Large–Scale Transposon Mutagenesis Reveals Type III Secretion Effector HopR1 Is a Major Virulence Factor in Pseudomonas syringae pv. actinidiae
Abstract
1. Introduction
2. Results
2.1. Isolation of Psa3 Mutants with Reduced Virulence
2.2. Identification of Genes Disrupted by Tn5 Insertions
2.3. Bacterial Growth of the Virulence Mutants in Plant Tissue
2.4. Hypersensitive Response Cell Death Assay with T3SS/T3E Mutants
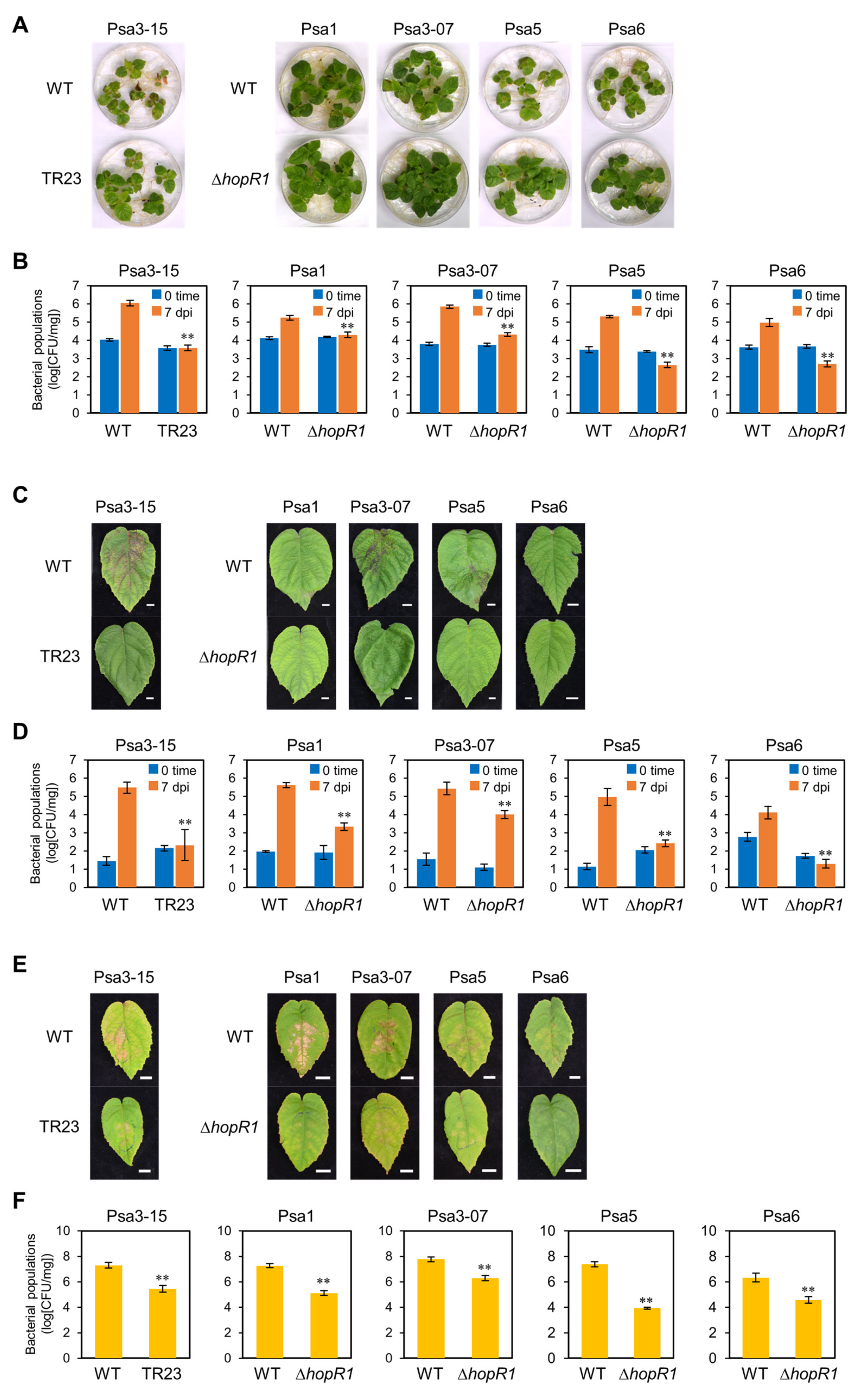
2.5. HopR1 Contributes to Psa3 Virulence
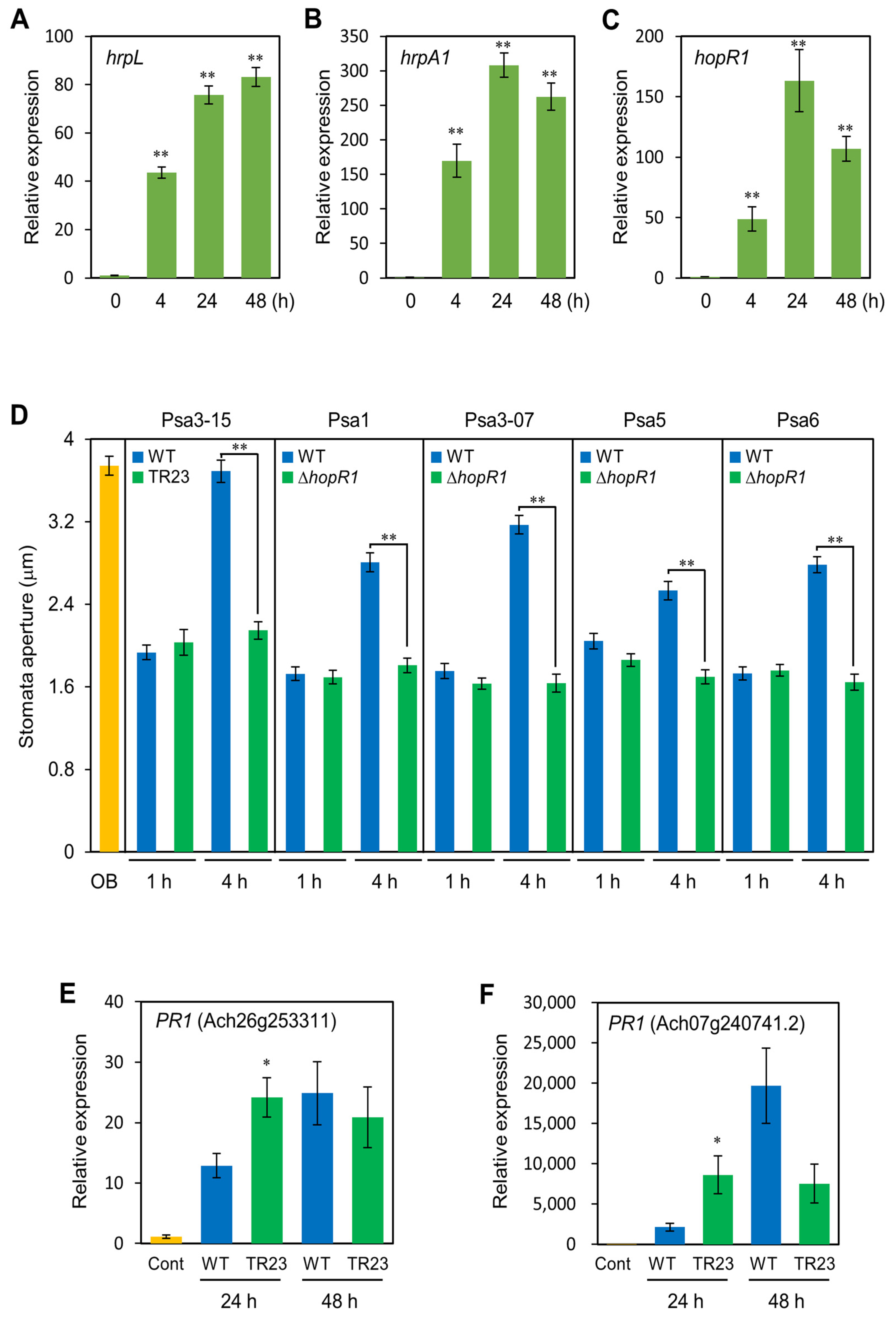
2.6. HopR1 Universally Contributes to Virulence in All Psa Biovars
| Bacterial Strain or Plasmid | Relevant Characteristics | Reference or Source |
|---|---|---|
| E. coli strain | ||
| DH5α | F−, λ−, φ80dlacZDM15, D(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17 (rK−, mK+), phoA, supE44, thi-1, gyrA96, relA1 | Takara Bio, Kusatsu, Japan |
| S17-1 | F−, thi, pro, hsdR, hsdM+, recA [chr::RP4-2-Tc::Km::Tn7] | [54] |
| P. syringae pv. actinidiae (Psa) | ||
| Psa1 | Psa biovar 1 wild-type, Nalr | MAFF 613022 |
| Psa1-ΔhopR1 | Psa biovar 1 ΔhopR1 mutant, Nalr | This study |
| Psa3-07 | Psa biovar 3 wild-type, Nalr | MAFF 212107 |
| Psa3-07-ΔhopR1 | Psa biovar 3 ΔhopR1 mutant, Nalr | This study |
| Psa3-15 | Psa biovar 3 wild-type, Nalr | MAFF 212115 |
| Psa3-VR series | Whole genome Tn5 transposon library, Nalr, Kmr, Cmr | This study |
| Psa5 | Psa biovar 5 wild-type, Nalr | MAFF 212056 |
| Psa5-ΔhopR1 | Psa biovar 5 ΔhopR1 mutant, Nalr | This study |
| Psa6 | Psa biovar 6 wild-type, Nalr | MAFF 212133 |
| Psa6-ΔhopR1 | Psa biovar 6 ΔhopR1 mutant, Nalr | This study |
| Plasmid | ||
| pBSLC1 | Transposon vector constructed by ligation of pBSL118 and pHSG396 at EcoR I site, Ampr, Kmr, Cmr | [55] |
2.7. HopR1 Regulates Stomatal-Based Defense and Defense-Related Gene Expression in Kiwifruits
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Bacterial Strains, Plasmids, and Growth Conditions
4.3. Generation of a Psa3 Genomic Tn5 Mutant Library
4.4. Plasmid Rescue of Transposon-Integrated Regions and Sequencing Analysis to Identify Insertion Sites
4.5. Generation of ∆hopR1 Mutants
4.6. Growth Curve Assay
4.7. Screening Methods
4.8. Bacterial Inoculation Methods
4.9. Real-Time Quantitative RT-PCR
4.10. Stomatal Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef]
- Scortichini, M.; Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G. Pseudomonas syringae pv. actinidiae: A re-emerging, multi-faceted, pandemic pathogen. Mol. Plant Pathol. 2012, 13, 631–640. [Google Scholar] [CrossRef]
- Takikawa, Y.; Serizawa, S.; Ichikawa, T.; Tsuyumu, S.; Goto, M. Pseudomonas syringae pv. actinidiae pv. nov.: The causal bacterium of canker of kiwifruit in Japan. Jpn. J. Phytopathol. 1989, 55, 437–444. [Google Scholar] [CrossRef]
- Fujikawa, T.; Sawada, H. Genome analysis of the kiwifruit canker pathogen Pseudomonas syringae pv. actinidiae biovar 5. Sci. Rep. 2016, 6, 21399. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, T.; Sawada, H. Genome analysis of Pseudomonas syringae pv. actinidiae biovar 6, which produces the phytotoxins, phaseolotoxin and coronatine. Sci. Rep. 2019, 9, 3836. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.; Sarojini, V. Pseudomonas syringae pv. actinidiae: Chemical control, resistance mechanisms and possible alternatives. Plant Pathol. 2013, 63, 1–11. [Google Scholar] [CrossRef]
- Nakajima, M.; Goto, M.; Hibi, T. Similarity between copper resistance genes from Pseudomonas syringae pv. actinidiae and P. syringae pv. tomato. J. Gen. Plant Pathol. 2002, 68, 68–74. [Google Scholar] [CrossRef]
- Serizawa, S.; Takikawa, Y.; Ichikawa, T.; Tsuyumu, S.; Goto, M. Occurrence of bacterial canker of kiwifruit in Japan: Description of symptoms, isolation of the pathogen and screening of bactericides. Jpn. J. Phytopathol. 1989, 55, 427–436. [Google Scholar] [CrossRef]
- Vanneste, J.; Voyle, M. Genetic basis of copper resistance in New Zealand strains of Pseudomonas syringae. N. Z. Plant Prot. 1989, 56, 109–112. [Google Scholar] [CrossRef]
- Kisaki, G.; Tanaka, S.; Ishihara, A.; Igarashi, C.; Morimoto, T.; Hamano, K.; Endo, A.; Sugita-Konishi, S.; Tabuchi, M.; Gomi, K.; et al. Evaluation of various cultivars of Actinidia species and breeding source Actinidia rufa for resistance to Pseudomonas syringae pv. actinidiae biovar 3. J. Gen. Plant Pathol. 2019, 84, 399–406. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef]
- Zipfel, C.; Felix, G. Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 2005, 8, 353–360. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef]
- Bednarek, P. Chemical warfare or modulators of defense responses—The function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 407–414. [Google Scholar] [CrossRef]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Daudi, A.; Butt, V.S.; Bolwell, G.P. Reactive oxygen species and their role in plant defense and cell wall metabolism. Planta 2012, 236, 765–779. [Google Scholar] [CrossRef]
- Sawinski, K.; Mersmann, S.; Robatzek, S.; Böhmer, M. Guarding the green: Pathways to stomatal immunity. Mol. Plant Microbe Interact. 2013, 26, 626–632. [Google Scholar] [CrossRef]
- Wang, S.; Sun, J.; Fan, F.; Tan, Z.; Zou, Y.; Lu, D. A Xanthomonas oryzae pv. oryzae effector.; XopR.; associates with receptor-like cytoplasmic kinases and suppresses PAMP-triggered stomatal closure. Sci. China Life Sci. 2016, 59, 897–905. [Google Scholar] [CrossRef]
- Underwood, W.; Melotto, M.; He, S.Y. Role of plant stomata in bacterial invasion. Cell Microbiol. 2007, 9, 1621–1629. [Google Scholar] [CrossRef]
- Ishiga, T.; Iida, Y.; Sakata, N.; Ugajin, T.; Hirata, T.; Taniguchi, S.; Hayashi, K.; Ishiga, Y. Acibenzolar-S-methyl activates stomatal-based defense against Pseudomonas cannabina pv. alisalensis in cabbage. J. Gen. Plant Pathol. 2020, 86, 48–54. [Google Scholar] [CrossRef]
- Ishiga, T.; Sakata, N.; Ugajin, T.; Ishiga, Y. Acibenzolar-S-methyl and probenazole activate stomatal-based defense at different times to control bacterial blight of cabbage. J. Gen. Plant Pathol. 2021, 87, 30–34. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Bourdais, G.; He, S.Y.; Robatzek, S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014, 202, 259–269. [Google Scholar] [CrossRef]
- Melotto, M.; Zhang, L.; Oblessuc, P.R.; He, S.Y. Stomatal defense a decade later. Plant Physiol. 2017, 174, 561–571. [Google Scholar] [CrossRef]
- Büttner, D.; He, S.Y. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009, 150, 656–1664. [Google Scholar] [CrossRef]
- Clarke, C.R.; Cai, R.; Studholme, D.J.; Guttman, D.S.; Vinatzer, B.A. Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc locus are common leaf colonizers equipped with an atypical type III secretion system. Mol. Plant Microbe Interact. 2010, 23, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Kvitko, B.H.; Park, D.H.; Velásquez, A.C.; Wei, C.F.; Russell, A.B.; Martin, G.B.; Schneider, D.J.; Collmer, A. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009, 5, e1000388. [Google Scholar] [CrossRef]
- Xin, X.F.; Nomura, K.; Aung, K.; Velásquez, A.C.; Yao, J.; Boutrot, F.; Chang, J.H.; Zipfel, C.; He, S.Y. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 2016, 539, 524–529. [Google Scholar] [CrossRef]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Brooks, D.M.; Hernández-Guzmán, G.; Kloek, A.P.; Alarcón-Chaidez, F.; Sreedharan, A.; Rangaswamy, V.; Peñaloza-Vázquez, A.; Bender, C.L.; Kunkel, B.N. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2004, 17, 162–174. [Google Scholar] [CrossRef]
- Schreiber, K.J.; Ye, D.; Fich, E.; Jian, A.; Lo, T.; Desveaux, D. A high-throughput forward genetic screen identifies genes required for virulence of Pseudomonas syringae pv. maculicola ES4326 on Arabidopsis. PLoS ONE 2012, 7, e41461. [Google Scholar] [CrossRef]
- Sakata, N.; Ishiga, T.; Saito, H.; Nguyen, V.T.; Ishiga, Y. Transposon mutagenesis reveals Pseudomonas cannabina pv. alisalensis optimizes its virulence factors for pathogenicity on different hosts. PeerJ 2019, 7, e7698. [Google Scholar] [CrossRef]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis thaliana-Pseudomonas syringae interaction. Arab. Book 2002, 1, e0039. [Google Scholar] [CrossRef]
- Bartoli, C.; Lamichhane, J.R.; Berge, O.; Guilbaud, C.; Varvaro, L.; Balestra, G.M.; Vinatzer, B.A.; Morris, C.E. A framework to gauge the epidemic potential of plant pathogens in environmental reservoirs: The example of kiwifruit canker. Mol. Plant Pathol. 2015, 16, 137–149. [Google Scholar] [CrossRef]
- Gao, X.; Huang, Q.; Zhao, Z.; Han, Q.; Ke, X.; Qin, H.; Huang, L. Studies on the infection, colonization, and movement of Pseudomonas syringae pv. actinidiae in kiwifruit tissues using a GFPuv-labeled strain. PLoS ONE 2016, 11, e0151169. [Google Scholar] [CrossRef]
- Ishiga, T.; Sakata, N.; Nguyen, V.T.; Ishiga, Y. Flood inoculation of seedlings on culture medium to study interactions between Pseudomonas syringae pv. actinidiae and kiwifruit. J. Gen. Plant Pathol. 2020, 27, 1–9. [Google Scholar] [CrossRef]
- Ishiga, Y.; Ishiga, T.; Uppalapati, S.R.; Mysore, K.S. Arabidopsis seedling flood-inoculation technique: A rapid and reliable assay for studying plant-bacterial interactions. Plant Methods. 2011, 7, 32. [Google Scholar] [CrossRef]
- Ishiga, Y.; Ishiga, T.; Ichinose, Y.; Mysore, K.S. Pseudomonas syringae flood-inoculation method in Arabidopsis. Bio-Protocol 2017, 7, e2106. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Ishiga, Y.; Wangdi, T.; Urbanczyk-Wochniak, E.; Ishiga, T.; Mysore, K.S.; Bender, C.L. Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: Phenotypic and gene expression analyses of the virulence function of coronatine. Mol. Plant Microbe Interact. 2008, 21, 383–395. [Google Scholar] [CrossRef]
- Chen, I.M.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019, 47, 666–677. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kvitko, B.H.; Ramos, A.R.; Morello, J.E.; Oh, H.S.; Collmer, A. Identification of harpins in Pseudomonas syringae pv. tomato DC3000.; which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 2007, 189, 8059–8072. [Google Scholar] [CrossRef]
- Oh, H.S.; Kvitko, B.H.; Morello, J.E.; Collmer, A. Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 2007, 189, 8277–8289. [Google Scholar] [CrossRef]
- Sawada, H.; Fujikawa, T. Genetic diversity of Pseudomonas syringae pv. actinidiae, pathogen of kiwifruit bacterial canker. Plant Pathol. 2019, 68, 1235–1248. [Google Scholar] [CrossRef]
- McCann, H.C.; Rikkerink, E.H.; Bertels, F.; Fiers, M.; Lu, A.; Rees-George, J.; Andersen, M.T.; Gleave, A.P.; Haubold, B.; Wohlers, M.W.; et al. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013, 9, e1003503. [Google Scholar] [CrossRef]
- Shimizu, R.; Taguchi, F.; Marutani, M.; Mukaihara, T.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. The ΔfliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol. Genet Genomics. 2003, 269, 21–30. [Google Scholar] [CrossRef]
- Sawada, T.; Eguchi, M.; Asaki, S.; Kashiwagi, R.; Shimomura, K.; Taguchi, F.; Matsui, H.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; et al. MexEF-OprN multidrug efflux pump transporter negatively controls N-acyl-homoserine lactone accumulation in Pseudomonas syringae pv. tabaci 6605. Mol. Genet Genomics. 2018, 293, 907–917. [Google Scholar] [CrossRef]
- Helmann, T.C.; Deutschbauer, A.M.; Lindow, S.E. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc. Natl. Acad. Sci. USA. 2019, 116, 18900–18910. [Google Scholar] [CrossRef]
- Patel, H.K.; Ferrante, P.; Xianfa, M.; Javvadi, S.G.; Subramoni, S.; Scortichini, M.; Venturi, V. Identification of loci of Pseudomonas syringae pv. actinidiae involved in lipolytic activity and their role in colonization of kiwifruit leaves. Phytopathology 2017, 107, 645–653. [Google Scholar] [CrossRef]
- Yao, J.; Allen, C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J Bacteriol. 2006, 188, 3697–3708. [Google Scholar] [CrossRef]
- Tumewu, S.A.; Matsui, H.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; Ichinose, Y. Requirement of γ-aminobutyric acid chemotaxis for virulence of Pseudomonas syringae pv. tabaci 6605. Microbes Environ. 2020, 35, ME20114. [Google Scholar] [CrossRef]
- Tumewu, S.A.; Ogawa, Y.; Okamoto, T.; Sugihara, Y.; Yamada, H.; Taguchi, F.; Matsui, H.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; et al. Cluster II che genes of Pseudomonas syringae pv. tabaci 6605.; orthologs of cluster I in Pseudomonas aeruginosa.; are required for chemotaxis and virulence. Mol. Genet. Genom. 2021, 296, 299–312. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, J.; Gao, X.; Zhang, D.; Zhang, J.; Wen, J.; Qin, H.; Guo, M.; Huang, L. Comparative genomics reveal pathogenicity-related loci in Pseudomonas syringae pv. actinidiae biovar 3. Mol. Plant Pathol. 2019, 20, 923–942. [Google Scholar] [CrossRef]
- Jayaraman, J.; Jones, W.T.; Harvey, D.; Hemara, L.M.; McCann, H.C.; Yoon, M.; Warring, S.L.; Fineran, P.C.; Mesarich, C.H.; Templeton, M.D. Variation at the common polysaccharide antigen locus drives lipopolysaccharide diversity within the Pseudomonas syringae species complex. Environ. Microbiol. 2020, 22, 5356–5372. [Google Scholar] [CrossRef]
- Schechter, L.M.; Vencato, M.; Jordan, K.L.; Schneider, S.E.; Schneider, D.J.; Collmer, A. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 2006, 19, 1180–1192. [Google Scholar] [CrossRef]
- Lee, J.; Teitzel, G.M.; Munkvold, K.; del Pozo, O.; Martin, G.B.; Michelmore, R.W.; Greenberg, J.T. Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol. 2012, 158, 1803–1818. [Google Scholar] [CrossRef]
- Jayaraman, J.; Yoon, M.; Applegate, E.R.; Stroud, E.A.; Templeton, M.D. AvrE1 and HopR1 from Pseudomonas syringae pv. actinidiae are additively required for full virulence on kiwifruit. Mol. Plant Pathol. 2020, 21, 1467–1480. [Google Scholar] [CrossRef]
- Jin, L.; Ham, J.H.; Hage, R.; Zhao, W.; Soto-Hernández, J.; Lee, S.Y.; Paek, S.M.; Kim, M.G.; Boone, C.; Coplin, D.L.; et al. Direct and indirect targeting of PP2A by conserved bacterial type III effector proteins. PLoS Pathog. 2016, 12, e1005609. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- McAtee, P.A.; Brian, L.; Curran, B.; van der Linden, O.; Nieuwenhuizen, N.J.; Chen, X.; Henry-Kirk, R.A.; Stroud, E.A.; Nardozza, S.; Jayaraman, J.; et al. Re-programming of Pseudomonas syringae pv. actinidiae gene expression during early stages of infection of kiwifruit. BMC Genom. 2018, 19, 822. [Google Scholar] [CrossRef]
- Kaji, R.; Yariuchi, R.; Fujii, Y.; Taniguchi, S.; Uji, Y.; Suzuki, G.; Kashihara, K.; Kisaki, G.; Suezawa, K.; Ohtani, M.; et al. Expression analysis of defense-related genes in bacterial canker-tolerant wild kiwifruit; Actinidia rufa. J. Gen. Plant Pathol. 2021, 87, 361–365. [Google Scholar] [CrossRef]
- Ferradás, Y.; Rey, L.; Martínez, Ó.; Rey, M.; González, M.V. Identification and validation of reference genes for accurate normalization of real-time quantitative PCR data in kiwifruit. Plant Physiol. Biochem 2016, 102, 27–36. [Google Scholar] [CrossRef]


| Classification | Mutant | Locus | Description | Gene Name | Virulence Score | Growth in MMMF |
|---|---|---|---|---|---|---|
| Amino acid metabolism and transport | TJ35 | IYO_RS04020 | Serine hydroxymethyltransferase | glyA-2 | 0 | - |
| TK40 | IYO_RS23580 | Ketol-acid reductoisomerase | 0 | - | ||
| TK18 | IYO_RS25805 | Homoserine O-acetyltransferase | metX | 1 | - | |
| TQ26 | IYO_RS19640 | 1-aminocyclopropane-1-carboxylate deaminase | 1.33 | + | ||
| TAk13 | IYO_RS29530 | Aminodeoxychorismate/anthranilate synthase component II | trpE | 3 | + | |
| Carbohydrate transport and metabolism | TAr22 | IYO_RS06400 | 6-phosphogluconolactonase | pgl | 0.67 | - |
| TBt16 | IYO_RS10680 | Transaldolase | tal | 1 | + | |
| TAc31 | IYO_RS02340 | Aminoglycoside phosphotransferase | 1.33 | - | ||
| TBc20 | IYO_RS23010 | GDP-mannose 4,6-dehydratase | gmd | 2 | + | |
| TAt22 | IYO_RS10265 | Aconitate hydratase | acnA | 2.67 | - | |
| Cell motility/Chemotaxis/Adhesion | TA15 | IYO_RS28990 | Filamentous hemagglutinin | 1 | + | |
| TS01 | IYO_RS09885 | Fagellar basal body rod protein FlgF | flgF | 1.33 | + | |
| TAh35 | IYO_RS13580 | Chemotaxis protein | 2 | + | ||
| TAg29 | IYO_RS23025 | Methyl-accepting chemotaxis protein | 2.67 | + | ||
| TAw30 | IYO_RS09875 | Flagellar hook protein FlgE | flgE | 2.67 | + | |
| TAv34 | IYO_RS26210 | Fimbrial protein | pilQ | 3.33 | + | |
| TV06 | IYO_RS18685 | Chemotaxis protein | 3.67 | + | ||
| DNA processing and modification | TN40 | IYO_RS11935 | Group II intron maturase | 1.67 | + | |
| TBw17 | IYO_RS29390 | Transposase | 2 | + | ||
| TAc15 | IYO_RS29385 | Integrase | 2.33 | + | ||
| TAz17 | IYO_RS21055 | DEAD/DEAH box helicase | 2.33 | + | ||
| TAl20 | IYO_RS00095 | Serine recombinase | 2.67 | + | ||
| TBj10 | IYO_RS05170 | Integrase | 2.67 | + | ||
| TZ39 | IYO_RS00125 | Type III restriction protein res subunit | 2.67 | + | ||
| TAj16 | IYO_RS27700 | Transposase | 3 | + | ||
| TW16 | IYO_RS16675 | Integrase | 3 | + | ||
| Glycosyl transferase | TAc09 | IYO_RS22975 | Glycosyl transferase | 2.33 | + | |
| TAm12 | IYO_RS22990 | Glycosyl transferase family 1 | 2.33 | + | ||
| Hypothetical protein | TBf09 | IYO_RS29775 | Hypothetical protein | 1.67 | + | |
| TY14 | IYO_RS19470 | Hypothetical protein | 1.67 | + | ||
| TBa33 | IYO_RS20550 | Hypothetical protein | 2 | + | ||
| TU31 | IYO_RS18120 | Hypothetical protein | 2.33 | + | ||
| TU37 | IYO_RS23615 | Hypothetical protein | 2.33 | + | ||
| TAt06 | IYO_RS07375 | Hypothetical protein | 2.67 | + | ||
| TAk12 | IYO_RS14405 | Hypothetical protein | 3 | + | ||
| TM22 | IYO_RS26985 | Hypothetical protein | 3.33 | + | ||
| Lipid metabolism and transport | TBj05 | IYO_RS15545 | Peptidyl-prolyl cis-trans isomerase | 3 | + | |
| TAj03 | IYO_RS22005 | 1-acyl-sn-glycerol-3-phosphate acyltransferase | 3.33 | + | ||
| TBf32 | IYO_RS00500 | Acetyltransferase | dsbB | 3.33 | + | |
| TBo40 | IYO_RS07835 | Acetyltransferase | 3.33 | + | ||
| Membrane | TC07 | IYO_RS00540 | Membrane protein | 2.33 | + | |
| TBp22 | IYO_RS20435 | Membrane protein | 3 | + | ||
| TC14 | IYO_RS07875 | Membrane protein | 2 | + | ||
| Nucleotide metabolism and transport | TBk15 | IYO_RS08710 | Phosphoribosylformylglycinamidine cyclo-ligase | purM | 0.67 | + |
| TV14 | IYO_RS24905 | 23S rRNA pseudouridine synthase D | rluD | 1 | + | |
| TX17 | IYO_RS20385 | FAD-dependent 5-carboxymethylaminomethyl-2-thiouridine(34) oxidoreductase MnmC | mnmC | 1.67 | + | |
| TAd31 | IYO_RS05655 | Pyrimidine utilization protein D | 2 | + | ||
| TBb34 | IYO_RS25270 | Ribonuclease R | vacB | 2 | + | |
| TAm23 | IYO_RS27525 | Cell division ATP-binding protein FtsE | ftsE | 3 | + | |
| TBh17 | IYO_RS19355 | Endonuclease/exonuclease/phosphatase | 3 | + | ||
| TBl30 | IYO_RS08955 | Cytidylate kinase | cmk | 3 | + | |
| TAr33 | IYO_RS18310 | 23S rRNA (guanine(2445)-N(2))/(guanine(2069)-N(7))- methyltransferase | 3.33 | + | ||
| TBv24 | IYO_RS21455 | tRNA 5-methoxyuridine(34)/uridine 5-oxyacetic acid(34) synthase CmoB | 3.33 | + | ||
| Others | TAz15 | IYO_RS16115 | Monooxygenase | 2 | + | |
| TBc29 | IYO_RS29540 | Coenzyme F390 synthetase | 2 | + | ||
| TY16 | IYO_RS15475 | N-acetyltransferase | 2 | + | ||
| TH14 | IYO_RS15735 | Alcohol dehydrogenase | 2.67 | + | ||
| Peptidase | TT37 | IYO_RS02615 | Peptidase M23 | 1.33 | + | |
| TAc18 | IYO_RS27100 | Peptidase M20 | 2 | + | ||
| TBt26 | IYO_RS12305 | Zn-dependent hydrolase | 3.33 | + | ||
| Signal transduction/ Transcriptional regulator | TR05 | IYO_RS11995 | LysR family transcriptional regulator | lysR | 0.67 | + |
| TX21 | IYO_RS04255 | Sensor domain-containing phosphodiesterase | 2 | + | ||
| TAa14 | IYO_RS06430 | DNA-binding response regulator | 2.67 | + | ||
| TAa40 | IYO_RS16240 | Two-component sensor histidine kinase | colS | 3.33 | + | |
| TZ32 | IYO_RS07365 | Hybrid sensor histidine kinase/response regulator | 3.33 | + | ||
| TA08 | IYO_RS04485 | DNA-binding transcriptional activator OsmE | osmE | 4 | + | |
| TE04 | IYO_RS21745 | Sigma-54 dependent transcriptional regulator/ response regulator | 4 | + | ||
| Sulfur metabolism | TC13 | IYO_RS00620 | Pyridine nucleotide-disulfide oxidoreductase | 3.67 | + | |
| Type III secretion system/Type III secretion effector | TAh32 | IYO_RS06840 | Type III secretion system export apparatus switch protein | hrcU | 0 | + |
| TAl39 | IYO_RS06775 | Sigma-54-dependent Fis family transcriptional regulator | hrpR | 0 | + | |
| TBi11 | IYO_RS06800 | Type III secretion inner membrane ring protein | hrcJ | 0 | + | |
| TD35 | IYO_RS06775 | Sigma-54-dependent Fis family transcriptional regulator | hrpR | 0 | + | |
| TJ40 | IYO_RS06855 | Type III secretion system export apparatus protein | hrcR | 0 | + | |
| TAw18 | IYO_RS06905 | Type III helper protein HrpK1 | hrpK1 | 0.67 | + | |
| TR23 | IYO_RS24135 | Type III effector HopR1 | hopR1 | 0.67 | + | |
| TBw35 | IYO_RS06765 | Type III effector AvrE1 | avrE1 | 2.33 | + | |
| TAd35 | IYO_RS06770 | Lytic transglycosylase | hrpH | 2.67 | + | |
| TBr13 | IYO_RS25475 | Type III effector HopAC1 | hopAC1 | 3 | + | |
| TM33 | IYO_RS28930 | Avirulence protein HopZ3 | hopZ3 | 3.67 | + | |
| Transporters | TAy25 | IYO_RS23000 | Sugar ABC transporter ATP-binding protein | 1 | - | |
| TP3 | IYO_RS16960 | Ectoine/hydroxyectoine ABC transporter permease subunit EhuC | 1.33 | + | ||
| TBo16 | IYO_RS15485 | OmpA family protein | 1.67 | + | ||
| TW19 | IYO_RS02405 | Autotransporting lipase, GDSL family | 1.67 | - | ||
| TL23 | IYO_RS11430 | AI-2E family transporter | 2.33 | + | ||
| TY11 | IYO_RS21760 | MFS transporter | 2.67 | + | ||
| TBr15 | IYO_RS01165 | ABC transporter ATP-binding protein | 3 | + | ||
| TBu09 | IYO_RS27805 | Sulfonate ABC transporter | ssuC | 3.33 | + | |
| Type IV secretion system | TBr28 | IYO_RS07305 | Protein disulfide-isomerase | dsbC | 0.33 | - |
| TAd01 | IYO_RS08235 | Conjugative coupling factor TraD, PFGI-1 class | 2.67 | + | ||
| Xenobiotics biodegradation and metabolism | TU11 | IYO_RS15180 | Dienelactone hydrolase | 1.33 | + | |
| TBd36 | IYO_RS16315 | (2Fe-2S)-binding protein | vanA | 3 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishiga, T.; Sakata, N.; Usuki, G.; Nguyen, V.T.; Gomi, K.; Ishiga, Y. Large–Scale Transposon Mutagenesis Reveals Type III Secretion Effector HopR1 Is a Major Virulence Factor in Pseudomonas syringae pv. actinidiae. Plants 2023, 12, 141. https://doi.org/10.3390/plants12010141
Ishiga T, Sakata N, Usuki G, Nguyen VT, Gomi K, Ishiga Y. Large–Scale Transposon Mutagenesis Reveals Type III Secretion Effector HopR1 Is a Major Virulence Factor in Pseudomonas syringae pv. actinidiae. Plants. 2023; 12(1):141. https://doi.org/10.3390/plants12010141
Chicago/Turabian StyleIshiga, Takako, Nanami Sakata, Giyu Usuki, Viet Tru Nguyen, Kenji Gomi, and Yasuhiro Ishiga. 2023. "Large–Scale Transposon Mutagenesis Reveals Type III Secretion Effector HopR1 Is a Major Virulence Factor in Pseudomonas syringae pv. actinidiae" Plants 12, no. 1: 141. https://doi.org/10.3390/plants12010141
APA StyleIshiga, T., Sakata, N., Usuki, G., Nguyen, V. T., Gomi, K., & Ishiga, Y. (2023). Large–Scale Transposon Mutagenesis Reveals Type III Secretion Effector HopR1 Is a Major Virulence Factor in Pseudomonas syringae pv. actinidiae. Plants, 12(1), 141. https://doi.org/10.3390/plants12010141







