Nitrogen Acquisition and Transport in the Ectomycorrhizal Symbiosis—Insights from the Interaction between an Oak Tree and Pisolithus tinctorius
Abstract
1. Introduction
2. Results
2.1. P. tinctorius Root Colonization. Effect on Plant Growth and N Concentration
2.2. Identification of Cork Oak Genes Involved in N Transport and Assimilation
2.3. Impact of ECM Symbiosis on the Expression of Genes Involved in N Transport and Assimilation
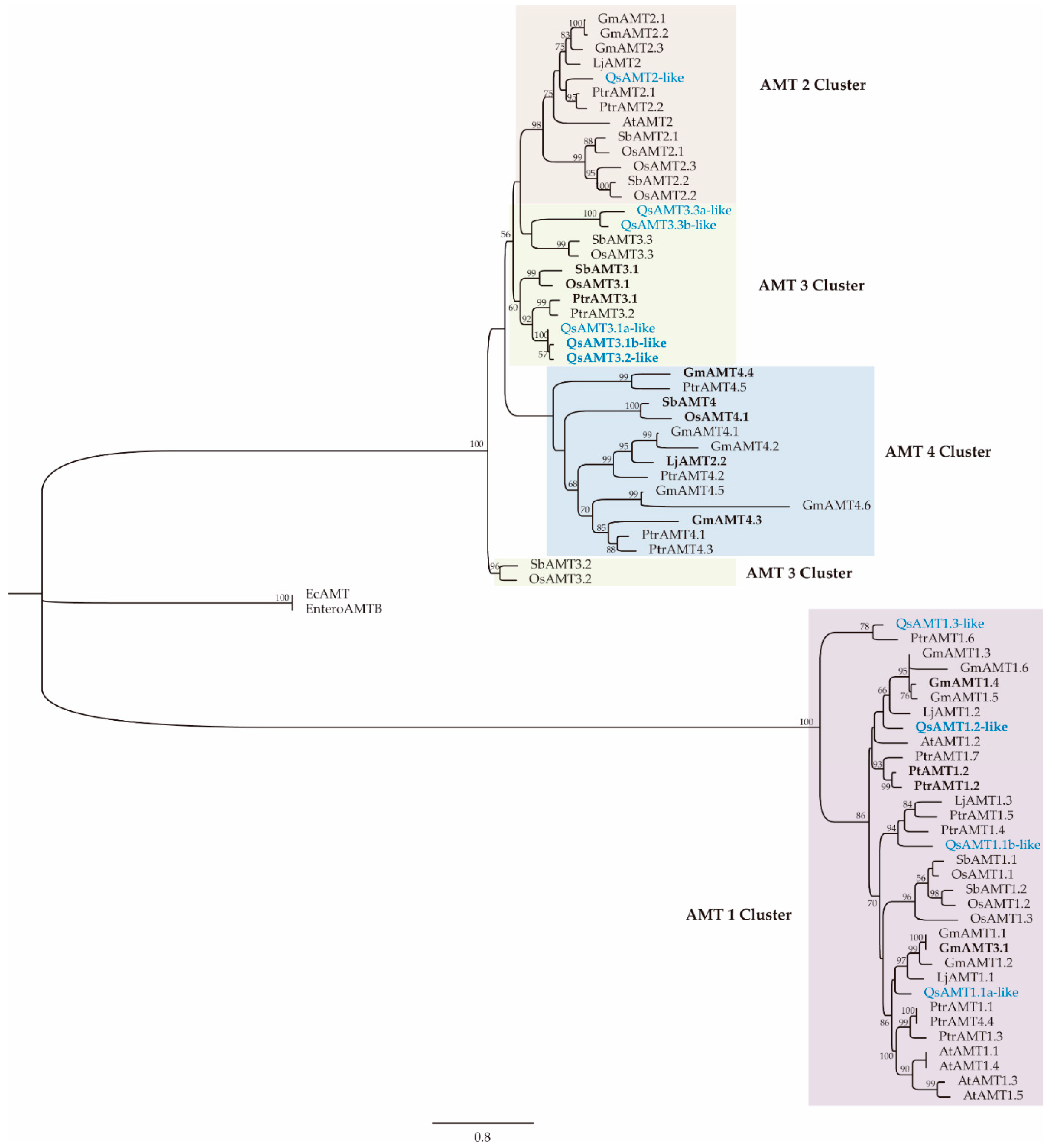
2.4. Phylogenetic Analysis of Cork Oak AMTs
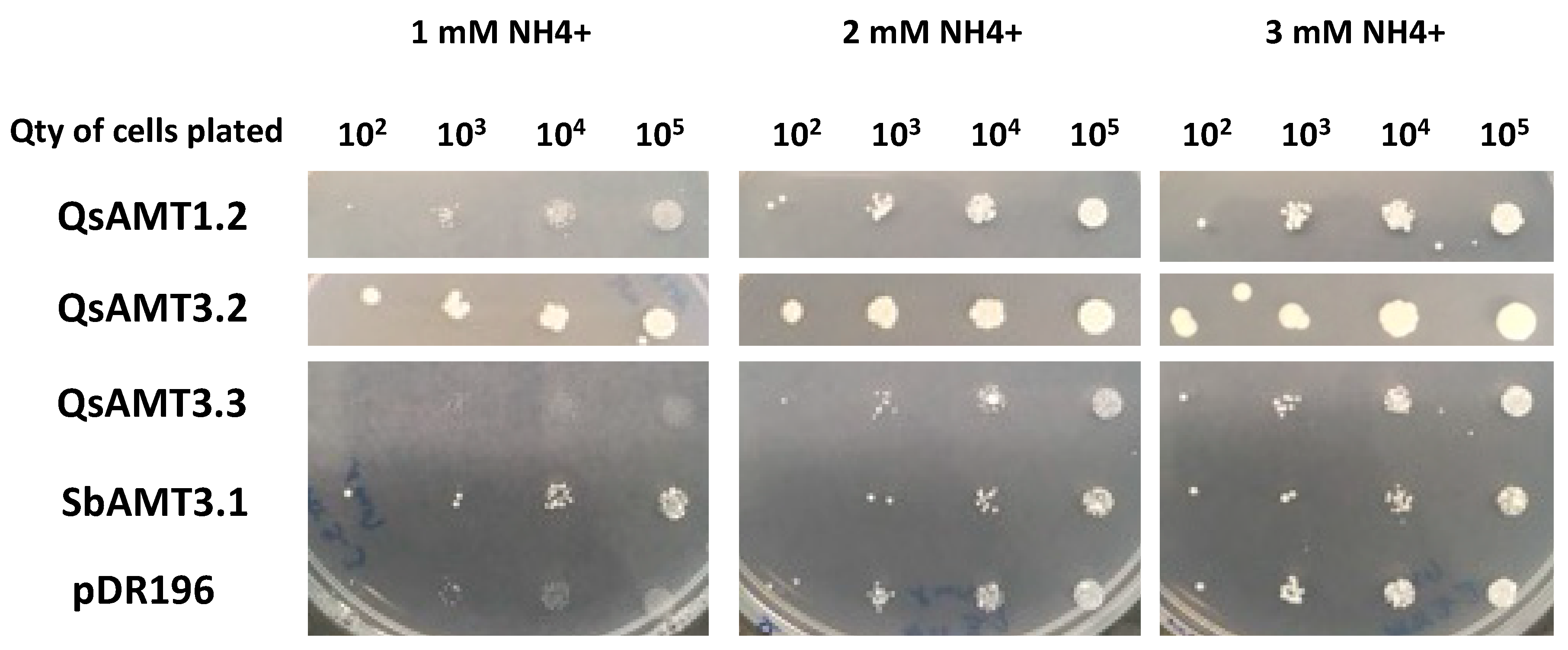
2.5. Heterologous Complementation of a Yeast Mutant Defective in NH4+ Uptake
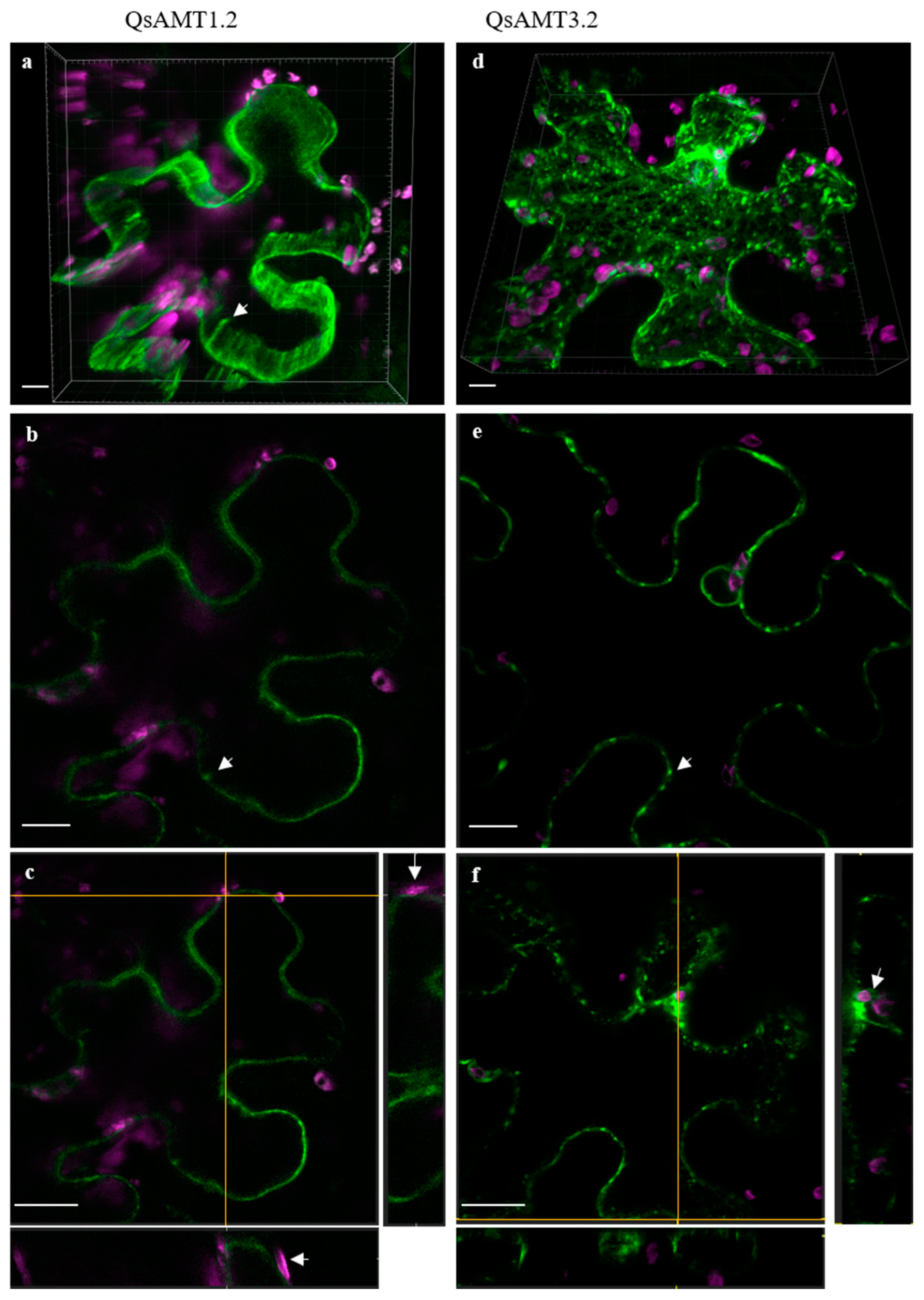
2.6. Subcellular Localization of Cork Oak AMTs
2.7. Homology Modeling of AMTs Upregulated in Cork Oak ECM Roots
3. Discussion
4. Materials and Methods
4.1. ECM Symbiosis Establishment and Plant Growth
4.2. Determination of N Concentration
4.3. Identification of Cork Oak Genes Involved in N Transport and Assimilation
4.4. Phylogenetic Analysis of Q. suber AMT Gene Family
4.5. Gene Expression Analysis by qPCR
4.6. Isolation of ECM Over-Expressed Q. suber AMT Genes and Functional Expression in Yeast
4.7. Subcellular Localization of Q. suber AMT Genes in N. benthamiana
4.8. Q. suber AMT Homology Modelling
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, S.E.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, Q.; Kraus, T.E.C.; Dahlgren, R.A.; Anastasio, C.; Zasoski, R.J. Contribution of Amino Compounds to Dissolved Organic Nitrogen in Forest Soils. Biogeochemistry 2002, 61, 173–198. [Google Scholar] [CrossRef]
- Brundrett, M. Diversity and Classification of Mycorrhizal Associations. Biol. Rev. Camb. Philos. Soc. 2004, 79, 473–495. [Google Scholar] [CrossRef] [PubMed]
- Stuart, E.K.; Plett, K.L. Digging Deeper: In Search of the Mechanisms of Carbon and Nitrogen Exchange in Ectomycorrhizal Symbioses. Front. Plant Sci. 2020, 10, 1658. [Google Scholar] [CrossRef]
- Courty, P.E.; Smith, P.; Koegel, S.; Redecker, D.; Wipf, D. Inorganic Nitrogen Uptake and Transport in Beneficial Plant Root-Microbe Interactions. CRC. Crit. Rev. Plant Sci. 2015, 34, 4–16. [Google Scholar] [CrossRef]
- Finlay, R.D.; Frostegård, A.; Sonnerfeldt, A.-M. Utilization of Organic and Inorganic Nitrogen Sources by Ectomycorrhizal Fungi in Pure Culture and in Symbiosis with Pinus Contorta Dougl. Ex Loud. New Phytol. 1992, 120, 105–115. [Google Scholar] [CrossRef]
- Govindarajulu, M.; Pfeffer, P.E.; Jin, H.; Abubaker, J.; Douds, D.D.; Allen, J.W.; Bücking, H.; Lammers, P.J.; Shachar-Hill, Y. Nitrogen Transfer in the Arbuscular Mycorrhizal Symbiosis. Nature 2005, 435, 819–823. [Google Scholar] [CrossRef]
- Leigh, J.; Hodge, A.; Fitter, A.H. Arbuscular Mycorrhizal Fungi Can Transfer Substantial Amounts of Nitrogen to Their Host Plant from Organic Material. New Phytol. 2009, 181, 199–207. [Google Scholar] [CrossRef]
- Chalot, M.; Blaudez, D.; Brun, A. Ammonia: A Candidate for Nitrogen Transfer at the Mycorrhizal Interface. Trends Plant Sci. 2006, 11, 263–266. [Google Scholar] [CrossRef]
- Couturier, J.; Montanini, B.; Martin, F.; Brun, A.; Blaudez, D.; Chalot, M. The Expanded Family of Ammonium Transporters in the Perennial Poplar Plant. New Phytol. 2007, 174, 137–150. [Google Scholar] [CrossRef]
- Müller, T.; Avolio, M.; Olivi, M.; Benjdia, M.; Rikirsch, E.; Kasaras, A.; Fitz, M.; Chalot, M.; Wipf, D. Nitrogen Transport in the Ectomycorrhiza Association: The Hebeloma Cylindrosporum-Pinus Pinaster Model. Phytochemistry 2007, 68, 41–51. [Google Scholar] [CrossRef]
- Müller, T.; Neuhäuser, B.; Ludewig, U.; Houdinet, G.; Zimmermann, S.D.; Courty, P.E.; Wipf, D. New Insights into HcPTR2A and HcPTR2B, Two High-Affinity Peptide Transporters from the Ectomycorrhizal Model Fungus Hebeloma Cylindrosporum. Mycorrhiza 2020, 30, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.E.; Sreedasyam, A.; Trivedi, G.; Podila, G.K.; Cseke, L.J.; Collart, F.R. Using next Generation Transcriptome Sequencing to Predict an Ectomycorrhizal Metabolome. BMC Syst. Biol. 2011, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Hortal, S.; Plett, K.L.; Plett, J.M.; Cresswell, T.; Johansen, M.; Pendall, E.; Anderson, I.C. Role of Plant-Fungal Nutrient Trading and Host Control in Determining the Competitive Success of Ectomycorrhizal Fungi. ISME J. 2017, 11, 2666–2676. [Google Scholar] [CrossRef] [PubMed]
- Näsholm, T.; Ekblad, A.; Nordin, A.; Giesler, R.; Högberg, M.; Högberg, P. Boreal Forest Plants Take up Organic Nitrogen. Nature 1998, 392, 914–917. [Google Scholar] [CrossRef]
- Nehls, U.; Plassard, C. Nitrogen and Phosphate Metabolism in Ectomycorrhizas. New Phytol. 2018, 220, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Selle, A.; Willmann, M.; Grunze, N.; Geßler, A.; Weiß, M.; Nehls, U. The High-Affinity Poplar Ammonium Importer PttAMT1.2 and Its Role in Ectomycorrhizal Symbiosis. New Phytol. 2005, 168, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Koegel, S.; Ait Lahmidi, N.; Arnould, C.; Chatagnier, O.; Walder, F.; Ineichen, K.; Boller, T.; Wipf, D.; Wiemken, A.; Courty, P.E. The Family of Ammonium Transporters (AMT) in Sorghum bicolor: Two AMT Members Are Induced Locally, but Not Systemically in Roots Colonized by Arbuscular Mycorrhizal Fungi. New Phytol. 2013, 198, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tienda, J.; Corrêa, A.; Azcón-Aguilar, C.; Ferrol, N. Transcriptional Regulation of Host NH4+ Transporters and GS/GOGAT Pathway in Arbuscular Mycorrhizal Rice Roots. Plant Physiol. Biochem. 2014, 75, 1–8. [Google Scholar] [CrossRef]
- Kobae, Y.; Tamura, Y.; Takai, S.; Banba, M.; Hata, S. Localized Expression of Arbuscular Mycorrhiza-Inducible Ammonium Transporters in Soybean. Plant Cell Physiol. 2010, 51, 1411–1415. [Google Scholar] [CrossRef]
- Guether, M.; Neuhäuser, B.; Balestrini, R.; Dynowski, M.; Ludewig, U.; Bonfante, P. A Mycorrhizal-Specific Ammonium Transporter from Lotus japonicus Acquires Nitrogen Released by Arbuscular Mycorrhizal Fungi. Plant Physiol. 2009, 150, 73–83. [Google Scholar] [CrossRef]
- Breuillin-Sessoms, F.; Floss, D.S.; Karen Gomez, S.; Pumplin, N.; Ding, Y.; Levesque-Tremblay, V.; Noar, R.D.; Daniels, D.A.; Bravo, A.; Eaglesham, J.B.; et al. Suppression of Arbuscule Degeneration in Medicago truncatula Phosphate Transporter4 Mutants Is Dependent on the Ammonium Transporter 2 Family Protein AMT2;3. Plant Cell 2015, 27, 1352–1366. [Google Scholar] [CrossRef] [PubMed]
- Castro-Rodríguez, V.; Assaf-Casals, I.; Pérez-Tienda, J.; Fan, X.; Avila, C.; Miller, A.; Cánovas, F.M. Deciphering the Molecular Basis of Ammonium Uptake and Transport in Maritime Pine. Plant Cell Environ. 2016, 39, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen Uptake, Assimilation and Remobilization in Plants: Challenges for Sustainable and Productive Agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Marini, A.-M.; Springael, J.-Y.; Frommer, W.B.; Andre, B. Cross-Talk between Ammonium Transporters in Yeast and Interference by the Soybean SAT1 Protein. Mol. Microbiol. 2000, 35, 378–385. [Google Scholar] [CrossRef]
- Thomas, G.H.; Mullins, J.G.L.; Merrick, M. Membrane Topology of the Mep/Amt Family of Ammonium Transporters. Mol. Microbiol. 2000, 37, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Koegel, S.; Mieulet, D.; Baday, S.; Chatagnier, O.; Lehmann, M.F.; Wiemken, A.; Boller, T.; Wipf, D.; Bernèche, S.; Guiderdoni, E.; et al. Phylogenetic, Structural, and Functional Characterization of AMT3;1, an Ammonium Transporter Induced by Mycorrhization among Model Grasses. Mycorrhiza 2017, 27, 695–708. [Google Scholar] [CrossRef]
- Wipf, D.; Benjdia, M.; Rikirsch, E.; Zimmermann, S.; Tegeder, M.; Frommer, W.B. An Expression CDNA Library for Suppression Cloning in Yeast Mutants, Complementation of a Yeast His4 Mutant, and EST Analysis from the Symbiotic Basidiomycete Hebeloma Cylindrosporum. Genome 2003, 46, 177–181. [Google Scholar] [CrossRef]
- Marini, A.-M.; Soussi-Boudekou, S.; Vissers, S.; Andre, B. A Family of Ammonium Transporters in Saccharomyces Cerevisiae. Mol. Cell. Biol. 1997, 17, 4282–4293. [Google Scholar] [CrossRef]
- Hess, D.C.; Lu, W.; Rabinowitz, J.D.; Botstein, D. Ammonium Toxicity and Potassium Limitation in Yeast. PLoS Biol. 2006, 4, 2012–2023. [Google Scholar] [CrossRef]
- Khademi, S.; O’connell, J., III; Remis, J.; Robles-Colmenares, Y.; Miercke, L.J.W.; Stroud, R.M. Mechanism of Ammonia Transport by Amt/MEP/Rh: Structure of AmtB at 1.35 Å. New Ser. 2004, 305, 1587–1594. [Google Scholar] [CrossRef]
- von Wittgenstein, N.J.; Le, C.H.; Hawkins, B.J.; Ehlting, J. Evolutionary Classification of Ammonium, Nitrate, and Peptide Transporters in Land Plants. BMC Evol. Biol. 2014, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, S.; Pérez-Tienda, J.; Ellerbeck, M.; Arnould, C.; Chatagnier, O.; Boller, T.; Schüßler, A.; Brachmann, A.; Wipf, D.; Ferrol, N.; et al. GintAMT3-a Low-Affinity Ammonium Transporter of the Arbuscular Mycorrhizal Rhizophagus Irregularis. Front. Plant Sci. 2016, 7, 679. [Google Scholar] [CrossRef] [PubMed]
- Bajgain, P.; Russell, B.; Mohammadi, M. Phylogenetic Analyses and In-Seedling Expression of Ammonium and Nitrate Transporters in Wheat. Sci. Rep. 2018, 8, 7082. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Orabi, E.A.; Baday, S.; Bernèche, S.; Lamoureux, G. Ammonium Transporters Achieve Charge Transfer by Fragmenting Their Substrate. J. Am. Chem. Soc. 2012, 134, 10419–10427. [Google Scholar] [CrossRef] [PubMed]
- López-Pedrosa, A.; González-Guerrero, M.; Valderas, A.; Azcón-Aguilar, C.; Ferrol, N. GintAMT1 Encodes a Functional High-Affinity Ammonium Transporter That Is Expressed in the Extraradical Mycelium of Glomus Intraradices. Fungal Genet. Biol. 2006, 43, 102–110. [Google Scholar] [CrossRef]
- Sebastiana, M.; da Silva, A.B.; Matos, A.R.; Alcântara, A.; Silvestre, S.; Malhó, R. Ectomycorrhizal Inoculation with Pisolithus tinctorius Reduces Stress Induced by Drought in Cork Oak. Mycorrhiza 2018, 28, 247–258. [Google Scholar] [CrossRef]
- Rodrigues, C.I.; Maia, R.; Máguas, C. Comparing Total Nitrogen And Crude Protein Content Of Green Coffee Beans (Coffea Spp.) From Different Geographical Origins. Coffe Sci. 2010, 5, 197–205. [Google Scholar]
- Ramos, A.M.; Usié, A.; Barbosa, P.; Barros, P.M.; Capote, T.; Chaves, I.; Simões, F.; Abreu, I.; Carrasquinho, I.; Faro, C.; et al. The Draft Genome Sequence of Cork Oak. Sci. Data 2018, 5, 180069. [Google Scholar] [CrossRef]
- The Arabidopsis Information Resource. Available online: https://www.arabidopsis.org/ (accessed on 20 March 2021).
- The Pant Genomics Resource. Available online: https://phytozome-next.jgi.doe.gov/ (accessed on 20 March 2021).
- Pfam. Available online: https://pfam.xfam.org/ (accessed on 15 May 2021).
- SignalP. Available online: https://services.healthtech.dtu.dk/service.php?SignalP (accessed on 15 May 2021).
- DeepTMHMM. Available online: https://dtu.biolib.com/DeepTMHMM/ (accessed on 15 May 2021).
- TargetP. Available online: https://services.healthtech.dtu.dk/service.php?TargetP-2.0 (accessed on 15 May 2021).
- Multiple Sequence Alignment by CLUSTALW. Available online: https://www.genome.jp/tools-bin/clustalw (accessed on 10 December 2021).
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 28 November 2022).
- Wan, C.Y.; Wilkins, T.A. A Modified Hot Borate Method Significantly Enhances the Yield of High-Quality RNA from Cotton (Gossypium Hirsutum L.). Anal. Biochem. 1994, 223, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Sebastiana, M.; Duarte, B.; Monteiro, F.; Malhó, R.; Caçador, I.; Matos, A.R. The Leaf Lipid Composition of Ectomycorrhizal Oak Plants Shows a Drought-Tolerance Signature. Plant Physiol. Biochem. 2019, 144, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Primer-BLAST. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 5 November 2021).
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, R.J.; Strasser, A.W.M.; Höner, C.B.; Hollenberg, C.P. An Efficient Transformation Procedure Enabling Long-Term Storage of Competent Cells of Various Yeast Genera. Yeast 1991, 7, 691–692. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, Transient Expression of Fluorescent Fusion Proteins in Tobacco Plants and Generation of Stably Transformed Plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- SWISS-MODEL. Available online: https://swissmodel.expasy.org/ (accessed on 12 February 2022).

| EcAmtB | QsAMT1.2-like | QsAMT3.2-like | QsAMT3.3-like (a) |
|---|---|---|---|
| Met23 | Ala61 | Gln40 | Gln41 |
| His168 | His211 | His205 | His201 |
| His318 | His378 | His359 | His355 |
| Leu114 | Ile147 | Leu151 | Leu147 |
| Leu208 | Leu256 | Leu245 | Leu241 |
| Ile266 | Leu327 | Ile303 | Met299 |
| Phe107 | Phe140 | Phe144 | Phe140 |
| Phe215 | Phe263 | Phe252 | Phe248 |
| Ile28 | Phe66 | Leu45 | Leu46 |
| Ser219 | Ser217 | Asp256 | Asp252 |
| Trp148 | Trp181 | Trp185 | Trp181 |
| Trp212 | Trp260 | Trp249 | Trp245 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sebastiana, M.; Serrazina, S.; Monteiro, F.; Wipf, D.; Fromentin, J.; Teixeira, R.; Malhó, R.; Courty, P.-E. Nitrogen Acquisition and Transport in the Ectomycorrhizal Symbiosis—Insights from the Interaction between an Oak Tree and Pisolithus tinctorius. Plants 2023, 12, 10. https://doi.org/10.3390/plants12010010
Sebastiana M, Serrazina S, Monteiro F, Wipf D, Fromentin J, Teixeira R, Malhó R, Courty P-E. Nitrogen Acquisition and Transport in the Ectomycorrhizal Symbiosis—Insights from the Interaction between an Oak Tree and Pisolithus tinctorius. Plants. 2023; 12(1):10. https://doi.org/10.3390/plants12010010
Chicago/Turabian StyleSebastiana, Mónica, Susana Serrazina, Filipa Monteiro, Daniel Wipf, Jérome Fromentin, Rita Teixeira, Rui Malhó, and Pierre-Emmanuel Courty. 2023. "Nitrogen Acquisition and Transport in the Ectomycorrhizal Symbiosis—Insights from the Interaction between an Oak Tree and Pisolithus tinctorius" Plants 12, no. 1: 10. https://doi.org/10.3390/plants12010010
APA StyleSebastiana, M., Serrazina, S., Monteiro, F., Wipf, D., Fromentin, J., Teixeira, R., Malhó, R., & Courty, P.-E. (2023). Nitrogen Acquisition and Transport in the Ectomycorrhizal Symbiosis—Insights from the Interaction between an Oak Tree and Pisolithus tinctorius. Plants, 12(1), 10. https://doi.org/10.3390/plants12010010











