Three Parts of the Plant Genome: On the Way to Success in the Production of Recombinant Proteins
Abstract
:1. Introduction
2. Specific Features of the Organization of Nuclear–Cytoplasmic Transcriptional and Translational Machinery of the Plant Cell
2.1. Organization of Nuclear-Cytoplasmic Molecular Machinery for Protein Biosynthesis
2.2. Nuclear–Cytoplasmic Transcriptional and Translational Machinery as the Tool for the Biosynthesis of Recombinant Proteins
2.2.1. Stable Expression of Recombinant Proteins in Transgenic Plants
2.2.2. Stable Expression of Recombinant Proteins in Cultured Plant Cells (in Bioreactors)
2.2.3. Transient Expression
2.2.4. Cell-Free Expression Systems for Recombinant Proteins
2.3. Assessment of Biosynthetic Potential of Nuclear-Cytoplasmic Transcriptional and Translational Machinery: Problems and Possible Solutions
3. Specific Features of the Organization of the Plastid Transcriptional and Translational Machinery in the Plant Cell and Its Use as a Tool for the Biosynthesis of Recombinant Proteins
3.1. Structure of the Plastid Genome
3.2. Advantages of the Plastid Transcriptional and Translational Machinery for Recombinant Protein Biosynthesis
3.3. Ways to Raise the Rate of Plastid Genome Transformation
3.3.1. Methods for Delivering Exogenous DNA to the Plastid Genome
3.3.2. Designing Vectors for Plastid Transformation
Effects of Species Specificity of Spacer Sequences and of Selection of an Integration Site in the Plastome on the Insertion Rate and Expression of Recombinant Genes
Using the CRISPR/Cas9 Genome-Editing System
Effects of Promoters and 5′UTR and 3′UTR Regulatory Elements on the Expression of Recombinant Genes in Plastids
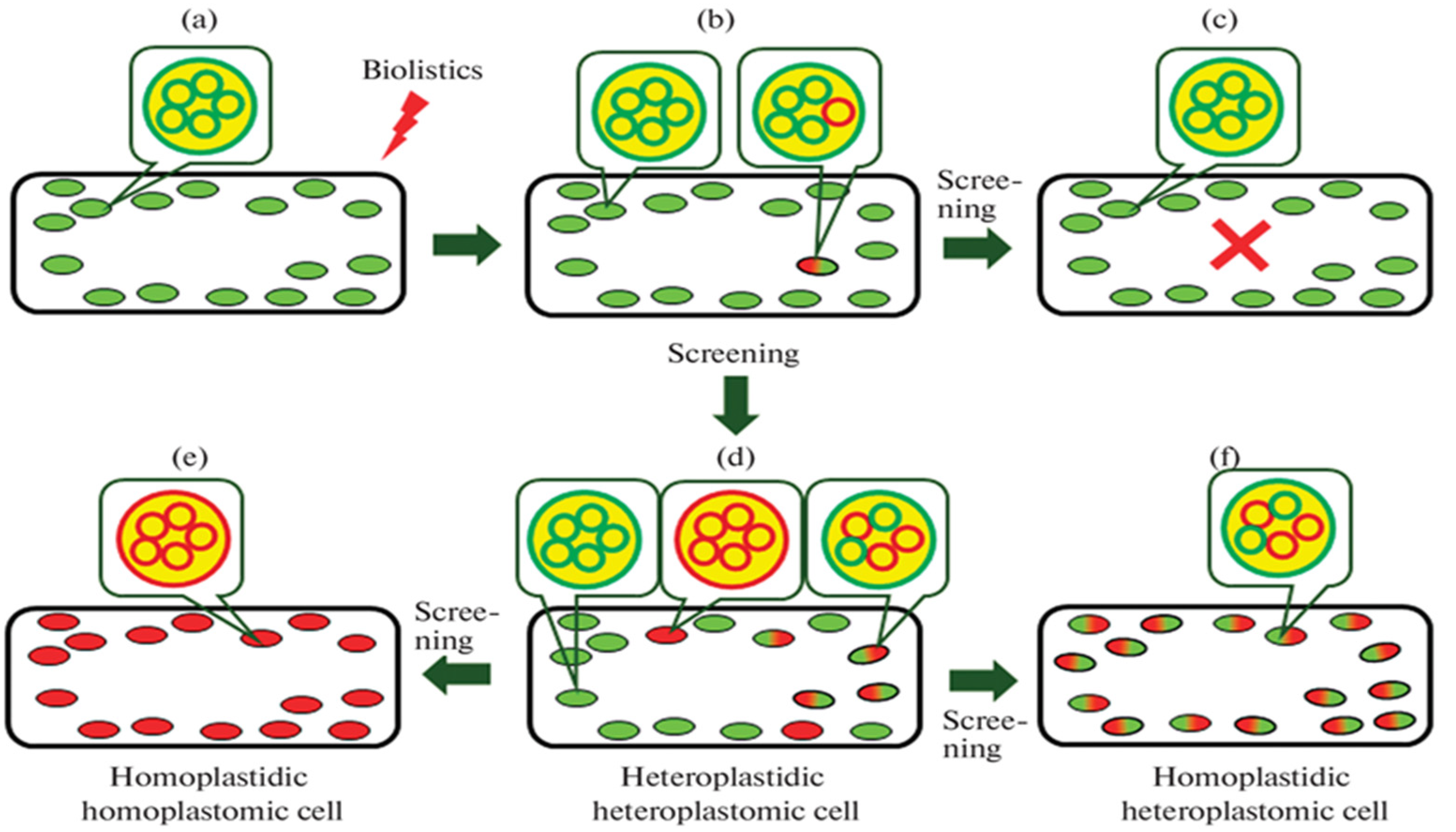
3.4. Methods for Constructing Homoplastomic and Homoplastidic Plants
3.4.1. Using Meristematic and Etiolated Callus Tissues to Transform Plastids
3.4.2. Selection and Selective Markers for Achieving Homoplastomy and a Homoplastidic State
3.5. Using Transplastomic Plants for Recombinant Protein Synthesis
4. The Mitochondrial Genome
4.1. Specific Features of the Organization and Function of the Plant Mitochondrial Genome
4.2. Prospects of Transcriptional and Translational Machinery of Plant Mitochondria for Recombinant Protein Biosynthesis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Plant-based Biologics and Expression Systems Global Market Report 2022: Profiles of Medicago, Leaf Expression Systems, Eleva, iBIO, PlantForm, G+Flas Life Sciences, Kentucky Bioprocessing and Angany. Available online: https://finance.yahoo.com/news/plant-based-biologics-expression-systems-101500030.html (accessed on 9 November 2022).
- Burnett, M.J.B.; Burnett, A.C. Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants People Planet. 2020, 2, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.researchandmarkets.com (accessed on 9 November 2022).
- Gill, K.S. Gene Distribution in Cereal Genomes; Gupta, P.K., Varshney, R.K., Eds.; Cereal Genomics; Kluwer Academic Publishers: Amsterdam, The Netherlands, 2004; pp. 361–384. [Google Scholar] [CrossRef]
- Dong, O.; Ronald, P. Genetic Engineering for Disease Resistance in Plants: Recent Progress and Future Perspectives. Plant Physiol. 2019, 180, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatehouse, A.M.R.; Ferry, N.; Edwards, M.G.; Bell, H.A. Insect-resistant biotech crops and their impacts on beneficial arthropods. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 1438–1452. [Google Scholar] [CrossRef] [Green Version]
- Wani, S.H.; Dutta, T.; Neelapu, N.R.R.; Surekha, C. Transgenic approaches to enhance salt and drought tolerance in plants. Plant Gene 2017, 11, 219–231. [Google Scholar] [CrossRef]
- Maghari, B.M.; Ardekani, A.M. Genetically modified foods and social concerns. Avicenna J. Med. Biotechnol. 2011, 3, 109–117, PMCID:PMC3558185. [Google Scholar] [PubMed]
- Paul, J.Y.; Khanna, H.; Kleidon, J.; Hoang, P.; Geijskes, J.; Daniells, J.; Zaplin, E.; Rosenberg, Y.; James, A.; Mlalazi, B.; et al. Golden bananas in the field: Elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 2017, 15, 520–532. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wohlhueter, R.; Zhang, H. Genetically modified foods: A critical review of their promise and problems. Food Sci. Hum. Wellness 2016, 5, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Liao, P.; Chen, X.; Wang, M.; Bach, T.J.; Chye, M.L. IMPROVED fruit α-tocopherol, carotenoid, squalene and phytosterol contents through manipulation of Brassica juncea 3-hydroxy-3-methylglutaryl-coa synthase1 in transgenic tomato. Plant Biotechnol. J. 2018, 16, 784–796. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Palve, A.; Joshi, C.; Srivastava, R.; Rukhsar, K. Crop biofortification for iron (Fe), zinc (Zn) and vitamin A with transgenic approaches. Heliyon 2019, 5, e01914. [Google Scholar] [CrossRef]
- Buyel, J.F.; Twyman, R.M.; Fischer, R. Very-large-scale production of antibodies in plants: The biologization of manufacturing. Biotechnol. Adv. 2017, 35, 458–665. [Google Scholar] [CrossRef]
- Chan, H.-T.; Daniell, H. Plant-made oral vaccines against human infectious diseases—Are we there yet? Plant Biotechnol. J. 2015, 13, 1056–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, J.L.; Waheed, M.T.; Lossl, A.G.; Martinussen, I.; Daniell, H. How can plant genetic engineering contribute to cost-effective fish vaccine development for promoting sustainable aquaculture? Plant Mol. Biol. 2013, 83, 33–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolotilin, I.; Topp, E.; Cox, E.; Devriendt, B.; Conrad, U.; Joensuu, J.; Stöger, E.; Warzecha, H.; McAllister, T.; Potter, A.; et al. Plant-based solutions for veterinary immunotherapeutics and prophylactics. Vet. Res. 2014, 45, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacDonald, J.; Doshi, K.; Dussault, M.; Hall, J.C.; Holbrook, L.; Jones, G.; Kaldis, A.; Klima, C.L.; Macdonald, P.; McAllister, T.; et al. Bringing plant-based veterinary vaccines to market: Managing regulatory and commercial hurdles. Biotechnol. Adv. 2015, 33, 1572–1581. [Google Scholar] [CrossRef] [Green Version]
- Schillberg, S.; Finnern, R. Plant molecular farming for the production of valuable proteins—Critical evaluation of achievements and future challenges. J. Plant Physiol. 2021, 258–259, 153359. [Google Scholar] [CrossRef]
- Park, K.Y.; Wi, S.J. Potential of Plants to Produce Recombinant Protein Products. J. Plant Biol. 2016, 59, 559–568. [Google Scholar] [CrossRef]
- Zagorskaya, A.A.; Deineko, E.V. Plant-Expression Systems: A New Stage in Production of Biopharmaceutical Preparations. Russ. J. Plant Physiol. 2021, 68, 17–30. [Google Scholar] [CrossRef]
- Gurusamy, P.D.; Schafer, H.; Ramamoorthy, S.; Wink, M. Biologically active recombinant human erythropoietin expressed in hairy root cultures and regenerated plantlets of Nicotiana tabacum L. PLoS ONE 2017, 12, e0182367. [Google Scholar] [CrossRef] [Green Version]
- Lenders, M.; Brand, E. Fabry disease: The current treatment landscape. Drugs 2021, 81, 635–645. [Google Scholar] [CrossRef]
- Limkul, J.; Misaki, R.; Kato, K.; Fujiyama, K. The combination of plant translational enhancers and terminator increase the expression of human glucocerebrosidase in Nicotiana benthamiana plants. Plant Sci. 2015, 240, 41–49. [Google Scholar] [CrossRef]
- Tan, Y.; Wang, K.Y.; Wang, N.; Li, G.; Liu, D. Ectopic expression of human acidic fibroblast growth factor 1 in the medicinal plant, Salvia miltiorrhiza, accelerates the healing of burn wounds. BMC Biotechnol. 2014, 9, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, C.; Du, X.; Wang, G.; Ji, J.; Jin, C.; Li, X. Expression of biologically active anti-thrombosis protein lumbrokinase in edible sunflower seed kernel. J. Plant Biochem. Biotechnol. 2014, 23, 257–265. [Google Scholar] [CrossRef]
- Reuter, L.J.; Bailey, M.J.; Joensuu, J.J.; Ritala, A. Scale-up of hydrophobin-assisted recombinant protein production in tobacco BY-2 suspension cells. Plant Biotechnol. J. 2014, 12, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.G.; Baek, M.Y.; Lee, E.K.; Kwon, T.H.; Yang, M.S. Expression of human growth hormone in transgenic rice cell suspension culture. Plant Cell Rep. 2008, 27, 885–891. [Google Scholar] [CrossRef]
- Ding, S.H.; Huang, L.Y.; Wang, Y.D.; Sun, H.C.; Xiang, Z.H. High-level expression of basic fibroblast growth factor in transgenic soybean seeds and characterization of its biological activity. Biotechnol. Lett. 2006, 28, 869–875. [Google Scholar] [CrossRef]
- Komarnytsky, S.; Borisjuk, N.V.; Borisjuk, L.G.; Alam, M.Z.; Raskin, I. Production of Recombinant Proteins in Tobacco Guttation Fluid. Plant Physiol. 2000, 124, 927–933. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.oryzogen.net (accessed on 9 November 2022).
- Available online: https://www.sigmaaldrich.com (accessed on 9 November 2022).
- Available online: https://www.thermofisher.com (accessed on 9 November 2022).
- Available online: https://www.orfgenetics.com (accessed on 9 November 2022).
- Ma, J.K.-C.; Drossard, J.; Lewis, D.; Altmann, F.; Boyle, J.; Christou, P.; Cole, T.; Dale, P.; van Dolleweerd, C.J.; Isitt, V.; et al. Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants. Plant Biotechnol. J. 2015, 13, 1106–1120. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Velázquez, A.; López-Quesada, A.; Ceballo-Cámara, Y.; Cabrera-Herrera, G.; Tiel-González, K.; Mirabal-Ortega, L.; Pérez-Martínez, M.; Pérez-Castillo, R.; Rosabal-Ayán, Y.; Ramos-González, O.; et al. Tobacco seeds as efficient production platform for a biologically active anti-HBsAg monoclonal antibody. Transgenic Res. 2015, 24, 897–909. [Google Scholar] [CrossRef]
- Sabalza, M.; Madeira, L.; van Dolleweerd, C.; Ma, J.K.; Capell, T.; Christou, P. Functional characterization of the recombinant HIV-neutralizing monoclonal antibody 2F5 produced in maize seeds. Plant Mol. Biol. 2012, 80, 477–488. [Google Scholar] [CrossRef]
- Paul, M.J.; Teh, A.Y.; Twyman, R.M.; Ma, J.K. Target product selection—Where can molecular pharming make the difference? Curr. Pharm. 2013, 19, 5478–5485. [Google Scholar] [CrossRef] [Green Version]
- Biesgen, C.; Hillebrand, H.; Herbers, K. Technical enzymes produced in transgenic plants. Phytochem. Rev. 2002, 1, 79–85. [Google Scholar] [CrossRef]
- Hood, E.; Witcher, D.; Maddock, S.; Meyer, T.; Baszczynski, C.; Bailey, M.; Flynn, P.; Register, J.; Marshall, L.; Bond, D.; et al. Commercial production of avidin from transgenic maize: Characterization of transformant, production, processing, extraction and purification. Mol. Breed. 1997, 3, 291–306. [Google Scholar] [CrossRef]
- Krishnan, A.; Woodard, S. TryZean™: An animal-free alternative to bovine trypsin. In Commercial Plant-Produced Recombinant Protein Products. Biotechnology in Agriculture and Forestry; Howard, J., Hood, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 69, pp. 43–63. [Google Scholar] [CrossRef]
- Bornke, F.; Broer, I. Tailoring plant metabolism for the production of novel polymers and platform chemicals. Curr. Opin. Plant Biol. 2010, 13, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Meng, Q.; Meng, K.; Zhang, W.; Zhou, X.; Luo, H.; Chen, R.; Yang, P.; Yao, B. Overexpression of a fungal beta-mannanase from Bispora sp. MEY-1 in maize seeds and enzyme characterization. PLoS ONE 2013, 8, e56146. [Google Scholar] [CrossRef] [Green Version]
- Available online: http://www.sigmaaldrich.com (accessed on 9 November 2022).
- Available online: http://www.agrenvec.com (accessed on 9 November 2022).
- Available online: http://www.orfgenetics.com (accessed on 9 November 2022).
- Available online: http://www.sifcosmetics.com (accessed on 9 November 2022).
- Available online: http://www.invitria.com (accessed on 9 November 2022).
- Available online: http://www.kbpllc.com (accessed on 9 November 2022).
- Available online: http://www.collplant.com (accessed on 9 November 2022).
- Available online: http://www.nbms.co.kr (accessed on 9 November 2022).
- Available online: http://gndp.cigb.edu.cu (accessed on 9 November 2022).
- Available online: http://www.syngenta.com (accessed on 9 November 2022).
- Available online: http://www.originseed.com.cn (accessed on 9 November 2022).
- Available online: http://www.nexgen.com (accessed on 9 November 2022).
- Wu, T.; Kerbler, S.M.; Fernie, A.R.; Zhang, Y. Plant cell cultures as heterologous bio-factories for secondary metabolite production. Plant Commun. 2021, 2, 100235. [Google Scholar] [CrossRef]
- Schillberg, S.; Raven, N.; Fischer, R.; Twyman, R.; Schiermeyer, A. Molecular farming of pharmaceutical proteins using plant suspension cell and tissue cultures. Curr. Pharm. Des. 2013, 19, 5531–5542. [Google Scholar] [CrossRef]
- Schiermeyer, A. Optimizing product quality in molecular farming. Curr. Opin. Biotechnol. 2020, 61, 15–20. [Google Scholar] [CrossRef]
- Dicker, M.; Tschofen, M.; Maresch, D.; König, J.; Juarez, P.; Orzaez, D.; Altmann, F.; Steinkellner, H.; Strasser, R. Transient Glyco-Engineering to Produce Recombinant IgA1 with Defined N- and O-Glycans in Plants. Front. Plant Sci. 2016, 7, 18. [Google Scholar] [CrossRef]
- Mercx, S.; Smargiasso, N.; Chaumont, F.; De Pauw, E.; Boutry, M.; Navarre, C. Inactivation of the β(1,2)xylosyl transferase and the α(1,3)fucosyltransferase genes in Nicotiana tabacum BY2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant specific glycans. Front. Plant Sci. 2017, 8, 403. [Google Scholar] [CrossRef] [Green Version]
- Hanania, U.; Ariel, T.; Tekoah, Y.; Fux, L.; Gubbay, Y.; Weiss, M.; Oz, D.; Azulay, Y.; Turbovsky, A.; Forster, Y.; et al. Establishment of a tobacco BY2 cell line devoid of plant specific xylose and fucose as a platform for the production of biotherapeutic proteins. Plant Biotechnol. J. 2017, 15, 1120. [Google Scholar] [CrossRef] [Green Version]
- Shaaltiel, Y.; Hashmueli, S.; Bartfeld, D.; Baum, G.; Ratz, T.; Mizrachi, E.; Forster, Y. System and Method for Production of Antibodies in Plant Cell Culture. Patent no.: US8119406B2, 21 February 2012. [Google Scholar]
- Moon, K.B.; Park, J.S.; Park, Y.I.; Song, I.J.; Lee, H.J.; Cho, H.S.; Jeon, J.H.; Kim, H.S. Development of Systems for the Production of Plant-Derived Biopharmaceuticals. Plants 2019, 9, 30. [Google Scholar] [CrossRef] [Green Version]
- Kwon, J.Y.; Yang, Y.S.; Cheon, S.H.; Nam, H.J.; Jin, G.H.; Kim, D.I. Bioreactor engineering using disposable technology for enhanced production of hCTLA4Ig in transgenic rice cell cultures. Biotechnol. Bioeng. 2013, 110, 2412–2424. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-F.; Tan, C.-C.; Yeh, J.-F.; Liu, H.-Y.; Liu, Y.-K.; Ho, S.-L.; Lu, C.-A. Efficient secretion of recombinant proteins from rice suspension-cultured cells modulated by the choice of signal peptide. PLoS ONE 2015, 10, e0140812. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-W.; Kim, N.-S. Production of functional recombinant cyclic citrullinated peptide monoclonal antibody in transgenic rice cell suspension culture. Transgenic. Res. 2019, 28, 177–188. [Google Scholar] [CrossRef]
- Islam, M.R.; Kim, N.S.; Jung, J.W.; Kim, H.B.; Han, S.C.; Yang, M.S. Spontaneous pepsin C-catalyzed activation of human pepsinogen C in transgenic rice cell suspension culture: Production and characterization of human pepsin C. Enzym. Microb. Technol. 2018, 108, 66–73. [Google Scholar] [CrossRef]
- Nam, H.J.; Kwon, J.Y.; Choi, H.Y.; Kang, S.H.; Jung, H.S.; Kim, D.I. Production and Purification of Recombinant Glucocerebrosidase in Transgenic Rice Cell Suspension Cultures. Appl. Biochem. Biotechnol. 2017, 181, 1401–1415. [Google Scholar] [CrossRef]
- Lu, C.-A.; Lim, E.-K.; Yu, S.-M. Sugar response sequence in the promoter of a rice α-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 1998, 273, 10120–10131. [Google Scholar] [CrossRef] [Green Version]
- Chung, N.D.; Kim, N.S.; Giap, D.V.; Jang, S.H.; Oh, S.M.; Jang, S.H.; Kim, T.G.; Jang, Y.S.; Yang, M.S. Production of functional human vascular endothelial growth factor(165) in transgenic rice cell suspension cultures. Enzyme Microb. Technol. 2014, 63, 58–63. [Google Scholar] [CrossRef]
- Tekoah, Y.; Shulman, A.; Kizhner, T.; Ruderfer, I.; Fux, L.; Nataf, Y.; Bartfeld, D.; Ariel, T.; Gingis–Velitski, S.; Hanania, U.; et al. Large-scale production of pharmaceutical proteins in plant cell culture—The protalix experience. Plant Biotechnol. J. 2015, 13, 1199–1208. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; Tello-Olea, M.A. Carrot Cells: A Pioneering Platform for Biopharmaceuticals Production. Mol. Biotechnol. 2015, 57, 3. [Google Scholar] [CrossRef]
- Hellwig, S.; Drossard, J.; Twyman, R.M.; Fischer, R. Plant cell cultures for the production of recombinant proteins. Nat. Biotechnol. 2004, 22, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.B.; Chandrasekar, B.; Mandal, M.K.; Kaschani, F.; Kaiser, M.; Both, L.; van der Hoorn, R.A.L.; Schiermeyer, A.; Abranches, R. Low protease content in Medicago truncatula cell cultures facilitates recombinant protein production. Biotechnol. J. 2018, 13, e1800050. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-K.; Falk, B.W.; Dandekar, A.M.; McDonald, K.A. Enhancement of Recombinant Protein Production in Transgenic Nicotiana benthamiana Plant Cell Suspension Cultures with Co-Cultivation of Agrobacterium Containing Silencing Suppressors. Int. J. Mol. Sci. 2018, 19, 1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukenik, S.C.; Karuppanan, K.; Li, Q.; Lebrilla, C.B.; Nandi, S.; McDonald, K.A. Transient Recombinant Protein Production in Glycoengineered Nicotiana benthamiana Cell Suspension Culture. Int. J. Mol. Sci. 2018, 16, 1205. [Google Scholar] [CrossRef] [Green Version]
- Donini, M.; Marusic, C. Current state-of-the-art in plant-based antibody production systems. Biotechnol. Lett. 2019, 41, 335–346. [Google Scholar] [CrossRef]
- Chen, Q.; Lai, H. Gene delivery into plant cells for recombinant protein production. Biomed. Res. Int. 2015, 3, 932161. [Google Scholar] [CrossRef] [Green Version]
- Tyurin, A.A.; Suhorukova, A.V.; Kabardaeva, K.V.; Goldenkova-Pavlova, I.V. Transient Gene Expression is an Effective Experimental Tool for the Research into the Fine Mechanisms of Plant Gene Function: Advantages, Limitations, and Solutions. Plants 2020, 9, 1187. [Google Scholar] [CrossRef]
- Norkunas, K.; Harding, R.; Dale, J.; Dugdale, B. Improving agroinfiltration-based transient gene expression in Nicotiana benthamiana. Plant Methods 2018, 14, 71. [Google Scholar] [CrossRef]
- Yamamoto, T.; Hoshikawa, K.; Ezura, K.; Okazawa, R.; Fujita, S.; Takaoka, M.; Mason, H.S.; Ezura, H.; Miura, K. Improvement of the transient expression system for production of recombinant proteins in plants. Sci. Rep. 2018, 8, 4755. [Google Scholar] [CrossRef] [Green Version]
- Vaquero, C.; Sack, M.; Chandler, M.J.; Drossard, J.; Schuster, F.; Monecke, M.; Schillberg, S.; Fischer, R. Transient expression of a tumor-specific single-chain fragmentand a chimeric antibody in tobacco leaves. Proc. Natl. Acad. Sci. USA 1999, 96, 11128–11133. [Google Scholar] [CrossRef] [Green Version]
- D’Aoust, M.A.; Couture, M.M.; Charland, N.; Trépanier, S.; Landry, N.; Ors, F.; Vézina, L.P. The production of hemagglutinin based virus-like particles in plants: A rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 2010, 8, 5–607. [Google Scholar] [CrossRef] [PubMed]
- Shoji, Y.; Chichester, J.A.; Bi, H.; Musiychuk, K.; de la Rosa, P.; Goldschmidt, L.; Horsey, A.; Ugulava, N.; Palmer, G.A.; Mett, V.; et al. Plant-expressed HA as a seasonal influenza vaccine candidate. Vaccine 2008, 26, 2930. [Google Scholar] [CrossRef]
- Shoji, Y.; Chichester, J.A.; Jones, M.; Manceva, S.D.; Damon, E.; Mett, V.; Musiychuk, K.; Bi, H.; Farrance, C.; Shamloul, M.; et al. Plant-based rapid production of recombinant subunit hemagglutinin vaccines targeting H1N1 and H5N1 influenza. Hum. Vaccin. 2011, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Na, W.; Park, N.; Yeom, M.; Song, D. Ebola outbreak in Western Africa 2014: What is going on with Ebola virus? Clin. Exp. Vaccine Res. 2015, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybicki, E.P. Plant molecular farming of virus-like nanoparticles as vaccines and reagents. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1587. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://medicago.com/en/our-science/our-vaccine-candidates/novovirus/ (accessed on 9 November 2022).
- Pillet, S.; Couillard, J.; Trépanier, S.; Poulin, J.-F.; Yassine-Diab, B.; Guy, B.; Ward, B.J.; Landry, N. Immunogenicity and safety of a quadrivalent plant-derived virus like particle influenza vaccine candidate—Two randomized Phase II clinical trials in 18 to 49 and ≥50 years old adults. PLoS ONE 2019, 14, e0216533. [Google Scholar] [CrossRef] [Green Version]
- Capell, T.; Twyman, R.M.; Armario-Najera, V.; Ma, J.K.; Schillberg, S.; Christou, P. Potential Applications of Plant Biotechnology against SARS-CoV-2. Trends Plant Sci. 2020, 25, 635–643. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; Comas-García, M.; Korban, S.S. Challenges and Opportunities for the Biotechnology Research Community during the Coronavirus Pandemic. Trends Biotechnol. 2020, 38, 823–824. [Google Scholar] [CrossRef]
- Hemmati, F.; Hemmati-Dinarvand, M.; Karimzade, M.; Rutkowska, D.; Eskandari, M.H.; Khanizadeh, S.; Afsharifar, A. Plant-Derived VLP: A Worthy Platform to Produce Vaccine against SARS-CoV-2. Biotechnol. Lett. 2022, 44, 45–57. [Google Scholar] [CrossRef]
- Maharjan, P.M.; Choe, S. Plant-Based COVID-19 Vaccines: Current Status, Design, and Development Strategies of Candidate Vaccines. Vaccines 2021, 9, 992. [Google Scholar] [CrossRef]
- Diego-Martin, B.; González, B.; Vazquez-Vilar, M.; Selma, S.; Mateos-Fernández, R.; Gianoglio, S.; Fernández-del-Carmen, A.; Orzáez, D. Pilot Production of SARS-CoV-2 Related Proteins in Plants: A Proof of Concept for Rapid Repurposing of Indoor Farms into Biomanufacturing Facilities. Front. Plant Sci. 2020, 11, 2101. [Google Scholar] [CrossRef]
- Kumar, M.; Kumari, N.; Thakur, N.; Bhatia, S.K.; Saratale, G.D.; Ghodake, G.; Mistry, B.M.; Alavilli, H.; Kishor, D.S.; Du, X.; et al. A Comprehensive Overview on the Production of Vaccines in Plant-Based Expression Systems and the Scope of Plant Biotechnology to Combat against SARS-CoV-2 Virus Pandemics. Plants 2021, 10, 1213. [Google Scholar] [CrossRef] [PubMed]
- Peyret, H.; Steele, J.F.C.; Jung, J.-W.; Thuenemann, E.C.; Meshcheriakova, Y.; Lomonossoff, G.P. Producing Vaccines against Enveloped Viruses in Plants: Making the Impossible, Difficult. Vaccines 2021, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Mamedov, T.; Yuksel, D.; Ilgın, M.; Gürbüzaslan, I.; Gulec, B.; Mammadova, G.; Ozdarendeli, A.; Yetiskin, H.; Kaplan, B.; Islam Pavel, S.T.; et al. Production and Characterization of Nucleocapsid and RBD Cocktail Antigens of SARS-CoV-2 in Nicotiana Benthamiana Plant as a Vaccine Candidate against COVID-19. Vaccines 2021, 9, 1337. [Google Scholar] [CrossRef] [PubMed]
- Rybicki, E.P. Plant-based vaccines against viruses. Virol. J. 2014, 11, 205–225. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.L.; Paruch, L.; Dobrica, M.-O.; Caras, I.; Tucureanu, C.; Onu, A.; Ciulean, S.; Stavaru, C.; Eerde, A.; Wang, Y.; et al. Lettuce-produced hepatitis C virus E1E2 heterodimer triggers immune responses in mice and antibody production after oral vaccination. Plant Biotech. J. 2017, 15, 1611–1621. [Google Scholar] [CrossRef] [Green Version]
- Naupu, P.N.; van Zyl, A.R.; Rybicki, E.P.; Hitzeroth, I.I. Immunogenicity of Plant-Produced Human Papillomavirus (HPV) Virus-Like Particles (VLPs). Vaccine 2020, 8, 740. [Google Scholar] [CrossRef]
- Kasinger, S.L.E.; Dent, M.W.; Mahajan, G.; Hamorsky, K.T.; Matoba, N. A novel anti-HIV-1 bispecific bNAb-lectin fusion protein engineered in a plant-based transient expression system. Plant Biotechnol. J. 2019, 17, 1646–1656. [Google Scholar] [CrossRef]
- Nessa, M.U.; Rahman, M.A.; Kabir, Y. Plant-Produced Monoclonal Antibody as Immunotherapy for Cancer. Biomed. Res. Int. 2020, 24, 3038564. [Google Scholar] [CrossRef]
- Buyel, J.F. Plants as sources of natural and recombinant anti-cancer agents. Biotechnol. Adv. 2018, 36, 506–520. [Google Scholar] [CrossRef]
- Gengenbach, B.B.; Keil, L.L.; Opdensteinen, P.; Müschen, C.R.; Melmer, G.; Lentzen, H.; Bührmann, J.; Buyel, J.F. Comparison of microbial and transient expression (tobacco plants and plant-cell packs) for the production and purification of the anticancer mistletoe lectin viscumin. Biotechnol. Bioeng. 2019, 116, 2236–2249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knodler, M.J.; Buyel, F. Plant-made immunotoxin building blocks: A roadmap for producing therapeutic antibody-toxin fusions. Biotechnol. Adv. 2021, 47, 107683. [Google Scholar] [CrossRef] [PubMed]
- Nirenberg, M.W.; Matthaei, H.J. The Dependence of Cell-Free Protein Synthesis In E. coli Upon Naturally Occurring or Synthetic Polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 15, 1588–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buntru, M.; Vogel, S.; Finnern, R.; Schillberg, S. Plant-Based Cell-Free Transcription and Translation of Recombinant Proteins. In Recombinant Proteins in Plants. Methods in Molecular Biology; Schillberg, S., Spiegel, H., Eds.; Humana: New York, NY, USA, 2022; Volume 2480. [Google Scholar] [CrossRef]
- Schillberg, S.; Raven, N.; Spiegel, H.; Rasche, S.; Buntru, M. Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front. Plant. Sci. 2019, 10, 720. [Google Scholar] [CrossRef] [Green Version]
- Buntru, M.; Vogel, S.; Spiegel, H.; Schillberg, S. Tobacco BY-2 cell-free lysate: An alternative and highly-productive plant-based in vitro translation system. BMC Biotechnol. 2014, 14, 37. [Google Scholar] [CrossRef] [Green Version]
- Buntru, M.; Vogel, S.; Stoff, K.; Spiegel, H.; Schillberg, S. A versatile coupled cell-free transcription-translation system based on tobacco BY-2 cell lysates. Biotechnol. Bioeng. 2015, 112, 867–878. [Google Scholar] [CrossRef]
- Available online: www.leniobio.com/alice (accessed on 9 November 2022).
- Havenith, H.; Kern, C.; Rautenberger, P.; Spiegel, H.; Szardenings, M.; Ueberham, E.; Lehmann, J.; Buntru, M.; Vogel, S.; Treudler, R.; et al. Combination of two epitope identification techniques enables the rational design of soy allergen Gly m 4 mutants. Biotechnol. J. 2017, 12, 1600441. [Google Scholar] [CrossRef]
- Huck, N.V.; Leissing, F.; Majovsky, P.; Buntru, M.; Aretz, C.; Flecken, M.; Müller, J.P.; Vogel, S.; Schillberg, S.; Hoehenwarter, W.; et al. Combined 15N-labeling and tandemMOAC quantifies phosphorylation of MAP kinase substrates downstream of MKK7 in Arabidopsis. Front. Plant Sci. 2017, 8, 2050. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, H.; Stöger, E.; Twyman, R.M.; Buyel, J.F. Current Status and Perspectives of the Molecular Farming Landscape. In Molecular Pharming: Applications, Challenges, and Emerging Areas; Allison, R., Kermode, L.J., Eds.; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2018; pp. 1–23. [Google Scholar] [CrossRef]
- Shanmugaraj, B.; Bulaon, C.J.I.; Malla, A.; Phoolcharoen, W. Biotechnological insights on the expression and production of antimicrobial peptides in plants. Molecules 2021, 26, 4032. [Google Scholar] [CrossRef]
- Fischer, R.; Schillberg, S.; Buyel, F.J.; Twyman, M.R. Commercial Aspects of Pharmaceutical Protein Production in Plants. Curr. Pharm. Des. 2013, 19, 5471–5477. [Google Scholar] [CrossRef] [Green Version]
- Sethi, L.; Kumari, K.; Dey, N. Engineering of Plants for Efficient Production of Therapeutics. Mol. Biotechnol. 2021, 63, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- Rozov, S.M.; Deineko, E.V. Strategies for Optimizing Recombinant Protein Synthesis in Plant Cells: Classical Approaches and New Directions. Mol. Biol. 2019, 53, 157–175. [Google Scholar] [CrossRef]
- Dong, O.X.; Ronald, P.C. Targeted DNA insertion in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2004834117. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Ishii, H.; Aoyagi, H.; Ohme-Takagi, M.; Tanaka, H. Comparative analyses of the gene expression profiles of Arabidopsis intact plant and cultured cells. Biotechnol. Lett. 2005, 27, 1097–1103. [Google Scholar] [CrossRef]
- Tanurdzic, M.; Vaughn, M.W.; Jiang, H.; Lee, T.J.; Slotkin, R.K.; Sosinski, B.; Thompson, W.F.; Doerge, R.W.; Martienssen, R.A. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008, 6, e302–e395. [Google Scholar] [CrossRef] [PubMed]
- Rozov, S.M.; Permyakova, N.V.; Sidorchuk, Y.V.; Deineko, E.V. Optimization of Genome Knock-In Method: Search for the Most Efficient Genome Regions for Transgene Expression in Plants. Int. J. Mol. Sci. 2022, 23, 4416. [Google Scholar] [CrossRef]
- Permyakova, N.V.; Marenkova, T.V.; Belavin, P.A.; Zagorskaya, A.A.; Sidorchuk, Y.V.; Uvarova, E.A.; Kuznetsov, V.V.; Rozov, S.M.; Deineko, E.V. Assessment of the Level of Accumulation of the dIFN Protein Integrated by the Knock-In Method into the Region of the Histone H3.3 Gene of Arabidopsis thaliana. Cells 2021, 8, 2137. [Google Scholar] [CrossRef]
- Kim, T.G.; Lee, H.J.; Jang, Y.S.; Shin, Y.J.; Kwon, T.H.; Yang, M.S. Co-expression of proteinase inhibitor enhances recombinant human granulocyte-macrophage colony stimulating factor production in transgenic rice cell suspension culture. Protein. Expr. Purif. 2008, 61, 117–121. [Google Scholar] [CrossRef]
- Niemer, M.; Mehofer, U.; Torres Acosta, J.A.; Verdianz, M.; Henkel, T.; Loos, A.; Strasser, R.; Maresch, D.; Rademacher, T.; Steinkellner, H.; et al. The human anti-HIVantibodies 2F5, 2G12, and PG9 differint heir susceptibility to proteolytic degradation: Down-regulation of endogenous serine and cysteine proteinase activities could improve antibody production in plant-based expression platforms. Biotechnol. J. 2014, 9, 493–500. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.S.; Kim, T.G.; Kim, O.H.; Ko, E.M.; Jang, Y.S.; Jung, E.S.; Kwon, T.H.; Yang, M.S. Improvement of recombinant hGM-CSF production by suppression of cysteine proteinase gene expression using RNA interference in a transgenic rice culture. Plant Mol. Biol. 2008, 68, 263–275. [Google Scholar] [CrossRef]
- Xu, J.; Ge, X.; Dolan, M.C. Towards high-yield production of pharmaceutical proteins with plant cell suspension cultures. Biotechnol. Adv. 2011, 29, 278–299. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Dolan, M.C.; Medrano, G.; Cramer, C.L.; Weathers, P.J. Green factory: Plants as bioproduction platforms for recombinant proteins. Biotechnol. Adv. 2012, 30, 1171–1184. [Google Scholar] [CrossRef]
- Rozov, S.M.; Permyakova, N.V.; Deineko, E.V. Main Strategies of Plant Expression System Glycoengineering for Producing Humanized Recombinant Pharmaceutical Proteins. Biochemistry 2018, 83, 215–232. [Google Scholar] [CrossRef]
- Pogson, B.J.; Ganguly, D.; Albrecht-Borth, V. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 2015, 1847, 1017–1024. [Google Scholar] [CrossRef] [Green Version]
- Mellor, S.B.; Behrendorff, J.B.Y.H.; Nielsen, A.Z.; Jensen, P.E.; Pribil, M. Non-photosynthetic plastids as hosts for metabolic engineering. Essays Biochem. 2018, 62, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farquhar, J.; Zerkle, A.L.; Bekker, A. Geological constraints on the origin of oxygenic photosynthesis. Photosynth. Res. 2011, 107, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Stroher, E.; Millar, A.H. Arabidopsis organelle isolation and characterization. Methods Mol. Biol. 2014, 1062, 551–572. [Google Scholar] [CrossRef]
- Palmer, J.D.; Osorio, B.; Aldrich, J.; Thompson, W.F. Chloroplast DNA evolution among legumes: Loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr. Genet. 1987, 11, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [Green Version]
- Barkan, A. Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011, 155, 1520–1532. [Google Scholar] [CrossRef] [Green Version]
- Prikryl, J.; Rojas, M.; Schuster, G.; Barkan, A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA 2011, 108, 415–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gawronski, P.; Jensen, P.E.; Karpinski, S.; Leister, D.; Scharff, L.B. Pausing of chloroplast ribosomes is induced by multiple features and is linked to the assembly of photosynthetic complexes. Plant Physiol. 2018, 176, 2557–2569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vries, J.; Archibald, J.M. Plastid genomes. Curr. Biol. 2018, 28, R336–R337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldenburg, D.J.; Bendich, A.J. DNA maintenance in plastids and mitochondria of plants. Front. Plant Sci. 2015, 6, 883. [Google Scholar] [CrossRef] [Green Version]
- Mehmood, F.; Abdullah, U.Z.; Shahzadi, I.; Ahmed, I.; Waheed, M.T.; Poczai, P.; Mirza, B. Plastid genomics of Nicotiana (Solanaceae): Insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 2020, 8, e9552. [Google Scholar] [CrossRef]
- Sakamoto, W.; Takami, T. Chloroplast DNA dynamics: Copy number, quality control and degradation. Plant Cell Physiol. 2018, 59, 1120–1127. [Google Scholar] [CrossRef]
- Rose, R.J. Sustaining Life: Maintaining Chloroplasts and Mitochondria and their Genomes in Plants. Yale J. Biol. Med. 2019, 92, 499–510. [Google Scholar]
- Zoschke, R.; Bock, R. Chloroplast Translation: Structural and Functional Organization, Operational Control, and Regulation. Plant Cell 2018, 30, 745–770. [Google Scholar] [CrossRef] [Green Version]
- Tadini, L.; Jeran, N.; Peracchio, C.; Masiero, S.; Colombo, M.; Pesaresi, P. The plastid transcription machinery and its coordination with the expression of nuclear genome: Plastid-Encoded Polymerase, Nuclear-Encoded Polymerase and the Genomes Uncoupled 1-mediated retrograde communication. Phil. Trans. R Soc. 2020, B375, 20190399. [Google Scholar] [CrossRef]
- Jarvis, P.; Lopez-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef]
- Bock, R. Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 2015, 66, 211–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, K.E.; Svab, Z.; Schaaf, D.J.; Hogan, P.S.; Stalker, D.M.; Maliga, P. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology 1995, 13, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, P.; Armarego-Marriott, T.; Bock, R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 2018, 49, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Mehmood, M.A.; Malik, S. Recombinant protein production in microalgae: Emerging trends. Protein Pept. Lett. 27, 105–110. [CrossRef]
- Dreesen, I.A.J.; Charpin-El Hamri, G.; Fussenegger, M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J. Biotechnol. 2010, 145, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Gregory, J.A.; Topol, A.B.; Doerner, D.Z.; Mayfield, S. Alga-produced cholera toxin-Pfs25 fusion proteins as oral vaccines. Appl. Environ. Microbiol. 2013, 79, 3917–3925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boynton, J.E.; Gillham, N.W.; Harris, E.H.; Hosler, J.P.; Johnson, A.M.; Jones, A.R.; Randolf-Anderson, B.L.; Robertson, D.; Klein, T.M.; Shark, K.B.; et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 1988, 240, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- Svab, Z.; Hajdukiewicz, P.; Maliga, P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA 1990, 87, 8526–8530. [Google Scholar] [CrossRef] [Green Version]
- Tamburino, R.; Marcolongo, L.; Sannino, L.; Ionata, E.; Scotti, N. Plastid Transformation: New Challenges in the Circular Economy Era. Int. J. Mol. Sci. 2022, 23, 15254. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Daniell, H. The engineered chloroplast genome just got smarter. Trends in Plant Sci. 2015, 20, 622–640. [Google Scholar] [CrossRef] [Green Version]
- Ruf, S.; Forner, J.; Hasse, C.; Kroop, X.; Seeger, S.; Schollbach, L.; Schadach, A.; Bock, R. High-efficiency generation of fertile transplastomic Arabidopsis plants. Nat. Plants 2019, 5, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Cutolo, E.A.; Mandalà, G.; Dall’Osto, L.; Bassi, R. Harnessing the Algal Chloroplast for Heterologous Protein Production. Microorganisms 2022, 10, 743. [Google Scholar] [CrossRef] [PubMed]
- Kumari, U.; Singh, R.; Ray, T.; Rana, S.; Saha, P.; Malhotra, K.; Daniell, H. Validation of leaf enzymes in the detergent and textile industries: Launching of a new platform technology. Plant Biotechnol. J. 2019, 17, 1167–1182. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.A.E.; Castillo, J.A.T.; Cruz, Q.R.; García, S.R.S. Biotechnological Applications of Plastid Foreign Gene Expression. In Plant Growth and Regulation-Alterations to Sustain Unfavorable Conditions; IntechOpen: Vienna, Austria, 2018. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Rijzaani, H.; Karcher, D.; Ruf, S.; Bock, R. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc. Natl. Acad. Sci. USA 2013, 110, E623–E632. [Google Scholar] [CrossRef] [Green Version]
- Kwak, S.Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, J.; Cao, J.; Zhao, Y.; Huang, J.; Zheng, Z.; Li, W.; Jang, S.; Qiao, J.; Xing, B.; et al. Opportunities for graphene, single-walled and multi-walled carbon nanotube applications in agriculture: A review. Crop. Des. 2022, 1, 100006. [Google Scholar] [CrossRef]
- Thagun, C.; Chuah, J.A.; Numata, K. Targeted gene delivery into various plastids mediated by clustered cell-penetrating and chloroplast-targeting peptides. Adv. Sci. 2019, 6, 1902064. [Google Scholar] [CrossRef] [Green Version]
- Hanson, M.R.; Gray, B.N.; Ahner, B.A. Chloroplast transformation for engineering of photosynthesis. J. Exp. Bot. 2013, 64, 731–742. [Google Scholar] [CrossRef]
- Dauvillee, D.; Hilbig, L.; Preiss, S.; Johanningmeier, U. Minimal extent of sequence homology required for homologous recombination at the psbA locus in Chlamydomonas reinhardtii chloroplasts using PCR-generated DNA fragments. Photosynth. Res. 2004, 79, 219–224. [Google Scholar] [CrossRef]
- Saski, C.; Lee, S.B.; Fjellheim, S.; Guda, C.; Jansen, R.K.; Luo, H.; Tomkins, J.; Rognli, O.A.; Daniell, H.; Clarke, J.L. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Appl. Genet. 2007, 115, 571–590. [Google Scholar] [CrossRef] [Green Version]
- Yarra, R. Plastome engineering in vegetable crops: Current status and future prospects. Mol. Biol. Rep. 2020, 47, 8061–8074. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Maliga, P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat. Biotechnol 1999, 17, 910–915. [Google Scholar] [CrossRef]
- Yoo, B.C.; Yadav, N.S.; Orozco, E.M.; Sakai, H. Cas9/gRNA-mediated genome editing of yeast mitochondria and Chlamydomonas chloroplasts. PeerJ 2020, 8, e8362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, N.; Xia, Y.; Zhan, Y.; Dan, J.; Yu, M.; Bu, X.; Cao, M. Improvement of Chloroplast Transformation Using CRISPR/Cas9. J. Biobased Mater. Bioenergy 2020, 14, 401–407. [Google Scholar] [CrossRef]
- Liere, K.; Börner, T. Transcription and transcriptional regulation in plastids. Top. Curr. Genet. 2007, 19, 121–174. [Google Scholar] [CrossRef]
- Hajdukiewicz, P.T.J.; Allison, L.A.; Maliga, P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997, 16, 4041–4048. [Google Scholar] [CrossRef]
- Bock, R. Engineering chloroplasts for high-level foreign protein expression. Chloroplast. Biotechnol. 2014, 1132, 93–106. [Google Scholar] [CrossRef]
- Ruhlman, T.; Verma, D.; Samson, N.; Daniell, H. The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 2010, 152, 2088–2104. [Google Scholar] [CrossRef]
- Bohne, A.-V.; Ruf, S.; Börner, T.; Bock, R. Faithful transcription initiation from a mitochondrial promoter in transgenic plastids. Nucleic Acids Res. 2007, 35, 7256–7266. [Google Scholar] [CrossRef] [Green Version]
- Eberhard, S.; Drapier, D.; Wollman, F.-A. Searching limiting steps in the expression of chloroplast-encoded proteins: Relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002, 31, 149–160. [Google Scholar] [CrossRef]
- Herz, S.; Füßl, M.; Steiger, S.; Koop, H.-U. Development of novel types of plastid transformation vectors and evaluation of factors controlling expression. Transgenic. Res. 2005, 14, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Oey, M.; Lohse, M.; Kreikemeyer, B.; Bock, R. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 2009, 57, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Badillo-Corona, J.A.; Karcher, D.; Gonzalez-Rabade, N.; Piepenburg, K.; Borchers, A.-M.I.; Maloney, A.P.; Kavanagh, T.A.; Gray, J.C.; Bock, R. High-level expression of HIV antigens from the tobacco and tomato plastid genomes. Plant Biotechnol. J. 2008, 6, 897–913. [Google Scholar] [CrossRef]
- Klein, R.R.; Mullet, J.E. Control of gene expression during higher plant chloroplast biogenesis. Protein synthesis and transcript levels of psbA, psaA-psaB, and rbcL in dark-grown and illuminated barley seedlings. J. Biol. Chem. 1987, 262, 4341–4348. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Ravin, N.V.; Kuznetsov, B.B.; Samigullin, T.H.; Antonov, A.S.; Kolganova, T.V.; Skryabin, K.G. Complete sequence of the duckweed (Lemna minor) chloroplast genome: Structural organization and phylogenetic relationships to other angiosperms. J. Mol. Evol. 2008, 66, 555–564. [Google Scholar] [CrossRef]
- Lin, C.S.; Chen, J.J.; Huang, Y.T.; Chan, M.T.; Daniell, H.; Chang, W.J.; Hsu, C.-T.; Liao, D.-C.; Wu, F.-H.; Lin, S.-Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 9040. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, B.; Cheng, F.; Ramchiary, N.; Choi, S.R.; Lim, Y.P.; Wang, X.-W. Sequencing of chloroplast genome using whole cellular DNA and Solexa sequencing technology. Front. Plant. Sci. 2012, 3, 234. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Yu, P.C.; Chang, W.J.; Yu, K.; Lin, C.S. Plastid transformation: How does it work? Can it be applied to crops? What can it offer? Int. J. Mol. Sci. 2020, 21, 4854. [Google Scholar] [CrossRef]
- Golczyk, H.; Greiner, S.; Wanner, G.; Weihe, A.; Bock, R.; Börner, T.; Herrmann, R.G. Chloroplast DNA in mature and senescing leaves: A reappraisal. Plant Cell 2014, 26, 847–854. [Google Scholar] [CrossRef] [Green Version]
- Greiner, S.; Golczyk, H.; Malinova, I.; Pellizzer, T.; Bock, R.; Börner, T.; Herrmann, R.G. Chloroplast nucleoids are highly dynamic in ploidy, number, and structure during angiosperm leaf development. Plant J. 2020, 102, 730–746. [Google Scholar] [CrossRef] [Green Version]
- Teske, D.; Peters, A.; Moöllers, A.; Fischer, M. Genomic profiling: The strengths and limitations of chloroplast genome-based plant variety authentication. J. Agric. Food Chem. 2020, 68, 14323–14333. [Google Scholar] [CrossRef] [PubMed]
- Tetsuaki Osafune, S.S.; Hase, E. Proplastids of dark-grown wax-rich cells of Euglena gracilis. Regul. Chloroplast Biog. 2012, 226, 361–366. [Google Scholar]
- Maliga, P. Engineering the plastid and mitochondrial genomes of flowering plants. Nat. Plants 2022, 8, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Devarumath, R.M.; Nerkar, G.A.; Farsangi, F.J.; Nikam, A.A.; Babu, K.H. Embracing biotechnology methods for crop improvement research in sugarcane. In Current Status of Sugarcane Research in India; Tiwari, A.K., Singh, A.K., Lal, M., Eds.; Nova Publishers: Hauppauge, NY, USA, 2015; pp. 33–53. [Google Scholar]
- Mustafa, G.; Khan, M.S. Transmission of Engineered Plastids in Sugarcane, a C4 Monocotyledonous Plant, Reveals that Sorting of Preprogrammed Progenitor Cells Produce Heteroplasmy. Plants 2021, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Maliga, P. Plastid transformation in flowering plants. In Genomomics of Chloroplasts Mitochondria; Bock, R., Knoop, V., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 393–414. [Google Scholar] [CrossRef]
- Kumar, S.; Dhingra, A.; Daniel, H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol. Biol. 2004, 56, 203–216. [Google Scholar] [CrossRef]
- Huang, F.C.; Klaus, S.M.J.; Herz, S.; Zou, Z.; Koop, H.U.; Golds, T.J. Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Mol. Genet. Genom. 2002, 268, 19–27. [Google Scholar] [CrossRef]
- Day, A.; Goldschmidt-Clermont, M. The chloroplast transformation toolbox: Selectable markers and marker removal. Plant Biotechnol. J. 2011, 9, 540–553. [Google Scholar] [CrossRef] [Green Version]
- Bock, R. Genetic engineering of the chloroplast: Novel tools and new applications. Curr. Opin. Biotechnol. 2014, 26, 7–13. [Google Scholar] [CrossRef]
- Borovinskaya, M.A.; Shoji, S.; Holton, J.M.; Fredric, K.; Cate, J.H.D. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem. Biol. 2007, 2, 545–552. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ruf, S.; Bock, R. Chloramphenicol acetyltransferase as selectable marker for plastid transformation. Plant Mol. Biol. 2011, 76, 443–451. [Google Scholar] [CrossRef]
- Barone, P.; Zhang, X.H.; Widholm, J.M. Tobacco plastid transformation using the feedback-insensitive anthranilate synthase [alpha]-subunit of tobacco (ASA2) as a new selectable marker. J. Exp. Bot. 2009, 60, 3195–3202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, D.; Daniell, H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007, 145, 1129–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef]
- Ye, G.N.; Colburn, S.; Xu, C.W.; Hajdukiewicz, P.T.J.; Staub, J.M. Persistence of unselected transgenic DNA during a plastid transformation and segregation approach to herbicide resistance. Plant Physiol. 2003, 133, 402–410. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, M.; Goto, M.; Hanai, M.; Shimizu, T.; Izawa, N.; Kanamoto, H.; Tomizawa, K.; Yokota, A.; Kobayashi, H. Selectable tolerance to herbicides by mutated acetolactate synthase genes integrated into the chloroplast genome of tobacco. Plant Physiol. 2008, 147, 1976–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufourmantel, N.; Dubald, M.; Matringe, M.; Canard, H.; Garcon, F.; Job, C.; Kay, E.; Wisniewski, J.P.; Ferullo, J.M.; Pelissier, B.; et al. Generation and characterization of soybean and marker-free tobacco plastid transformants over-expressing a bacterial 4-hydroxyphenylpyruvate dioxygenase which provides strong herbicide tolerance. Plant Biotechnol. J. 2007, 5, 118–133. [Google Scholar] [CrossRef]
- Okuzaki, A.; Tsuda, M.; Konagaya, K.I.; Tabei, Y. A novel strategy for promoting homoplasmic plastid transformant production using the barnase–barstar system. Plant Biotechnol. 2020, 37, 223–232. [Google Scholar] [CrossRef]
- Hartley, R.W. Barnase and barstar: Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J. Mol. Biol. 1988, 202, 913–915. [Google Scholar] [CrossRef] [Green Version]
- Abe, K.; Oshima, M.; Akasaka, M.; Konagaya, K.; Nanasato, Y.; Okuzaki, A.; Taniguchi, Y.; Tanaka, J.; Tabei, Y. Development and characterization of transgenic dominant male sterile rice toward an outcross-based breeding system. Breed. Sci. 2018, 68, 248–257. [Google Scholar] [CrossRef] [Green Version]
- Dyo, Y.M.; Purton, S. The algal chloroplast as a synthetic biology platform for production of therapeutic proteins. Microbiology 2018, 164, 113–121. [Google Scholar] [CrossRef]
- Zhou, F.; Karcher, D.; Bock, R. Identification of a plastid Intercistronic Expression Element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 2007, 52, 961–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Medina, R.; Goffinet, B. 350 my of mitochondrial genome stasis in mosses, an early land plant lineage. Mol. Biol. Evol. 2014, 31, 2586–2591. [Google Scholar] [CrossRef] [Green Version]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.Q.; Liao, X.Z.; Zhang, X.N.; Tembrock, L.R.; Broz, A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J. Syst. Evol. 2022, 60, 160–168. [Google Scholar] [CrossRef]
- Sloan, D.B.; Warren, J.M.; Williams, A.M.; Wu, Z.; Abdel-Ghany, S.E.; Chicco, A.J.; Havird, J.C. Cytonuclear integration and co-evolution. Nat. Rev. Genet. 2018, 19, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Wu, Z.; Sharbrough, J. Correction of persistent errors in Arabidopsis reference mitochondrial genomes. Plant Cell 2018, 30, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and Recombination of the Bacterial-Sized Multichromosomal Mitochondrial Genome of Cucumber. Plant Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preuten, T.; Cincu, E.; Fuchs, J.; Zoschke, R.; Liere, K.; Börner, T. Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010, 64, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Eckert-Ossenkopp, U.; Schmiedeberg, I.; Brandt, P.; Unseld, M.; Brennicke, A.; Schuster, W. Physical mapping of the mitochondrial genome of Arabidopsis thaliana by cosmid and YAC clones. Plant J. 1994, 6, 447–455. [Google Scholar] [CrossRef]
- Backert, S.; Lurz, R.; Oyarzabal, O.A.; Börner, T. High content, size and distribution of single-stranded DNA in the mitochondria of Chenopodium album (L.). Plant Mol. Biol. 1997, 33, 1037–1050. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Kühn, K. DNA-binding proteins in plant mitochondria: Implications for transcription. Mitochondrion 2014, 19, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Weihe, A. The Transcription of Plant Organelle Genomes. In Molecular Biology and Biotechnology of Plant Organelles: Chloroplasts and Mitochondria; Daniell, H., Chase, C., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 213–237. [Google Scholar] [CrossRef]
- Bonnefoy, N.; Remacle, C.; Fox, T.D. Genetic transformation of Saccharomyces cerevisiae and Chlamydomonas reinhardtii mitochondria. Methods Cell Biol. 2007, 80, 525–548. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, L.; Chen, J. Mitochondrial DNA heteroplasmy in Candida glabrata after mitochondrial transformation. Eukaryot 2010, 9, 806–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazama, T.; Okuno, M.; Watari, Y.; Yanase, S.; Koizuka, C.; Tsuruta, Y.; Susaya, H.; Toyoda, A.; Itoh, T.; Tsutsumi, N.; et al. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat. Plants 2019, 5, 722–730. [Google Scholar] [CrossRef]
- Arimura, S.I.; Ayabe, H.; Sugaya, H.; Okuno, M.; Tamura, Y.; Tsuruta, Y.; Watari, Y.; Yanase, S.; Yamauchi, T.; Itoh, T.; et al. Targeted gene disruption of ATP synthases 6-1 and 6-2 in the mitochondrial genome of Arabidopsis thaliana by mitoTALENs. Plant J. 2020, 104, 1459–1471. [Google Scholar] [CrossRef]
- Nakazato, I.; Okuno, M.; Zhou, C.; Itoh, T.; Tsutsumi, N.; Takenaka, M.; Arimura, S.I. Targeted base editing in the mitochondrial genome of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2022, 119, e2121177119. [Google Scholar] [CrossRef]
- Koulintchenko, M.; Konstantinov, Y.; Dietrich, A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003, 22, 1245–1254. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, N.; Handa, H.; Cosset, A.; Koulintchenko, M.; Konstantinov, Y.; Lightowlers, R.N.; Dietrich, A.; Weber-Lotfi, F. DNA delivery to mitochondria: Sequence specificity and energy enhancement. Pharm. Res. 2011, 28, 2871–2882. [Google Scholar] [CrossRef]
- Tarasenko, T.A.; Klimenko, E.S.; Tarasenko, V.I.; Koulintchenko, M.V.; Dietrich, A.; Weber-Lotfi, F.; Konstantinov, Y.M. Plant mitochondria import DNA via alternative membrane complexes involving various VDAC isoforms. Mitochondrion 2021, 60, 43–58. [Google Scholar] [CrossRef]
- Mileshina, D.; Koulintchenko, M.; Konstantinov, Y.; Dietrich, A. Transfection of plant mitochondria and in organello gene integration. Nucleic Acids Res. 2011, 39, e115. [Google Scholar] [CrossRef] [Green Version]
- Tarasenko, T.A.; Tarasenko, V.I.; Koulintchenko, M.V.; Klimenko, E.S.; Konstantinov, Y.M. DNA Import into Plant Mitochondria: Complex approach for in organello and in vivo studies. Biochemistry 2019, 84, 817–828. [Google Scholar] [CrossRef] [PubMed]
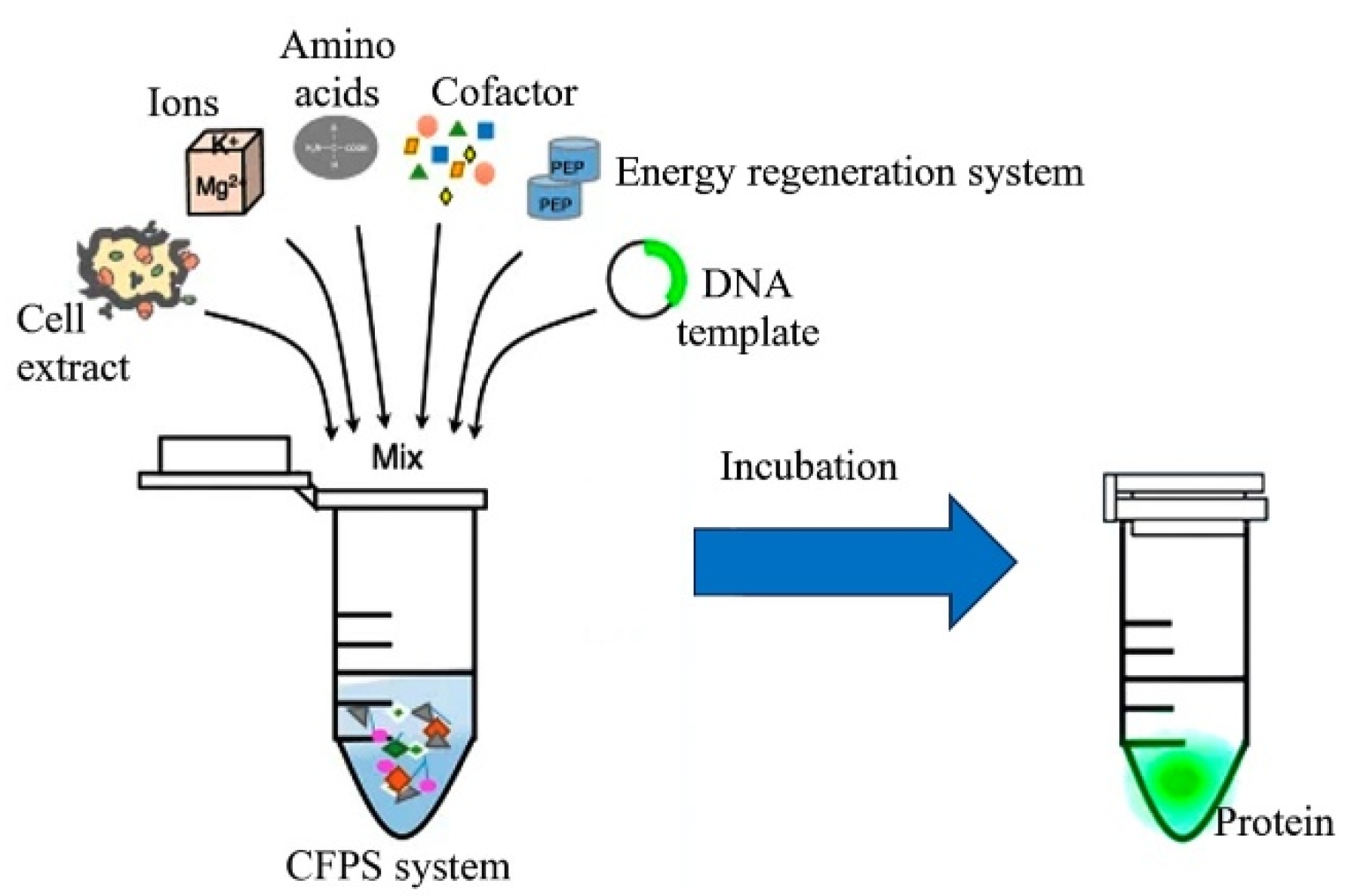
| Plant Species | Recombinant Protein | Productivity | Reference |
|---|---|---|---|
| Nicotiana tabacum | Recombinant human erythropoietin (rhEPO) | 66.75 pg/mg TSP | [21] |
| Physcomitrella patens | α-Galactosidase A | 0.5 mg/mL TSP | [22] |
| N. benthamiana | Human glucocerebrosidase | 68 μg/g FW | [23] |
| Salvia miltiorrhiza | Human acidic fibroblast growth factor 1 (FGF-1) | 272 ng/g FW | [24] |
| Helianthus annuus | Lumbrokinase (LK) | 5.1 g/kg seed | [25] |
| N. tabacum | Hydrophobin | 16.5% of TSP | [26] |
| Oryza sativa | Human growth hormone (hGH) | 57 mg/L of medium TSP | [27] |
| Glycine max | Fibroblast growth factor (bFGF) | 2.3% of TSP | [28] |
| N. tabacum | Placental alkaline phosphatase (SEAP) | 3% of TSP | [29] |
| O. sativa | Human serum albumin, epidermal growth factor, recombinant lactoferrin, basic fibroblast growth factor, insulin-like growth factor-1 LR3, lysozyme, vascular endothelial growth factor, α-1 antitrypsin, keratinocyte growth factor, and fibronectin | – | [30] |
| O. sativa | Transferrin and lysozyme | – | [31] |
| N. tabacum | B lymphocyte activating factor and bone morphogenic protein 7 | ||
| Zea mays | TrypZean® | ||
| Hordeum vulgare | Leukemia inhibitory factor | – | [32] |
| Human growth factors, cytokines, MESOkine (animal-like growth factors) | – | [33] | |
| N. tabacum | Monoclonal antibody 2G12 | 10–25 μg/g FW | [34] |
| Anti-HBsAg | 6.5 mg/g seeds | [35] | |
| Z. mays | Monoclonal antibody 2F5 | 0.61 ± 0.28 μg/g seed extract | [36] |
| Product | Company | Application | Plant Species | Reference |
|---|---|---|---|---|
| Trypsin, avidin, and endo-1,4-β-d-glucanase | ProdiGene/Sigma-Aldrich (St. Louis, MO, USA) | Technical reagents | Maize (seeds) | [43] |
| Cellobiohydrolase I | Infinite Enzymes/Sigma-Aldrich (St. Louis, MO, USA) | Technical reagent | Maize (seeds) | |
| Growth factors, cytokines, thioredoxin, and TIMP-2 | Agrenvec (Madrid, Spain) | Research reagents | Tobacco (leaves, transient expression) | [44] |
| Growth factors and cytokines | ORF Genetics (Kópavogur, Iceland) | Research reagents | Barley (seeds) | [45] |
| Epithelial growth factor | Sif Cosmetics (Kópavogur, Iceland) | Cosmetics | Barley (seeds) | [46] |
| Albumin, lactoferrin, lysozyme, transferrin, and insulin | Ventria Bioscience/InVitria (Fort Collins, CO, USA) | Research reagents | Rice (seeds) | [47] |
| Aprotinin | Kentucky BioProcessing (Owensboro, KY, USA) | Research reagent | Tobacco (leaves, transient expression) | [48] |
| Collagen | CollPlant (Rehovot, Israel) | Research reagent, tissue culture, and therapeutic | Tobacco | [49] |
| Trypsin, enterokinase, growth factors, and cytokines | Natural Biomaterials (Wanju-gun, Republic of Korea) | Research reagents and cosmetic ingredients | Rice (seeds) | [50] |
| Antibody | Center for Genetic Engineering and Biotechnology (Havana, Cuba) | Reagent for purification of hepatitis B vaccine | Transgenic tobacco | [51] |
| α-Amylase | Syngenta (Wilmington, DE, USA) | Reagent for bioethanol production | Maize seeds | [52] |
| Phytase | Origin Agritech (Beijing, China) | Feed | Maize seeds | [53] |
| Growth factors | NexGen (Suwon, Republic of Korea) | Tissue culture reagent | Tobacco leaves, transient expression | [54] |
| Plant | Protein | Productivity | Reference |
|---|---|---|---|
| O. sativa | Human cytotoxic T-lymphocyte antigen 4-immunoglobulin (hCTLA4Ig) | 43.7 mg/L | [63] |
| Human granulocyte macrophage-colony stimulating factor (hGM-CSF) | 31.7 mg/L | [64] | |
| Cyclic citrullinated peptide (CCP) antibody | 22.9 mg/L | [65] | |
| Human pepsinogen C (hPGC) | 18 mg/L | [66] | |
| Glucocerebrosidase (GDC) | – | [67] | |
| D. carota | β-Glucocerebrosidase and α-galactosidase A | – | [68] |
| N. tabacum (BY-2) | Actin inhibition resistance (AIR) DNase and antitumor necrosis factor | – | [68] |
| IgG4 | 25 mg/L | [61] |
| Insertion Sites | Promoter/5′UTR/Terminator (3′UTR) | Expression Efficiency, % of TSP |
|---|---|---|
| trnI/trnA | PpsbA/TpsbA | 32–38 and 17–26 |
| trnI/trnA | Prrn/TpsbA | 45.3 |
| trnI/trnA | TrbcL | 10.0 |
| trnI/trnA | Prrn/TpsbA | 0.85–1.0 |
| trnI/trnA | PpsbA/TpsbA | 5.16–9.27 |
| trnI/trnA | PpsbA/TpsbA | 0.2–6.0 |
| trnV/rps12, 7 | Prrn/TrbcL | >7.0 |
| rbcL/accD | PpsbA/rbcL 3′ TrbcL | 5.0 |
| rps7, 12/trnV | Prrn/T7g10/Trps16 | >10.0 |
| rbcL/accD | Prrn/TpsbA | 2.0–3.0 |
| trnfM/trnG | Prrn/T7g10/rbc3′ | 0.8–1.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozov, S.M.; Zagorskaya, A.A.; Konstantinov, Y.M.; Deineko, E.V. Three Parts of the Plant Genome: On the Way to Success in the Production of Recombinant Proteins. Plants 2023, 12, 38. https://doi.org/10.3390/plants12010038
Rozov SM, Zagorskaya AA, Konstantinov YM, Deineko EV. Three Parts of the Plant Genome: On the Way to Success in the Production of Recombinant Proteins. Plants. 2023; 12(1):38. https://doi.org/10.3390/plants12010038
Chicago/Turabian StyleRozov, Sergey M., Alla A. Zagorskaya, Yuri M. Konstantinov, and Elena V. Deineko. 2023. "Three Parts of the Plant Genome: On the Way to Success in the Production of Recombinant Proteins" Plants 12, no. 1: 38. https://doi.org/10.3390/plants12010038
APA StyleRozov, S. M., Zagorskaya, A. A., Konstantinov, Y. M., & Deineko, E. V. (2023). Three Parts of the Plant Genome: On the Way to Success in the Production of Recombinant Proteins. Plants, 12(1), 38. https://doi.org/10.3390/plants12010038






