Jiang, L.; Zhuang, P.; Zhang, L.; Zheng, Y.; Baskin, J.M.; Baskin, C. Geographic variation in seed traits within and among forty-two species of Rhododendron (Ericaceae) on the Tibetan plateau: Relationships with altitude, habitat, plant height, and phylogeny. Ecol. Evol. 2014, 4, 1913–1923. [Google Scholar] [CrossRef]
- Antonelli, A. Biogeography: Drivers of bioregionalization. Nat. Ecol. Evol. 2017, 1, 0114. [Google Scholar] [CrossRef]
- Shirk, A.J.; Landguth, E.L.; Cushman, S.A. A comparison of individual-based genetic distance metrics for landscape genetics. Mol. Ecol. Resour. 2017, 17, 1308–1317. [Google Scholar] [CrossRef]
- Allen, A.P.; Gillooly, J.F.; Savage, V.M.; Brown, J.H. Kinetic effects of temperature on rates of genetic divergence and speciation. Proc. Natl. Acad. Sci. USA 2006, 103, 9130–9135. [Google Scholar] [CrossRef] [Green Version]
- Strasburg, J.L.; Sherman, N.A.; Wright, K.M.; Moyle, L.C.; Willis, J.H.; Rieseberg, L.H. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Phil. Trans. R. Soc. B 2012, 367, 364–373. [Google Scholar] [CrossRef] [Green Version]
- Lira-Medeiros, C.F.; Parisod, C.; Fernandes, R.A.; Mata, C.S.; Cardoso, M.A.; Ferreira, P.C.G. Epigenetic variation in mangrove plants occurring in contrasting natural environments. PLoS ONE 2010, 5, e10326. [Google Scholar] [CrossRef]
- Latzel, V.; Allan, E.; Silveira, A.B.; Colot, V.; Fischer, M.; Bossdorf, O. Epigenetic diversity increases the productivity and stability of plant populations. Nat. Commu. 2013, 4, 2875. [Google Scholar] [CrossRef]
- Hogg, E.H.; Barr, A.G.; Black, T.A. A simple soil moisture index for representing multi-year drought impacts on aspen productivity in the western Canadian interior. Agric. For. Meteorol. 2013, 178, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Depardieu, C.; Girardin, M.P.; Nadeau, S.; Lenz, P.; Bousquet, J.; Isabel, N. Adaptive genetic variation to drought in a widely distributed conifer suggests a potential for increasing forest resilience in a drying climate. New Phytol. 2020, 227, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Abid, G.; Mingeot, D.; Muhovski, Y.; Mergeai, G.; Aouida, M.; Abdelkarim, S.; Aroua, I.; El Ayed, M.; M’hamdi, M.; Sassi, K.; et al. Analysis of DNA methylation patterns associated with drought stress response in faba bean (Vicia faba L.) using methylation-sensitive amplification polymorphism (MSAP). Environ. Exp. Bot. 2017, 142, 34–44. [Google Scholar] [CrossRef]
- Jackson, P.; Basnayake, J.; Inman-Bamber, G.; Lakshmanan, P.; Natarajan, S.; Stokes, C. Genetic variation in transpiration efficiency and relationships between whole plant and leaf gas exchange measurements in Saccharum spp. and related germplasm. J. Exp. Bot. 2016, 67, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Bayliss, S.L.J.; Mueller, L.O.; Ware, I.M.; Schweitzer, J.A.; Bailey, J.K. Plant genetic variation drives geographic differences in atmosphere-plant-ecosystem feedbacks. Plant-Environ. Interact. 2020, 1, 166–180. [Google Scholar] [CrossRef]
- Fensholt, R.; Sandholt, I.; Rasmussen, M.S. Evaluation of MODIS LAI, fPAR and the relation between fAPAR and NDVI in a semiarid environment using in situ measurements. Remote Sens. Environ. 2004, 91, 490–507. [Google Scholar] [CrossRef]
- Violle, C.; Enquist, B.J.; McGill, B.J.; Jiang, L.; Albert, C.H.; Hulshof, C.; Jung, V.; Messier, J. The return of the variance: Intraspecific variability in community ecology. Trends Ecol. Evol. 2012, 27, 244–252. [Google Scholar] [CrossRef]
- Li, Y.-S.; Chang, C.-T.; Wang, C.-N.; Thomas, P.; Chung, J.-D.; Hwang, S.-Y. The contribution of neutral and environmentally dependent processes in driving population and lineage divergence in Taiwania (Taiwania cryptomerioides). Front. Plant Sci. 2018, 9, 1148. [Google Scholar] [CrossRef] [Green Version]
- Pettorelli, N. The Normalized Difference Vegetation Index; Oxford University Press: Oxford, UK, 2013; ISBN 9780199693160. [Google Scholar]
- Körner, C. The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 2007, 22, 569–574. [Google Scholar] [CrossRef]
- Hof, A.R.; Jansson, R.; Nilsson, C. The usefulness of elevation as a predictor variable in species distribution modelling. Ecol. Model. 2012, 246, 86–90. [Google Scholar] [CrossRef]
- Palazzi, E.; Mortarini, L.; Terzago, S.; von Hardenberg, J. Elevation-dependent warming in global climate model simulations at high spatial resolution. Clim. Dyn. 2019, 52, 2685–2702. [Google Scholar] [CrossRef] [Green Version]
- Nakazato, T.; Bogonovich, M.; Moyle, L.C. Environmental factors predict adaptive phenotypic differentiation within and between two wild Andean tomatoes. Evolution 2008, 62, 774–792. [Google Scholar] [CrossRef]
- Nakazato, T.; Warren, D.L.; Moyle, L.C. Ecological and geographic modes of species divergence in wild tomatoes. Am. J. Bot. 2010, 97, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Amatulli, G.; Domisch, S.; Tuanmu, M.N.; Parmentier, B.; Ranipeta, A.; Malczyk, J.; Jetz, W. A suite of global, cross-scale topographic variables for environmental and biodiversity modeling. Sci. Data 2018, 5, 180040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, J.J.; Doyle, J.L. Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Schulz, B.; Eckstein, R.L.; Durka, W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol. Ecol. Resour. 2013, 134, 642–653. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing Vienna, Austria. 2020. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 6 January 2022).
- Herrera, C.M.; Medrano, M.; Bazaga, P. Comparative epigenetic and genetic spatial structure of the perennial herb Helleborous foetidus: Isolation by environment, isolation by distance, and functional trait divergence. Am. J. Bot. 2017, 104, 1195–1204. [Google Scholar] [CrossRef] [Green Version]
- Bonin, A.; Bellemain, E.; Bronken, E.P.; Pompanon, F.; Brochmann, C.; Taberlet, P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004, 13, 3261–3273. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; ISBN 9780231886710. [Google Scholar]
- Zhivotovsky, L.A. An R Package for Bayesian Multilevel Models Using Stan. Mol. Ecol. 1999, 8, 907–913. [Google Scholar] [CrossRef]
- Vekemans, X.; Beauwens, T.; Lemaire, M.; Roldán-Ruiz, I. Data from amplified fragment length polymorphism (AFLP) markers Hijmans139–151. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Brown, A.H.; Feldman, M.W.; Nevo, E. Multilocus structure of natural populations of Hordeum spontaneum. Genetics 1980, 96, 523–536. [Google Scholar] [CrossRef]
- Agapow, P.M.; Burt, A. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 2001, 1, 101–102. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef] [Green Version]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Soft. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 2nd ed.; SAGE Publications Inc.: Newbury Park, CA, USA, 2011; ISBN 9781544336473. [Google Scholar]
- Lenth, R. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.3.1. 2018. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 6 January 2022).
- Naimi, B.; Hamm, N.A.S.; Groen, T.A.; Skidmore, A.K.; Toxopeus, A.G. Where is positional uncertainty a problem for species distribution modelling? Ecography 2014, 37, 191–203. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L. Vegan: Community Ecology Package. R Package Version 2.4-2. 2017. Available online: https://CRAN.R-project.org/package=vegan (accessed on 15 January 2018).
- Herve, M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics. R Package Version 0.9-69. 2018. Available online: https://CRAN.R-project.org/package=RVAideMemoire (accessed on 2 April 2018).
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [Green Version]
- Jeffreys, H. Theory of Probability; Oxford University Press: Oxford, UK, 1961; ISBN 9780198503682. [Google Scholar]
- Foll, M. Bayescan 2.1 User Manual. 2012. Available online: https://cmpg.unibe.ch/software/BBayeSca/files/BayeScan2.1_manual.pdf (accessed on 8 March 2014).
- Dray, S.; Dufour, A.-B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.B. Method for combining non-independent, one-sided tests of significance. Biometrics 1975, 31, 987–992. [Google Scholar] [CrossRef]
- Dray, S.; Bauman, D.; Blanchet, F.G.; Borcard, D.; Clappe, S.; Guenard, G.; Jombart, T.; Larocque, G.; Legendre, P.; Madi, N.; et al. Adespatial: Multivariate Multiscale Spatial Analysis. R Package Version 0.3-7. 2019. Available online: https://cran.r-project.org/web/packages/adespatial/index.html (accessed on 21 February 2020).
- Herrera, C.M.; Bazaga, P. Untangling individual variation in natural populations: Ecological, genetic and epigenetic correlates of long-term inequality in herbivory. Mol. Ecol. 2011, 20, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.N.; Bemmels, J.B. Time to get moving: Assisted gene flow of forest trees. Evol. Appl. 2016, 9, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Rey, O.; Eizaguirre, C.; Angers, B.; Baltazar-Soares, M.; Sagonas, K.; Prunier, J.G.; Blanchet, S. Linking epigenetics and biological conservation: Towards a conservation epigenetic perspective. Funct. Ecol. 2019, 34, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Su, H.-J. Studies on the climate and vegetation types of the natural forests in Taiwan (II): Altitudinal vegetation zones in relation to temperature gradient. Quart. J. Chinese Forest. (In Chinese with English Summary). 1984, 17, 57–73. [Google Scholar]
- Li, C.-F.; Chytrý, M.; Zelený, D.; Chen, M.-Y.; Chen, T.-Y.; Chiou, C.-R.; Hsia, Y.-J.; Liu, H.-Y.; Yang, S.-Z.; Yeh, C.-L.; et al. Classification of Taiwan forest vegetation. Appl. Veg. Sci. 2013, 16, 698–719. [Google Scholar] [CrossRef]
- Huete, A.R.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Chang, C.-T.; Wang, S.-F.; Vadeboncoeur, M.A.; Lin, T.-C. Relating vegetation dynamics to temperature and precipitation at monthly and annual timescales in Taiwan using MODIS vegetation indices. Int. J. Remote Sens. 2014, 35, 598–620. [Google Scholar] [CrossRef]
- Chang, C.-T.; Lin, T.-C.; Lin, N.-H. Estimating the critical load and the environmental and economic impact of acid deposition in Taiwan. J. Geogr. Sci. 2009, 56, 39–58. [Google Scholar]
- Wei, T.; Simko, V. R. Package "corrplot": Visualization of a Correlation Matrix (Version 0.84). 2017. Available online: https://github.com/taiyun/corrplot (accessed on 6 January 2019).
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Zhivotovsky, L.A. Estimating population structure in diploids with multilocus dominant DNA markers. Mol. Ecol. 1999, 8, 907–913. [Google Scholar] [CrossRef]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. qvalue: Q-Value Estimation for False Discovery Rate Control. R Package Version 2.14.1. 2019. Available online: http://github.com/StoreyLab/qvalue. (accessed on 8 January 2019).
- Menon, S. ArcGIS 10.3. 2014, The Next Generation of GIS is Here. Environmental Systems Research Institute, Inc., CA, USA. Available online: http://www.esri.com/software/arcgis (accessed on 8 January 2019).
- Open Government Data Providing Organization in Taiwan. Available online: http://data.gov.tw/node/35430 (accessed on 11 January 2020).
- Breheny, P.; Burchett, W. Visualization of Regression Models Using visreg. R. J. 2017, 9, 56–71. [Google Scholar] [CrossRef]
- Kassambara, A. 2014, easyGgplot2: Perform and Customize Easily a Plot with ggplot2. R Package Version 1.0.0.9000. Available online: http://www.sthda.com (accessed on 13 February 2022).
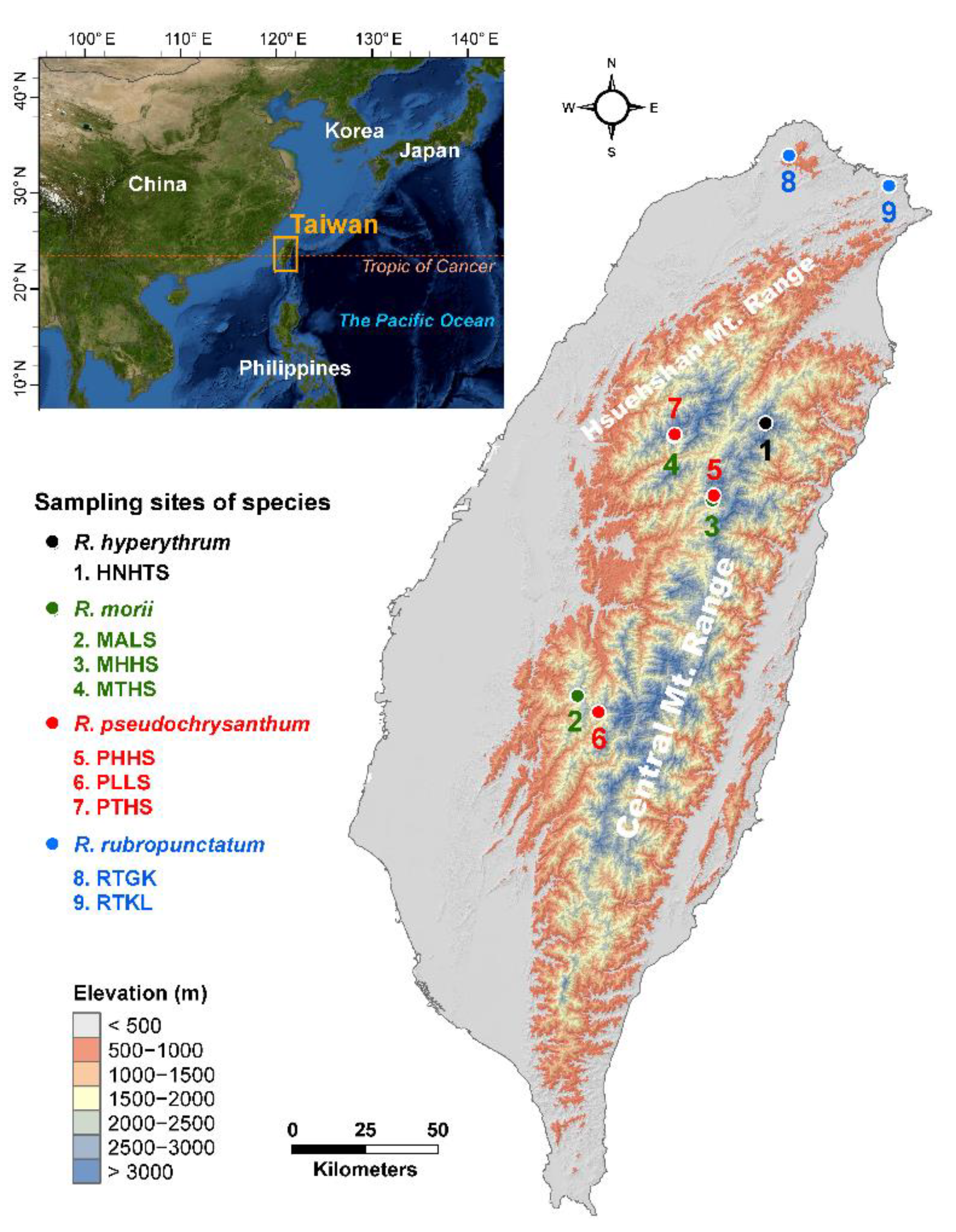
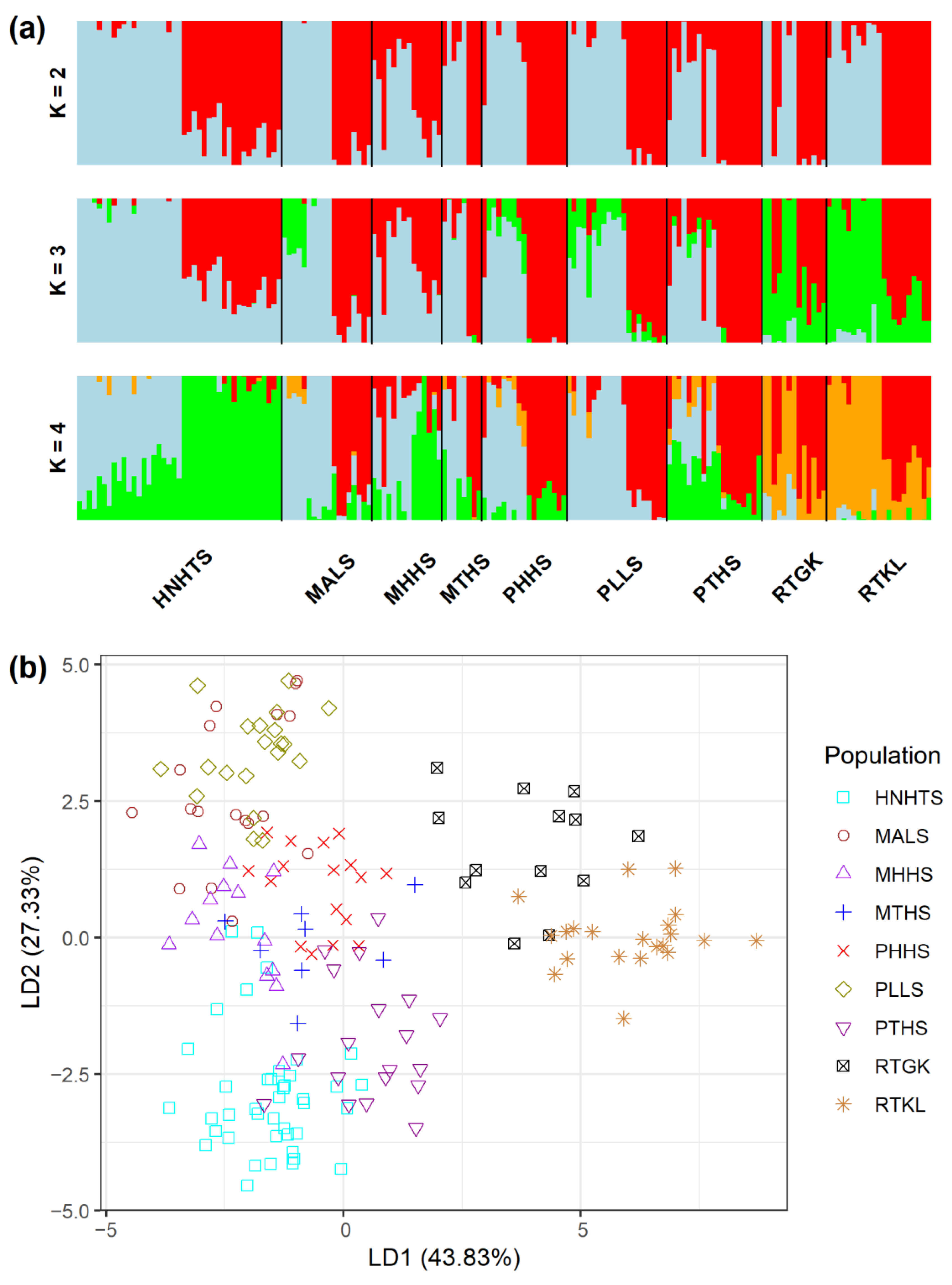
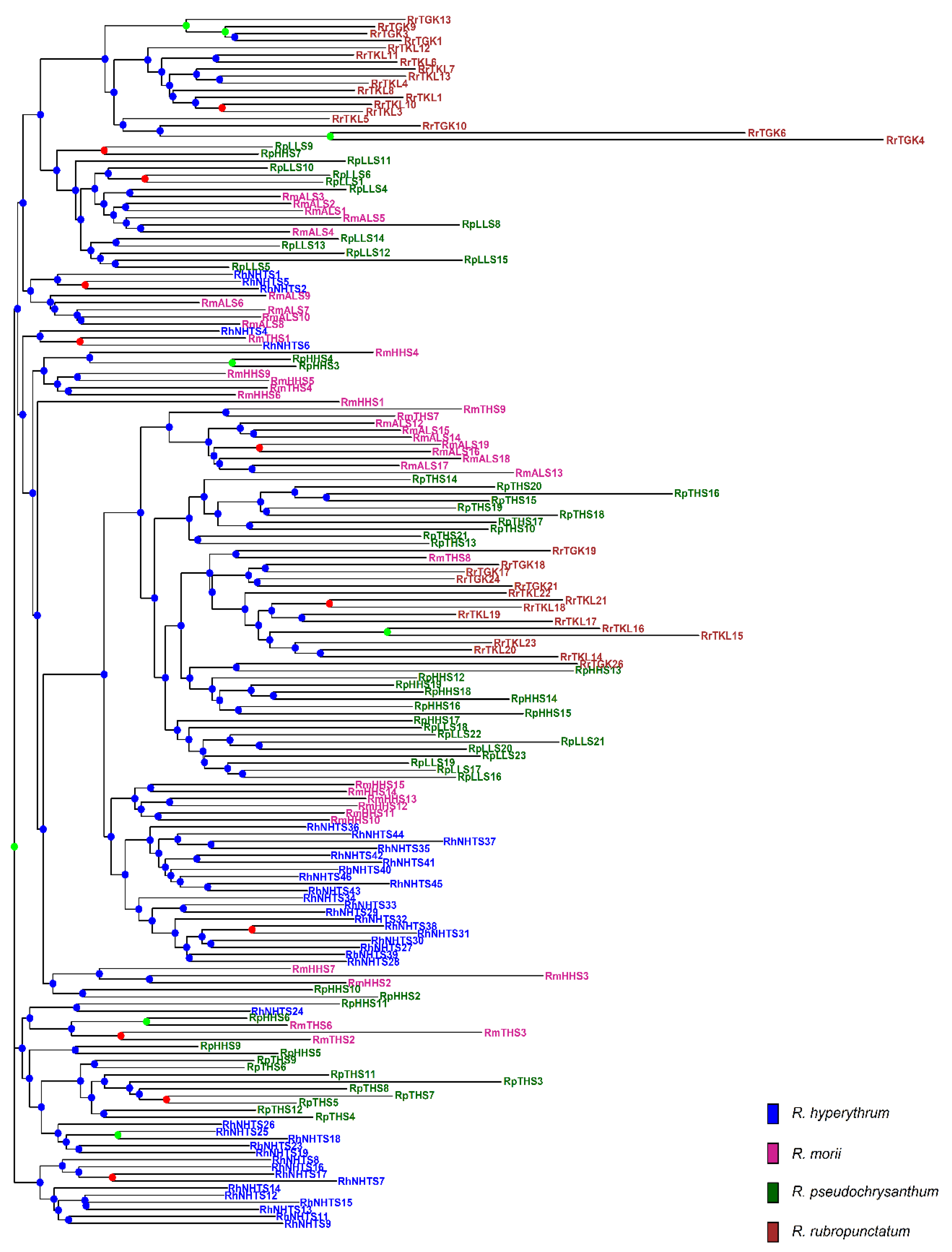

| Species Population | Longitude Latitude | Altitude (m) | N | %p | uHE (SE) | IA (p) | rD (p) |
|---|---|---|---|---|---|---|---|
| R. hyperythrum | |||||||
| Nanhutashan (HNHTS) | 121.4381 24.3575 | 3500 | 41 (45) | 63.5 | 0.2074 (0.009) | 2.037 (0.001) | 0.008 (0.001) |
| R. morii | |||||||
| Alishan (MALS) | 120.8006 23.51111 | 2100 | 18 | 65.1 | 0.2160 (0.009) | 3.926 (0.001) | 0.017 (0.001) |
| Hohuanshan (MHHS) | 121.2575 24.11944 | 2800 | 14 (15) | 57.3 | 0.2152 (0.009) | 2.172 (0.001) | 0.010 (0.001) |
| Tahsueshan (MTHS) | 121.1281 24.31861 | 3085 | 8 (9) | 60.4 | 0.2260 (0.010) | 2.969 (0.001) | 0.016 (0.001) |
| R. pseudochrysanthum | |||||||
| Lulinshan (PLLS) | 120.8719 23.46139 | 2862 | 20 | 69.8 | 0.2393 (0.009) | 2.539 (0.001) | 0.009 (0.001) |
| Hohuanshan (PHHS) | 121.2619 24.13417 | 3400 | 17 (20) | 64.3 | 0.2218 (0.009) | 3.601 (0.001) | 0.151 (0.001) |
| Tahsueshan (PTHS) | 121.1303 24.32361 | 3121 | 19 (20) | 56.0 | 0.2038 (0.010) | 2.024 (0.001) | 0.009 (0.001) |
| R. rubropunctatum | |||||||
| Tsaigongken (RTGK) | 121.5217 25.18972 | 886 | 13 | 59.4 | 0.2439 (0.010) | 4.881 (0.001) | 0.020 (0.001) |
| Tsankuangliao (RTKL) | 121.8633 25.09444 | 630 | 21 (23) | 62.5 | 0.2161 (0.010) | 3.384 (0.001) | 0.015 (0.001) |
| Total | 171 (132) | ||||||
| Average | 17 (22) | 62.03 (4.26) | 0.2211 (0.009) | ||||
| Source of Variation | Degree of Freedom | Sum of Squares | Percent Variation | Φ Statistics (p) |
|---|---|---|---|---|
| Total Data | ||||
| Between species | 3 | 692.09 | 5.20 | ΦCT = 0.0520 (0.012) |
| Between populations within species | 5 | 516.28 | 9.48 | ΦSC = 0.1000 (0.001) |
| Within populations | 162 | 6083.74 | 85.32 | ΦST = 0.1468 (0.001) |
| Total | 170 | 7292.12 | 100 | |
| Outlier Data | ||||
| Between species | 3 | 178.32 | 22.83 | ΦCT = 0.2283(0.011) |
| Between populations within species | 5 | 81.32 | 23.91 | ΦSC = 0.3099 (0.001) |
| Within populations | 162 | 326.50 | 53.25 | ΦST = 0.4675(0.001) |
| Total | 170 | 586.140 | 100 |
| Outlier Genetic/Epigenetic Variation | Category of Environmental Variables | Adjusted R2 | Cumulative Adjusted R2 | F Value (p) |
|---|---|---|---|---|
| AFLP | ||||
| Bioclimate | ||||
| BIO1 | 0.1576 | 0.1576 | 32.81 (0.001) | |
| BIO2 | 0.1070 | 0.2646 | 25.59 (0.001) | |
| BIO12 | 0.0429 | 0.3076 | 11.42 (0.001) | |
| Topology | ||||
| Elevation | 0.1394 | 0.1394 | 28.54 (0.001) | |
| Aspect | 0.0518 | 0.1912 | 11.83 (0.001) | |
| Slope | 0.0431 | 0.2343 | 10.45 (0.001) | |
| Ecology | ||||
| PET | 0.1150 | 0.1150 | 23.09 (0.001) | |
| CLO | 0.0848 | 0.1998 | 18.90 (0.001) | |
| NDVI | 0.0497 | 0.2495 | 12.13 (0.001) | |
| RH | 0.0458 | 0.2953 | 11.85 (0.001) | |
| WSmean | 0.0245 | 0.3197 | 6.97 (0.001) | |
| MSAP-m | ||||
| Bioclimate | ||||
| BIO1 | 0.1925 | 0.1925 | 32.22 (0.001) | |
| BIO2 | 0.0382 | 0.2307 | 7.46 (0.001) | |
| Topology | ||||
| Elevation | 0.1272 | 0.1272 | 20.09 (0.001) | |
| Aspect | 0.0336 | 0.1608 | 6.21 (0.001) | |
| Slope | 0.0200 | 0.1804 | 4.08 (0.001) | |
| Ecology | ||||
| PET | 0.1664 | 0.1664 | 27.16 (0.001) | |
| NDVI | 0.0350 | 0.2014 | 6.70 (0.001) | |
| MSAP-u | ||||
| Bioclimate | ||||
| BIO1 | 0.3446 | 0.3446 | 69.89 (0.001) | |
| BIO2 | 0.1720 | 0.5166 | 47.25 (0.001) | |
| BIO12 | 0.0236 | 0.5402 | 7.63(0.001) | |
| Topology | ||||
| Elevation | 0.1772 | 0.1772 | 29.22 (0.001) | |
| Aspect | 0.0613 | 0.2386 | 11.47 (0.001) | |
| Slope | 0.0463 | 0.2848 | 9.35 (0.001) | |
| Ecology | ||||
| NDVI | 0.3518 | 0.3518 | 72.09 (0.001) | |
| PET | 0.0996 | 0.4514 | 24.69 (0.001) |
| Mantel Test Mantel r (p) | Partial Mantel Test Mantel r (p) | |||||
|---|---|---|---|---|---|---|
| G vs. E | G vs. D | E vs. D | G vs. E|D | |||
| Total Data | ||||||
| AFLP | 0.3634 (0.001) | 0.4070 (0.001) | 0.7175 (0.001) | 0.1123 (0.001) | ||
| MSAP-m | 0.0300 (0.256) | −0.003 (0.481) | 0.8167 (0.001) | 0.0560 (0.062) | ||
| MSAP-u | 0.2844 (0.001) | 0.2658 (0.001) | 0.8167 (0.001) | 0.1210 (0.001) | ||
| Outlier Data | ||||||
| AFLP | 0.5286 (0.001) | 0.4959 (0.001) | 0.2858 (0.001) | |||
| MSAP-m | 0.3306 (0.001) | 0.3003 (0.001) | 0.1551 (0.001) | |||
| MSAP-u | 0.2545 (0.001) | 0.2553 (0.001) | 0.0825 (0.002) | |||
| MMRR | ||||||
| G vs. E | G vs. D | E vs. D | G vs. E|D | |||
| R2 | βD(p) | βE(p) | ||||
| Total Data | ||||||
| AFLP | 0.2800 (0.001) | 0.3014 (0.001) | 0.6443 (0.001) | 0.1902 | 0.2068 (0.001) | 0.1467 (0.001) |
| MSAP-m | −0.0230 (0.664) | −0.0400 (0.435) | 0.9431 (0.001) | 0.0046 | −0.1643 (0.001) | 0.1319 (0.010) |
| MSAP-u | 0.1925 (0.001) | 0.1653 (0.001) | 0.9431 (0.001) | 0.0509 | −0.1467 (0.001) | 0.3307 (0.001) |
| Outlier Data | ||||||
| AFLP | 0.4678 (0.001) | 0.4469 (0.001) | 0.2897 | 0.2488 (0.001) | 0.3074 (0.001) | |
| MSAP-m | 0.3247 (0.001) | 0.3024 (0.001) | 0.1074 | −0.0175 (0.576) | 0.3412 (0.001) | |
| MSAP-u | 0.2158 (0.001) | 0.1885 (0.001) | 0.0493 | −0.1351 (0.005) | 0.3431 (0.001) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.-J.; Li, Y.-S.; Chang, C.-T.; Chung, J.-D.; Hwang, S.-Y. Adaptive Divergence without Distinct Species Relationships Indicate Early Stage Ecological Speciation in Species of the Rhododendronpseudochrysanthum Complex Endemic to Taiwan. Plants 2022, 11, 1226. https://doi.org/10.3390/plants11091226
Cao J-J, Li Y-S, Chang C-T, Chung J-D, Hwang S-Y. Adaptive Divergence without Distinct Species Relationships Indicate Early Stage Ecological Speciation in Species of the Rhododendronpseudochrysanthum Complex Endemic to Taiwan. Plants. 2022; 11(9):1226. https://doi.org/10.3390/plants11091226
Chicago/Turabian StyleCao, Jia-Jia, Yi-Shao Li, Chung-Te Chang, Jeng-Der Chung, and Shih-Ying Hwang. 2022. "Adaptive Divergence without Distinct Species Relationships Indicate Early Stage Ecological Speciation in Species of the Rhododendronpseudochrysanthum Complex Endemic to Taiwan" Plants 11, no. 9: 1226. https://doi.org/10.3390/plants11091226
APA StyleCao, J.-J., Li, Y.-S., Chang, C.-T., Chung, J.-D., & Hwang, S.-Y. (2022). Adaptive Divergence without Distinct Species Relationships Indicate Early Stage Ecological Speciation in Species of the Rhododendronpseudochrysanthum Complex Endemic to Taiwan. Plants, 11(9), 1226. https://doi.org/10.3390/plants11091226







